Abstract
Lipid droplets (LDs), also called adiposomes, are found in many eukaryotic cells, and are highly upregulated in lipid-storage cells, such as adipocytes. The mechanism by which adiposomes and their component neutral lipids are degraded is an important health issue with the rapidly spreading epidemic of obesity. Recently, a novel triglyceride lipase (adipose triglyceride lipase (ATGL)) that catalyses the initial step in triglyceride hydrolysis in adipocyte LDs was identified. Here, we show that ATGL also functions in non-adipocyte cells, and has an important role in LD degradation in these cells. Overexpression of wild-type ATGL causes a marked decrease in LD size, whereas a catalytically inactive mutant retains the ability to localize to LDs, but is unable to decrease their size. Depletion of ATGL by RNA interference leads to a significant increase in the size of LDs. These results show that ATGL has an important role in LD/adiposome turnover in mammalian cells.
Keywords: adipose triglyceride lipase, lipid droplet, adiposome, patatin domain, adiposome triglyceride lipase
Introduction
Lipid droplets (LDs) are the main storage organelles for triglycerides in eukaryotic cells (Brown, 2001; Haemmerle et al, 2003; Londos et al, 2005). R. Anderson and co-workers (Liu et al, 2004) have proposed the name ‘adiposome' for LDs to reflect the fact that they are not merely inert storage bodies, but rather are dynamic organelles that are intimately linked to membrane transport pathways in the cell. We will use the two terms interchangeably here. Triglycerides are the principal energy reserve in cells, and, in mammals, excessive storage of triglycerides is directly linked to obesity (Haemmerle et al, 2003; Londos et al, 2005). Despite the fact that obesity and related disorders such as metabolic syndrome have become leading health problems, little is known about the molecular mechanisms of triglyceride turnover. The best-studied pathway of triglyceride degradation in adipocyte LDs is that mediated by hormone-sensitive lipase (HSL; Haemmerle et al, 2003; Yeaman, 2004). For the past 30 years, it has been widely accepted that HSL is the principal adiposome degradation lipase acting in adipocytes. It thus came as a great surprise when HSL knockout mice failed to become obese and cells from these mice were shown to possess triglyceride lipase activity (Haemmerle et al, 2003). These results indicated that other lipases are also involved in LD degradation in mammalian cells. At this time, several LD proteomic studies were published, and two groups reported the identification of a novel protein with homology to plant lipases in LDs (Liu et al, 2004; Umlauf et al, 2004). Although its biochemical function was not known at the time the proteomic studies were published, three groups have recently shown that this protein, known as adipose triglyceride lipase (ATGL), has triglyceride lipase activity (Jenkins et al, 2004; Villena et al, 2004; Zimmermann et al, 2004; Zechner et al, 2005). The discovery of a second family of mammalian triglyceride lipases resolves the conundrum posed by the HSL-null mice (Raben & Baldassare, 2005). It is clear from the above-mentioned studies that ATGL has a crucial role in LD degradation in adipocytes, but whether it is involved in this process in other cell types is not known. With this aim, we modulated the level of ATGL in non-adipocyte cells. We found that overexpression of ATGL in HeLa cells causes a decrease in the average size of adiposomes, whereas knockdown of ATGL by RNA interference leads to an increase in their size. This indicates that ATGL has an important role in adiposome degradation, not only in adipocytes, but also in other mammalian cell types.
