Abstract
We report the structure of the flock house virus B2 protein, a potent suppressor of RNA interference (RNAi) in animals and plants. The B2 protein is a homodimer in solution and contains three α-helices per monomer. Chemical shift perturbation shows that an antiparallel arrangement of helices (α2/α2′) forms an elongated binding interface with double-stranded RNA (dsRNA). This implies a novel mode of dsRNA recognition and provides insights into the mechanism of RNAi suppression by B2.
Keywords: RNAi, viral suppressor, dsRNA recognition
Introduction
RNA silencing is thought to have originated from an ancient cellular defence mechanism against viral and other endogenous double-stranded RNAs (dsRNAs; Plasterk, 2002; Roth et al, 2004; Voinnet, 2005). This antiviral response is mediated by the cellular RNA interference (RNAi) machinery and targets viral transcripts for degradation using short interfering RNAs (siRNAs). To counteract this host cell defence mechanism, many viruses have evolved proteins that suppress RNA silencing. Flock house virus (FHV) belongs to Nodaviridae, a family of RNA viruses with a genome comprising two positive sense RNAs (Johnson et al, 2001). Recently, it was shown that FHV replication, which involves dsRNA molecules as intermediates, triggers a potent antiviral silencing response in Caenorhabditis elegans. This response requires the Argonaute protein rde-1, which is essential for RNAi mediated by siRNAs, but not by microRNAs (Lu et al, 2005). The FHV B2 protein counteracts the silencing of viral gene expression and acts as a broad-spectrum RNAi inhibitor (Li et al, 2002). The B2 protein of a related Nodavirus family member, Nodamura virus (NoV), which is infectious for insect and mammalian hosts, has been shown to block RNAi in mammalian cells (Sullivan & Ganem, 2005). Both the FHV and NoV B2 proteins bind to siRNAs and longer dsRNAs, and may therefore sequester siRNAs and prevent the processing of long dsRNAs into siRNAs by the host RNaseIII-like enzyme Dicer (Lu et al, 2005; Sullivan & Ganem, 2005). This mode of inhibition of RNAi is distinct from the strategy used by other viruses. For example, the p19 protein of tombusviruses specifically binds to siRNAs and recognizes their characteristic size (Vargason et al, 2003; Ye et al, 2003), thus sequestering the products of Dicer and suppressing the silencing effect. The molecular basis of this novel mechanism of RNAi suppression by the 106-residue FHV B2 protein is unknown (Lu et al, 2005).
Results
We have determined the three-dimensional structure of the FHV B2 protein using heteronuclear NMR methods. NMR spectra show that the 30 carboxy-terminal residues of full-length B2 are unstructured and highly flexible in solution (supplementary Fig S1 online). For this reason, we solved the structure of a region comprising residues 1–72 of B2.
The B2 protein is a symmetric dimer in solution. The structure is defined well by the NMR data (Fig 1A; Table 1). Each monomer consists of three α-helices (α1: residues 4–23; α2: residues 32–61; α3: residues 64–70), which are arranged in a triangular manner such that the amino- and C terminus of each monomer are in close proximity (Fig 1B,C). Hydrophobic residues stabilize an antiparallel interaction between helices α1 and α2, which are connected by an extended linker between Gly 24 and Pro 31 (Fig 1B). Helix α2 is very elongated and spans 30 residues corresponding to eight helical turns, whereas helix α3 is short and oriented orthogonal to the first two helices.
Figure 1.
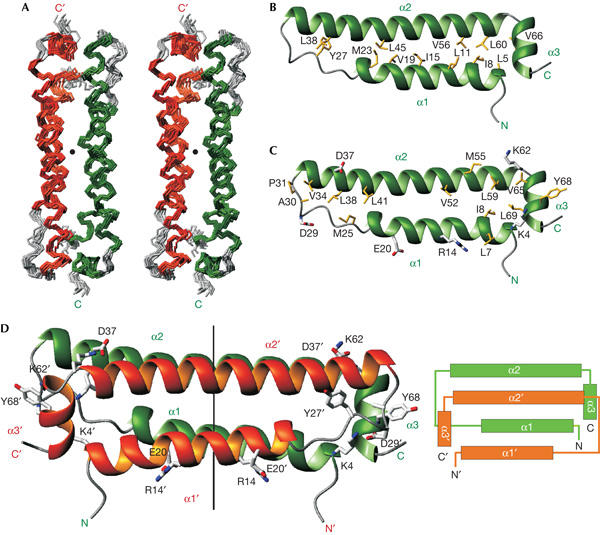
Solution structure of the flock house virus B2 protein (residues 1–72). (A) Stereo view of the ensemble of the ten lowest energy NMR structures of the flock house virus B2 dimer. Secondary structure elements are coloured in green and orange for the two subunits, respectively. The C2 symmetry axis is indicated by a black dot. (B) Ribbon representation of the B2 monomer, in a view rotated by two 90° rotations around the x and y axis compared with (A). The subunit is coloured as in (A). Side chains of hydrophobic residues involved in intra-monomer interactions are shown and labelled. (C) Ribbon representation of the B2 monomer; view and colouring are as in (B). Side chains of hydrophobic residues and of charged residues involved in inter-monomer electrostatic contacts contributing to the dimer interface are shown and labelled. (D) Ribbon representation of the B2 dimer. Subunits are coloured as in (A). The C2 symmetry axis is shown, as are aromatic residues and side chains of residues involved in electrostatic interactions. The topology of the B2 dimer is shown on the right.
Table 1.
Structural statistics for the flock house virus B2 dimer
| 〈SA〉* | 〈SA〉water-ref | |
|---|---|---|
| Number of nuclear Overhauser effect-derived distance restraints (restraints per monomer) | ||
| All (unambiguous/ambiguous) | 2,202/34 | |
| Intra-monomer (unambiguous/ambiguous) | 2,073/15 | |
| Inter-monomer (unambiguous/ambiguous) |
129/0 |
|
| R.m.s.d. (Å) from experimental distance restraints‡ | ||
| R.m.s.d. (unambiguous) | 0.019±0.003 | 0.051±0.010 |
| R.m.s.d. (ambiguous) | 0.030±0.009 | 0.037±0.009 |
| Hydrogen bonds (2 × 38 per monomer) |
0.032±0.002 |
0.036±0.004 |
| R.m.s.d. (deg) from experimental torsion restraints§ | ||
| R.m.s.d. (55 φ/ψ per monomer) |
0.42±0.03 |
0.82±0.05 |
| Q-factor for experimental residual dipolar coupling restraints∥ | ||
| 1DHN (22 per monomer) |
0.035±0.003 |
0.041±0.004 |
| Coordinate precision (Å) residues 1–71¶ | ||
| N, Cα, C′ (dimer) | 0.35±0.06 | 0.39±0.05 |
| All heavy atoms (dimer) | 0.76±0.08 | 0.80±0.06 |
| N, Cα, C′ (monomer A) | 0.31±0.05 | 0.37±0.04 |
| All heavy atoms (monomer A) |
0.74±0.07 |
0.79±0.06 |
| Structural quality# | ||
| Bad contacts | 3.2±1.7 | 0±0 |
| Ramachandran plot | ||
| Percentage in the most favoured region | 94.1±1.7 | 97.0±1.0 |
| Percentage in the additionally allowed region |
5.9±1.7 |
2.6±1.1 |
| *〈SA〉 is an ensemble of ten lowest-energy solution structures (out of 100 calculated) of the B2 dimer before water refinement. The CNS Erepel function was used to simulate van der Waals interactions with an energy constant of 25.0 kcal mol−1 Å−4 using ‘PROLSQ' van der Waals radii (Linge et al, 2003); r.m.s.d. for bond lengths, bond angles and improper dihedral angles are 0.00294±0.00008 Å, 0.52±0.01° and 0.558±0.008°. 1 kcal=4.18 kJ. Non-crystallographic symmetry restraints with an energy constant of 100 kcal mol−1 Å−2 were used to enforce the dimer symmetry. | ||
| ‡Distance restraints were used with a soft square-well potential using an energy constant of 50 kcal mol−1 Å2. For hydrogen bonds, distance restraints with bounds of 1.8–2.3 Å (H–O) and 2.8–3.3 Å (N–O) were derived for slowly exchanging amide protons. In the 〈SA〉 structures, 0.2±0.6 distance restraints were violated by more than 0.3 Å. | ||