Results and Discussion
We set out to determine the role of ATGL in non-adipocyte cells. ATGL localizes to LDs in mouse 3T3-L1 adipocytes (Zimmermann et al, 2004), but overexpressed ATGL was reported to localize to the cytoplasm in non-adipocyte (COS-7) cells (Villena et al, 2004). We raised antibodies against ATGL to examine the subcellular localization of the endogenous protein in HeLa cells by immunofluorescence analysis. Cells were co-stained with antibodies to TIP47 (tail-interacting protein of 47 kDa), a member of the perilipin/ADRP/TIP47 (PAT) family of abundant LD-associated proteins (Wolins et al, 2001; Hickenbottom et al, 2004; Londos et al, 2005). Endogenous ATGL showed a pattern of small rings that colocalized well with TIP47 (Fig 1A, upper panel). Nile Red is a dye selective for neutral lipids such as triglycerides and cholesterol esters that make up the core of adiposomes. When cells were co-stained with anti-ATGL antiserum and Nile Red, the rings of ATGL staining surrounded the Nile-Red-stained central cores of the adiposomes (Fig 1A, lower panel). The size of adiposomes in cells grown in culture can be regulated by the amount of fatty acid present in the medium. ATGL and TIP47 localization were examined in HeLa cells grown in the presence of oleic acid (Fig 1B, upper panel). Both proteins colocalized around the rims of the adiposomes, which were larger and more numerous than those in untreated cells (Fig 1B). When cells were co-stained with anti-ATGL antibodies and BODIPY 493/503 dye (which, like Nile Red, binds to the neutral lipid core of adiposomes), the ATGL staining surrounded the BODIPY-stained central core of the droplets (Fig 1B, lower panel). These results indicate that endogenous ATGL is localized to the external surface of adiposomes. We also found that ATGL localizes to adiposomes in COS-7 cells (not shown), indicating that ATGL is not only found on LDs in adipocytes, but also in non-adipocyte cells. Previous studies have found that mouse ATGL messenger RNA is highly expressed in adipose tissue, and is also present in other mouse tissues (Villena et al, 2004; Zimmermann et al, 2004), providing further evidence for the conclusion that ATGL function is not restricted to adipocytes alone, but present in other cell types as well.
Figure 1.
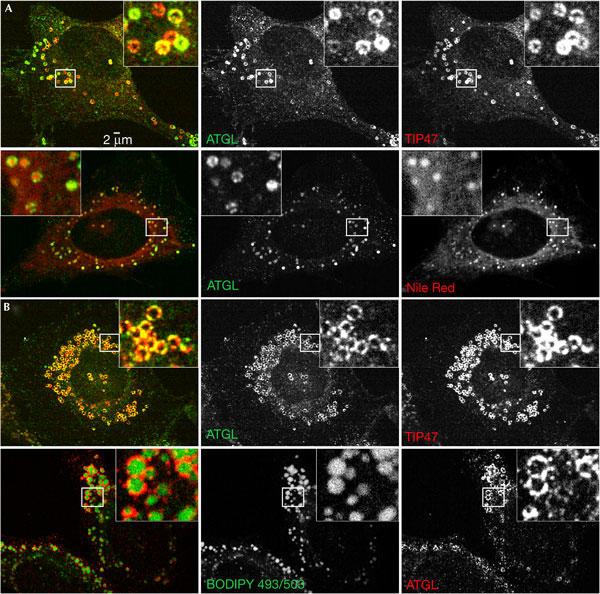
Endogenous ATGL localizes to adiposomes. HeLa cells were immunostained with antibodies against ATGL and either anti-TIP47 antibodies or the Nile Red (or BODIPY 493/503) dye. Cells were incubated overnight with 62.5 μM oleic acid complexed to albumin (OA/BSA), where indicated. (A) Cells grown in normal low-lipid-containing medium show colocalization of ATGL (green) with TIP47 (red; upper panel) and colocalization of ATGL (green) with Nile Red (red; lower panel). (B) Cells grown overnight in the presence of medium supplemented with 62.5 μM OA/BSA show colocalization of ATGL (green) with TIP47 (red; upper panel) and ATGL (red) with BODIPY 493/503 dye (green; lower panel).
To test whether ATGL has the capacity to affect adiposome turnover, we overexpressed a green fluorescent protein (GFP)-tagged version of the protein in HeLa cells. Cells overexpressing ATGL–GFP showed a marked reduction in the size of adiposomes compared with neighbouring untransfected controls (Fig 2A, upper panel). The average diameter of adiposomes in cells transfected with wild-type ATGL was approximately 0.50±0.09 μm, whereas LD diameter was 1.08±0.14 μm in untransfected cells (Fig 2D). This change in diameter corresponds to a tenfold decrease in the volume of LDs in cells overexpressing ATGL. When ATGL–GFP-transfected cells were grown in the presence of oleic acid to induce LD formation, overexpression of ATGL–GFP caused an even larger decrease in the size of adiposomes (Fig 2A, lower panel). The average adiposome diameter was 1.74±0.36 μm for untransfected cells compared with 0.47±0.14 μm for ATGL-transfected cells (Fig 2D), corresponding to a 50-fold decrease in volume. Similar results were observed in COS-7 cells (E.S. & C.L.J., unpublished data).
Figure 2.
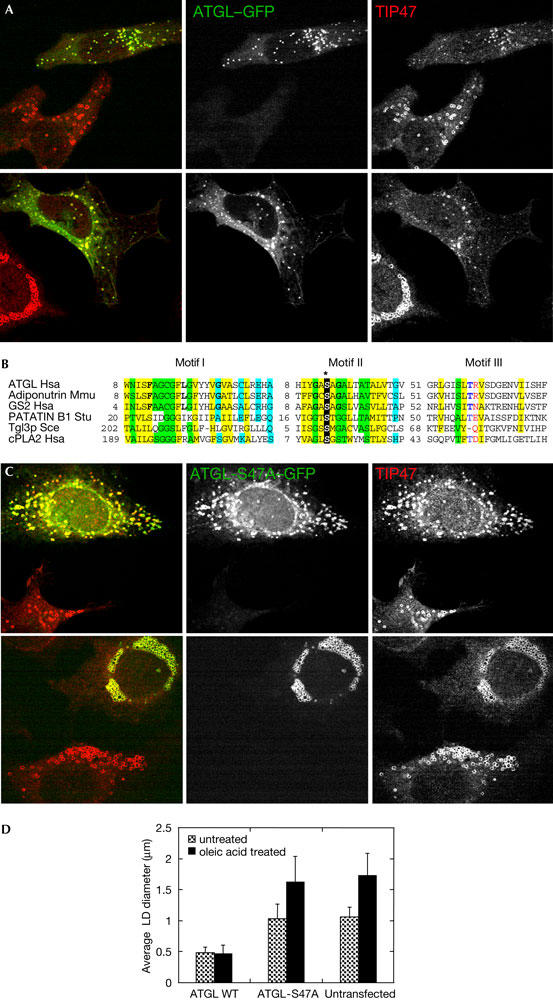
Overexpression of ATGL causes a decrease in the size of adiposomes. HeLa cells were transiently transfected with plasmid expressing ATGL–GFP (green fluorescent protein) or ATGL-S47A–GFP and analysed by immunofluorescence 24 h after transfection. (A) ATGL–GFP (green) and TIP47 (red) in cells growing under normal conditions (upper panel), or incubated with 400 μM oleic acid complexed to albumin (OA/BSA; lower panel). (B) Sequence alignment of a portion of the catalytic domain of ATGL and related lipases showing the catalytic serine (*; for details, see supplementary Fig S1 online). (C) ATGL-S47A–GFP (green) and TIP47 (red) in cells growing under regular conditions (upper panel), or incubated with 400 μM OA/BSA (lower panel). (D) Quantification of the results shown in (A,C). Data shown are mean±standard deviation. An average of 120 lipid droplets (LDs) was measured for each point. WT, wild type.
To test whether degradation of adiposomes was due to the lipase activity of ATGL, we overexpressed a catalytically inactive mutant. The enzymes of the ATGL family show significant homology to other lipases and serine esterases, including phospholipase A2 enzymes (Jenkins et al, 2004; Villena et al, 2004; Zimmermann et al, 2004). An invariant serine residue in the core of the catalytic domain of cytosolic phospholipase A2 (cPLA2) has been shown to be essential for its enzyme activity in vitro (Huang et al, 1996). This serine is conserved in ATGL (Fig 2B; see also supplementary Fig S1 online); hence, we constructed an ATGL-S47A mutant. We verified that an ATGL mutant lacking S47 has reduced lipase activity (supplementary Fig S2 online), and examined the size of adiposomes in cells expressing ATGL-S47A. The diameter of adiposomes, visualized by TIP47 staining, was essentially the same in untransfected and ATGL-S47A-transfected cells (Fig 2C, upper panel). Under conditions that stimulate adiposome formation, ATGL-S47A colocalized with TIP47 around the rim of the adiposomes. The diameter of adiposomes in ATGL-S47A-transfected cells was indistinguishable from the untransfected control cells (Fig 2C, lower panel). Quantification of these results is shown in Fig 2D. These results show that ATGL lipase activity is essential for adiposome shrinkage by ATGL.