| §Dihedral angle restraints derived from TALOS (Cornilescu et al, 1999) were applied to φ, ψ backbone angles using energy constants of 200 kcal mol−1 rad−2. No dihedral angle restraint was violated by more than 5°. | ||
| ∥Quality factor for the RDC refinement (Cornilescu et al, 1998). Residual dipolar couplings were applied with a final energy constant of 0.2 kcal mol−1 Hz−2 for an alignment tensor with an axial component of 22.7 Hz and a rhombicity of 0.56. | ||
| ¶Coordinate precision is given as the Cartesian coordinate r.m.s.d. of the ten lowest-energy structures in the NMR ensemble with respect to their mean structure. | ||
| #Structural quality was analysed using PROCHECK (Laskowski et al, 1996). | ||
The dimer is formed by a head-to-tail interaction of the two monomers and involves all three helices (Fig 1C,D). The dimer interface is stabilized by hydrophobic interactions mainly involving aliphatic side chains in helices α1 and α2, as evidenced by inter-monomer nuclear Overhauser effects (NOEs) for residues Leu 7, Ile 8, Met 25, Val 34, Leu 38, Leu 41, Val 52, Met 55, Leu 59, Val 65 and Leu 69 (Fig 1C). An interaction involving Tyr 68 and Ala 30/Pro 31 from separate subunits also contributes to the dimer interface, whereas the only other aromatic residue, Tyr 27, is involved in intra-monomer contacts only (Fig 1B). Three inter-monomer electrostatic interactions (Lys 4–Asp 29, Arg 14–Glu 20 and Lys 62–Asp 37) additionally stabilize the dimer interface (Fig 1C,D).
The dimeric conformation of the B2 protein is confirmed by gel filtration (data not shown) and NMR relaxation measurements (Fig 2). Both the average H–N residual dipolar coupling (RDC) values (Tjandra & Bax, 1997) and the average ratio of 15N T1 and T2 relaxation times (T1/T2; Tjandra et al, 1997; red lines in Fig 2) in helices α1 and α2 are similar and very different from those in helix α3. This is consistent with the approximately parallel and orthogonal orientations of α1/α2 and α3, respectively, and with the elongated structure of the B2 dimer. The dimerization buries a large surface area (1,500 Å2 or 25% of the total surface area) in each monomer. Searches in the Protein Data Bank with Dali (Holm & Sander, 1997) or MSDfold (Krissinel & Henrick, 2004) did not show any close structural homologues, indicating that the B2 dimer represents a novel fold. However, the topology of the four-helical bundle formed by α1, α2, α1′ and α2′ in B2 resembles the small dimeric Repressor of primer (Rop) protein (also see below; Banner et al, 1987). In contrast to B2, in which all four helices are almost parallel to each other, Rop forms a canonical left-handed four-helical bundle, as reflected in the rather large r.m.s.d. of the atomic backbone coordinates (3.7 Å) between the B2 and Rop dimers.
Figure 2.
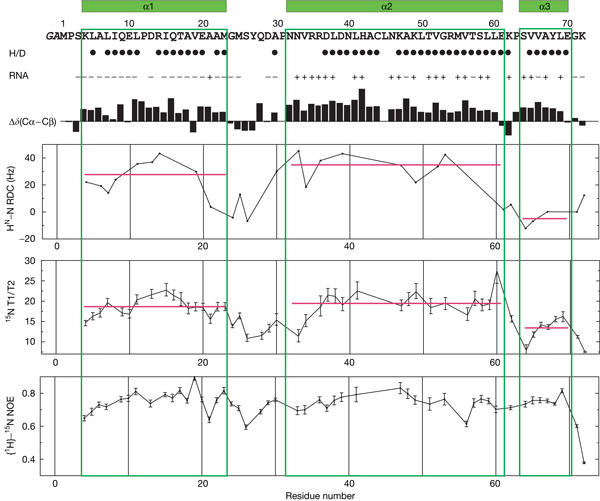
Summary of NMR data of the flock house virus B2 dimer. The secondary structure is shown above the sequence. Solvent-protected amide protons (slow H/D exchange in NMR measurements) are indicated by filled circles. Residues with and without chemical shift perturbation following double-stranded RNA binding are marked by + and −, respectively. Secondary chemical shifts, Δδ(Cα–Cβ), are indicated by black bars. 1H–15N residual dipolar couplings (RDCs) and 15N T1, T2 relaxation data are consistent with the elongated fold of the B2 dimer. Heteronuclear {1H}–15N nuclear Overhauser effect (NOE) data indicate that the protein is very rigid and that there are no regions of increased backbone mobility.
A recent study showed that recombinant glutathione S-transferase-tagged B2 binds to long dsRNA molecules or siRNAs of diverse sequences, but not to single-stranded siRNAs (Lu et al, 2005), indicating that B2 binds dsRNA in a size- and sequence-independent manner. We used NMR titration experiments to further characterize the RNA interaction of the B2 dimer. Addition of a 21-mer siRNA duplex induces large changes in the 1H, 15N correlation spectra of B2 (Fig 3A), with signals of the free form disappearing and reappearing at new positions (supplementary Fig S2 online). The slow exchange on the NMR chemical shift timescale between free and bound states is indicative of tight binding with a dissociation constant in the nanomolar range. The titration curve (Fig 3B) and the linewidths in the NMR spectra indicate a 1:1 stoichiometry for the B2 dimer:RNA complex in solution.
Figure 3.

Binding of flock house virus B2 (1–72) to double-stranded RNA. (A) Chemical shift perturbation after addition of a 21-mer short interfering RNA (siRNA) duplex to a 2H-, 13C-, 15N-labelled flock house virus (FHV) B2 dimer. The chemical shifts are monitored in 1H, 15N correlation spectra. The spectrum of the free protein is shown in black and the spectrum after addition of a twofold molar excess of siRNA is shown in red. The dimer concentration was 0.25 mM. (B) Titration curves of double-stranded RNA (dsRNA) binding to FHV B2. The fraction of bound protein (fbound) is plotted as a function of ligand concentration. fbound is defined by the signal intensities, fbound=intbound/(intbound+intfree), where intbound and intfree are the peak intensities of the RNA-bound and free NMR signals (supplementary Fig S2 online). The inflection point of the titration curve at equimolar dimer–RNA ratio indicates a 1:1 stoichiometry for the B2 dimer–RNA complex. (C) Left: surface representation of the B2 dimer coloured according to chemical shift changes induced following the addition of siRNA (see (A)). Residues that are affected/not affected by the RNA addition are shown in magenta/white. Residues that could not be analysed unambiguously because of signal overlap are shown in light yellow. The same orientation as in Fig 1A is shown. Middle: the same surface coloured in blue and red according to positive and negative electrostatic potential, respectively. Right: a ribbon representation in the same orientation. Side chains of residues discussed in the text are shown. (D) Proposed interface of the B2 dimer with dsRNA. The B2 dimer is shown rotated by 90° around the y axis compared with (C). Side chains of lysine and arginine residues in the RNA-binding surface are shown. Distances between these residues and the total length of the RNA-binding surface are indicated.
The chemical shift perturbations map to the long helices α2/α2′ and also involve residues in α3/α3′, spanning a region of 45 Å in length (Fig 3C,D). Notably, the α2,α2′ helices are highly positively charged, and many lysine and arginine residues are located in this surface. Arg 54, which has been shown to be crucial for dsRNA binding (Lu et al, 2005), is located at the centre of this binding interface. Arg 54 and Lys 47 in this region are separated with a spacing of approximately 10 Å (Fig 3C,D), which corresponds to the distance of phosphates across the major groove in an A-form dsRNA conformation. Thus, the exposed arginine and lysine residues may mediate non-sequence-specific electrostatic contacts with the phosphate–sugar backbone of the dsRNA stem. The 1:1 stoichiometry for the B2 dimer:RNA complex and the size of the RNA-binding surface extending across the whole length of the B2 dimer, corresponding to approximately 17-bp A-form dsRNA, indicate that the α2,α2′ helices bind parallel to the stem of the dsRNA helix. Such a mode of dsRNA binding does not recognize the unique 3′ and 5′ ends of an siRNA and is consistent with the ability of B2 to bind to 21-nt siRNA duplexes as well as to dsRNAs of 44–100 nt (Lu et al, 2005).