We noted that some cells overexpressing ATGL seemed to have less than normal or even no TIP47 staining on adiposomes. We examined cells overexpressing ATGL or the catalytically inactive ATGL-S47A mutant for the level of TIP47 on adiposomes. Interestingly, overexpression of wild-type ATGL significantly inhibits association of TIP47 with adiposomes (Table 1). The catalytically inactive ATGL-S47A mutant has a weaker effect on TIP47 displacement from adiposomes (Table 1). When cells were treated with oleic acid, the displacement of TIP47 from adiposomes by either ATGL or ATGL-S47A was less pronounced (Table 1). TIP47 has been shown to have a role in adiposome formation, associating rapidly with newly forming adiposomes (Wolins et al, 2001, 2005). These results are consistent with the idea that TIP47 associates with adiposomes during their formation, and that high levels of ATGL displace TIP47 during the process of adiposome degradation.
Table 1.
ATGL overexpression displaces TIP47 from adiposomes
| Normala | Decreasedb | Absentc | |
|---|---|---|---|
| ATGL–GFP (n=103d) | 20.4 | 21.4 | 58.2 |
| ATGL–GFP, OA (n=105) | 48.6 | 35.2 | 16.2 |
| ATGL-S47A–GFP (n=101) | 49.5 | 36.6 | 13.9 |
| ATGL-S47A–GFP, OA (n=105) | 79.0 | 20.0 | 1.0 |
aPercentage of ATGL- or ATGL-S47A-expressing cells having the same level of TIP47 staining on lipid droplets as untransfected control cells.
bPercentage of ATGL- or ATGL-S47A-expressing cells having fainter staining for TIP47 on LDs as compared with untransfected cells.
cPercentage of ATGL- or ATGL-S47A-expressing cells with no TIP47-stained LDs.
dn indicates the number of cells scored.
We have established that overexpression of ATGL causes a decrease in the size of adiposomes. For further proof of ATGL involvement in adiposome degradation, we tested the effect of knockdown of ATGL expression on adiposome size. HeLa cells were treated with a pool of short interfering RNAs (siRNAs) directed against ATGL and were prepared for immunofluorescence analysis using anti-TIP47 antibodies (Fig 3A). The siRNA treatment was effective at inhibiting ATGL expression. When ATGL–GFP was co-transfected with siRNAs against ATGL, less than 5% of ATGL–GFP was present in these cells compared with cells co-transfected with a control siRNA unrelated to ATGL (Fig 3C). Strikingly, HeLa cells treated with siRNA directed against ATGL had an average adiposome diameter of 1.43±0.21 μm compared with 1.02±0.14 μm for cells treated with the control siRNA (Fig 3D). This change in diameter corresponds to an approximately threefold increase in the volume of LDs in ATGL-depleted cells. In cells treated with oleic acid and ATGL siRNA, we also observed accumulation of larger LDs compared with the control (Fig 3B). The average LD size in oleic acid-treated cells was 1.70±0.18 μm compared with 2.09±0.24 μm in cells also treated with ATGL siRNA (Fig 3D). This difference in diameter corresponds to an approximately twofold difference in LD volume. These results indicate that endogenous ATGL in HeLa cells is involved in adiposome degradation, a conclusion that has important implications. First, the other human ATGL homologues (see supplementary Fig S3 online) do not have a completely overlapping function in HeLa cells, as they cannot maintain the steady-state adiposome size in the absence of ATGL. Second, ATGL function is not restricted to adipocytes, as removal of endogenous ATGL in HeLa cells results in an increase in adiposome size.
Figure 3.