Discussion
The structure of the B2 dimer and the RNA-binding surface mapped by the NMR data are very different from other dsRNA-binding motifs such as the dsRNA-binding domain (dsRBD; Fierro-Monti & Mathews, 2000) or dsRNA-binding zinc-fingers (Lu et al, 2003). In the dsRBD, residues of an N-terminal helix and those in loop regions interact with the RNA stem. In the fifth zinc-finger of TFIIIA, a short helix contacts the major groove backbone. Other proteins involved in RNAi also use different modes of RNA recognition (Lingel & Sattler, 2005). The viral p19 protein is, like B2, a dimer but uses an extended β-sheet surface and a small helical motif for binding and measuring the characteristic length of siRNAs (Vargason et al, 2003; Ye et al, 2003). Interestingly, a four-helical bundle fold is found in the homodimeric Rop protein, a regulator of plasmid copy number in Escherichia coli (Banner et al, 1987). The predicted binding interface with kissing RNA hairpins comprises an antiparallel pair of the first helix of each monomer (Predki et al, 1995), somewhat resembling the B2 dimeric RNA-binding interface. The N-terminal domain of the influenza virus NS1 protein is a symmetric homodimer formed by two three-helix monomers (Chien et al, 1997; Liu et al, 1997). Although the NS1 dimer binds dsRNA (Chien et al, 2004), the six-helical fold is distinct from that of the FHV B2 dimer. Thus, both the structural elements and the size of the protein–RNA interfaces used by the B2 dimer indicate an unprecedented mode of dsRNA binding.
While this manuscript was under review, the crystal structure of a B2 dimer bound to a palindromic 18 bp RNA duplex was described (Chao et al, 2005). Both the monomer and the dimer folds are very similar to our solution structure of the free B2 protein (backbone coordinate r.m.s.d. 1.2/1.3 Å for monomer/dimer). A slightly different orientation of the α3 helices is potentially linked to electrostatic contacts of residues in the preceding linker with the RNA. Notably, the overall high similarity of the structures shows that RNA binding does not induce large conformational changes in the protein. In the protein–RNA complex, the α2/α2′ helices of one B2 dimer are oriented almost parallel to the helical axis of the A-form RNA. Thus, the binding site and stoichiometry are consistent with our NMR data (Fig 3). As expected, the positively charged residues exposed from the RNA-binding surface (Fig 3D) interact with the phosphate–ribose backbone in the RNA helix.
In summary, we describe the structure of the FHV B2 (1–72) protein and characterize its interaction with dsRNA. Our studies show that the helical B2 dimer adopts a novel fold used for dsRNA binding. From biochemical studies (Lu et al, 2005; Sullivan & Ganem, 2005) and consistent with the structure of a B2 dimer–dsRNA complex (Chao et al, 2005), the RNA binding mode does not discriminate between siRNA duplexes and long dsRNAs, and is thus consistent with a role of B2 in the suppression of RNAi upstream of Dicer (Lu et al, 2005).
Methods
Sample preparation. FHV B2 complementary DNA (Swiss-Prot entry P68831) was amplified by PCR using a random-primed S2 cell cDNA as a template and cloned into the pETM11 vector, which is a derivative of the pET24-d vector (Novagen, Darmstadt, Germany). The recombinant protein was expressed in E. coli (BL21-DE3 strain) for 14 h at 25°C after induction by 0.5 mM isopropyl-β-D-thiogalactoside. For uniform 15N and 15N, 13C labelling, cells were grown in M9 minimal medium supplemented with 15NH4Cl without or with 13C6-labelled glucose, respectively. For the preparation of 2H-, 13C-, 15N-labelled sample, the cells were grown in minimal medium prepared with 90% D2O and supplemented with 10% 2H/15N/13C-rich medium (Silantes, Munich, Germany). Cell lysates were incubated with Ni-NTA Superflow beads (Qiagen, Hilden, Germany). After elution, the fusion protein was cleaved overnight by tobacco etch virus protease. After a second affinity purification step using Ni-NTA beads, the buffer was exchanged with NMR buffer (50 mM NaPi, pH 6.3, 50 mM NaCl, 1 mM dithiothreitol) and the protein preparation was concentrated to 0.2–1 mM for NMR experiments. For measurements in D2O, the protein was lyophilized and redissolved in D2O.