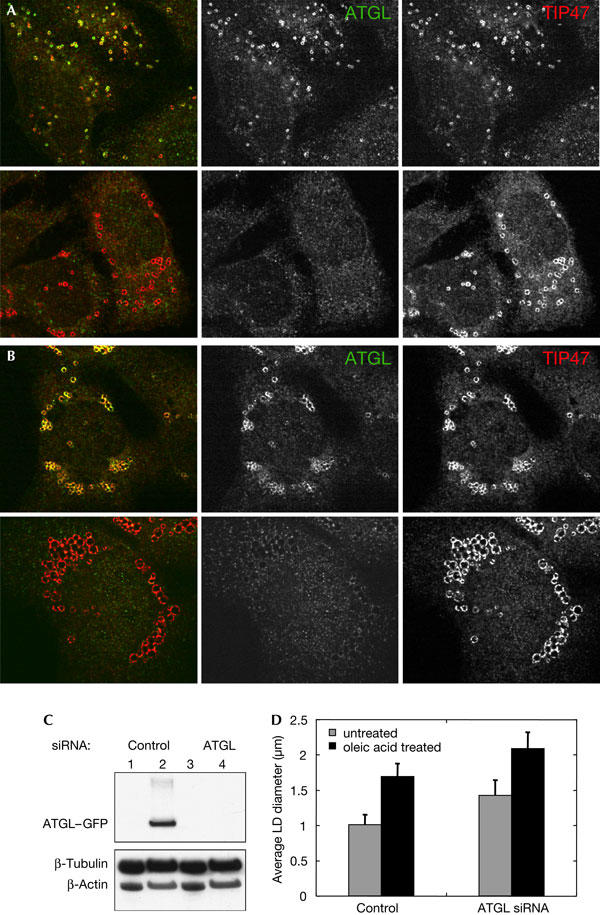
Depletion of ATGL by RNA interference increases the size of lipid droplets. (A) HeLa cells were treated with ATGL short interfering RNA (siRNA; lower panel) or control unrelated siRNA (upper panel) for 72 h, and were then analysed by immunofluorescence microscopy. Overlay images are shown in the left column, staining for endogenous ATGL (green) is in the centre and TIP47 (red) is on the right. (B) Same as (A), except that cells were incubated with 400 μM oleic acid complexed to BSA before fixation. (C) Immunoblot analysis of ATGL expression. HeLa cells were transfected with control siRNA (lanes 1,2) or ATGL siRNA (lanes 3,4). siRNAs were transfected either alone (lanes 1,3) or in combination with a plasmid expressing ATGL–GFP (green fluorescent protein; lanes 2,4). ATGL–GFP was detected using an anti-ATGL antibody. Tubulin and actin were used as loading controls. (D) Quantification of the results shown in (A,B). Data shown are mean±standard deviation. An average of 350 lipid droplets (LDs) was measured for each point.
As ATGL functions in human cells other than adipocytes, we were curious about whether other eukaryotic organisms have triglyceride lipases acting on adiposomes. The yeast Saccharomyces cerevisiae possesses adiposomes (Mullner & Daum, 2004), and it has been reported in a proteomic analysis of these organelles that a triglyceride lipase, Tgl3p, is associated with them (Athenstaedt & Daum, 2003). Tgl3p was shown to colocalize with adiposomes in yeast, and overexpression of Tgl3p led to a decrease in the ratio of triglyceride to protein in LDs. Deletion of Tgl3p had the opposite effect, leading to an increase in the level of triglyceride compared with that of protein in LDs (Athenstaedt & Daum, 2003). Hence, a eukaryote as divergent from humans as the yeast S. cerevisiae possesses a triglyceride lipase that is involved in adiposome degradation.
Both ATGL and Tgl3p are related to a large group of α/β hydrolases containing a patatin domain, as determined by sequence similarity searches (Altschul et al, 1997). We reconstructed a phylogenetic tree to determine the evolutionary relationship between ATGL and Tgl3p (Fig 4). The result shows that, early in eukaryotic evolution, a duplication event gave rise to the ATGL and Tgl3p families. Patatin and calcium-independent phospholipase A2 (iPLA2) are well-characterized lipases that are members of subfamilies that arose from duplication events after the Tgl3p–ATGL family split (Fig 4). In addition to the patatin, iPLA2, ATGL and Tgl3p families, there is a fifth lipase family in the patatin superfamily that includes human neuropathy target esterase (NTE). The NTE family has a distinct evolutionary origin compared with that of the other four families of lipases, as can be seen from its deep embedding in the branch of bacterial lipases (Fig 4). This is consistent with the distinct domain architecture of the NTE lipases (Lush et al, 1998). There are five human members of the ATGL family of lipases, as has been described previously (Zechner et al, 2005). We constructed a tree showing the evolutionary relationships among these lipases (supplementary Fig S3 online), which shows that ATGL is more closely related to adiponutrin (also known as iPLA2ɛ) than to GS2 (also known as iPLA2η), both of which have been shown to possess triglyceride lipase activity in vitro, like ATGL (Jenkins et al, 2004).