The sequence of the siRNA used for the titrations was as follows: sense GCA GCA CGA CUU CUU CAA GTT and antisense CUU GAA GAA GUC GUG CUG CTT.
NMR spectroscopy. NMR spectra were acquired at 22°C on Bruker DRX500, DRX600 and DRX900 spectrometers equipped with cryogenic triple-resonance probes. Spectra were processed with NMRPipe (Delaglio et al, 1995) and analysed using NMRVIEW (Johnson & Blevins, 1994). The 1H, 13C and 15N chemical shifts were assigned by standard methods (Sattler et al, 1999). Distance restraints were derived from 15N- or 13C-resolved three-dimensional NOESY. HN–N RDCs were measured using a spin-state-selective 1H, 15N correlation experiment in a dilute liquid crystalline medium (Rückert & Otting, 2000). Restraints for the backbone angles φ and ψ were derived from TALOS (Cornilescu et al, 1999). Slowly exchanging amide protons were identified from 1H, 15N correlation experiments after redissolving lyophilized protein in D2O. Stereospecific assignments of leucine and valine methyl groups were obtained using a 10% fractionally 13C-labelled sample, as described (Neri et al, 1989).
For NMR titrations, increasing amounts of the 21-mer siRNA were added to a 0.25 mM solution of 2H-, 15N-labelled B2 dimer up to a twofold molar excess. Chemical shifts were monitored in two-dimensional 1H, 15N HSQC experiments. The sigmoidal shape of the binding curves (Fig 3B) may result from slow to intermediate exchange between free and bound conformations on the NMR chemical shift timescale (Feeney et al, 1979), but could also indicate a more complex binding behaviour.
15N relaxation (T1, T2 and heteronuclear {1H}–15N NOE) was measured on a 15N-labelled B2 sample at 295 K and 500 MHz 1H Larmor frequency, as described (Farrow et al, 1994). The different T1/T2 ratios for helices α1 and α2 compared with helix α3 are consistent with the elongated structure of the B2 dimer, and indicate a considerable degree of anisotropy of the rotational diffusion tensor (Tjandra et al, 1997). Analysis of the data using TENSOR (Dosset et al, 2000) indicates a prolate diffusion tensor with an axial ratio of 1:1.6 and an overall tumbling correlation time of 15.5 ns. The relaxation data fit well to the elongated dimer structure, although the average T2 values (trimmed mean T2=42 ms) are shorter than expected, indicating further exchange broadening for many amide protons.
Structure calculation. The experimentally determined distance and dihedral and dipolar coupling restraints (Table 1) were applied in a simulated annealing protocol using ARIA (Linge et al, 2001) and CNS (Brünger et al, 1998). Non-crystallographic symmetry restraints were used to enforce the dimer symmetry. In a first step, all NOEs were entered with a fourfold ambiguity, that is, two intra-monomer and two inter-molecular assignments. After convergence, the NOEs were assigned unambiguously (when possible) as inter- or intra-monomer contacts. Symmetrically unambiguous and ambiguous peak lists were created and used in a final structure calculation for refined distance calibrations. The final ensemble of NMR structures was refined in a shell of water molecules (Linge et al, 2003). Structural quality was analysed using PROCHECK (Laskowski et al, 1996). Ribbon and surface representations were prepared with MOLMOL (Koradi et al, 1996) and GRASP (Nicholls et al, 1991), respectively.
Protein Data Bank accession numbers. The coordinates and NMR data for the B2 dimer structure have been deposited at the Protein Data Bank under accession number 2B9Z.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank M. Nilges (Institut Pasteur, Paris) for help with structure calculations and C. Mackereth and D. Thomas for a critical reading of the manuscript. High-field NMR measurement time granted by the German Research Foundation (DFG) at the Biomolecular Magnetic Resonance Centre in Frankfurt (Germany) is acknowledged. This study was supported by the European Molecular Biology Organization (EMBO), the DFG, the European Union (STREP FSG-V-RNA) and the Human Frontier Science Program Organization.
Competing interests statement The authors declare that they have no competing financial interests.
References
- Banner DW, Kokkinidis M, Tsernoglou D (1987) Structure of the ColE1 rop protein at 1.7 Å resolution. J Mol Biol 196: 657–675 [DOI] [PubMed] [Google Scholar]
- Brünger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR (2005) Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol (Epub ahead of print) doi:10.1038/nsmb1005 [DOI] [PubMed] [Google Scholar]
- Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT (1997) A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol 4: 891–895 [DOI] [PubMed] [Google Scholar]
- Chien CY, Xu Y, Xiao R, Aramini JM, Sahasrabudhe PV, Krug RM, Montelione GT (2004) Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry 43: 1950–1962 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Marquardt JL, Ottiger M, Bax A (1998) Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc 120: 6836–6837 [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX Pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Dosset P, Hus JC, Blackledge M, Marion D (2000) Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR 16: 23–28 [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003 [DOI] [PubMed] [Google Scholar]
- Feeney J, Batchelor JG, Albrand JP, Roberts GCK (1979) The effects of intermediate exchange processes on the estimation of equilibrium constants by NMR. J Magn Reson 33: 519–529 [Google Scholar]
- Fierro-Monti I, Mathews MB (2000) Proteins binding to duplexed RNA: one motif, multiplefunctions. Trends Biochem Sci 25: 241–246 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1997) Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res 25: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA (1994) NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Johnson KN, Johnson KL, Dasgupta R, Gratsch T, Ball LA (2001) Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J Gen Virol 82: 1855–1866 [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW (2002) Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Linge JP, O'Donoghue SI, Nilges M (2001) Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol 339: 71–90 [DOI] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M (2003) Refinement of protein structures in explicit solvent. Proteins 50: 496–506 [DOI] [PubMed] [Google Scholar]
- Lingel A, Sattler M (2005) Novel modes of protein–RNA recognition in the RNAi pathway. Curr Opin Struct Biol 15: 107–115 [DOI] [PubMed] [Google Scholar]
- Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM (1997) Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol 4: 896–899 [DOI] [PubMed] [Google Scholar]
- Lu D, Searles MA, Klug A (2003) Crystal structure of a zinc-finger–RNA complex reveals two modes of molecular recognition. Nature 426: 96–100 [DOI] [PubMed] [Google Scholar]
- Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW (2005) Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436: 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D, Szyperski T, Otting G, Senn H, Wüthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28: 7510–7516 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Funct Genet 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Plasterk RH (2002) RNA silencing: the genome's immune system. Science 296: 1263–1265 [DOI] [PubMed] [Google Scholar]
- Predki PF, Nayak LM, Gottlieb MB, Regan L (1995) Dissecting RNA–protein interactions: RNA–RNA recognition by Rop. Cell 80: 41–50 [DOI] [PubMed] [Google Scholar]
- Roth BM, Pruss GJ, Vance VB (2004) Plant viral suppressors of RNA silencing. Virus Res 102: 97–108 [DOI] [PubMed] [Google Scholar]
- Rückert M, Otting G (2000) Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc 122: 7793–7797 [Google Scholar]
- Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog NMR Spectrosc 34: 93–158 [Google Scholar]
- Sullivan CS, Ganem D (2005) A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol 79: 7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N, Bax A (1997) Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science 278: 1111–1114 [DOI] [PubMed] [Google Scholar]
- Tjandra N, Garrett DS, Gronenborn AM, Bax A, Clore GM (1997) Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy. Nat Struct Biol 4: 443–449 [DOI] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet 6: 206–220 [DOI] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426: 874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