Figure 4.
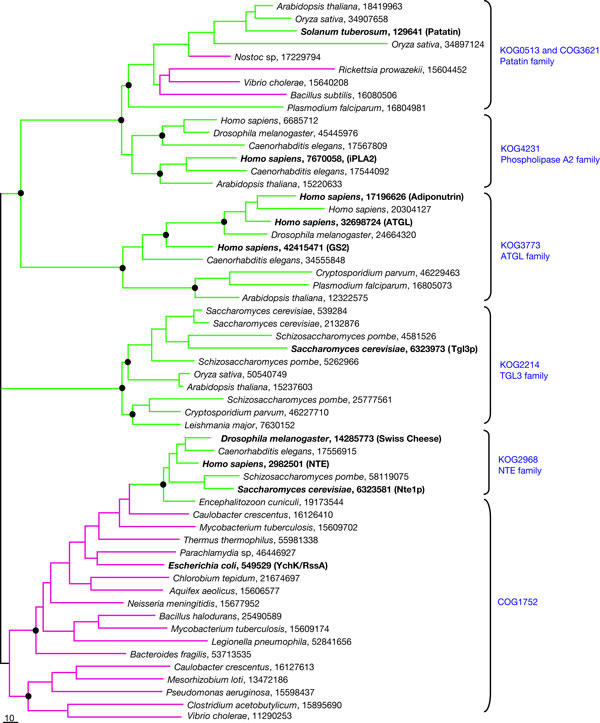
Phylogenetic tree for lipases of the patatin group. Maximum-likelihood unrooted tree was built using the MOLPHY program. The same program was used to compute bootstrap probabilities. Each terminal node of the tree is labelled by the scientific name of the organism in which the protein is encoded and the numeric GenBank identifier (GI). The names for experimentally characterized proteins are indicated in parentheses and are shown in bold type. Those main branches that were supported by bootstrap probability greater than 70% are marked by black circle. Principal protein families are named by a typical representative and by a KOG/COG number from the database at NCBI (http://www.ncbi.nlm.nih.gov/COG/new/). Branches and nodes for bacterial sequences are highlighted in magenta and those for eukaryotic sequences in green.
There is a significant amount of sequence similarity in the catalytic domains of Tgl3p, ATGL, iPLA2 and other members of the patatin superfamily, including the catalytic serine we mutated in ATGL (Fig 2B; supplementary Fig S1 online). As a further test of the functional similarity between Tgl3p and ATGL, we expressed yeast Tgl3p in HeLa cells. Remarkably, the yeast lipase was targeted efficiently to HeLa cell LDs (Fig 5). Although the expression level of Tgl3p in HeLa cells was not as high as that for ATGL, probably owing to problems of expression of the heterologous protein, in some cells it seemed that Tgl3p expression led to reduction in the size of adiposomes (Fig 5). Thus, despite their divergence, the Tgl3p and ATGL families of lipases are functionally similar. Interestingly, during eukaryotic evolution, the Tgl3p orthologues were apparently lost in animals, and ATGL-like genes were lost in fungi (Fig 4).
Figure 5.
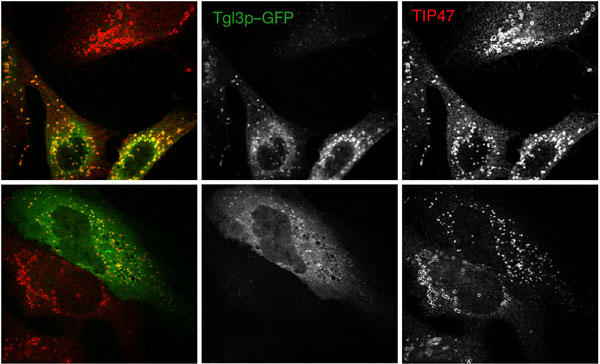
Yeast Tgl3p expressed in HeLa cells localizes to lipid droplets. HeLa cells were transfected with a plasmid encoding Saccharomyces cerevisiae Tgl3p fused to green fluorescent protein (GFP). At 24 h after transfections, cells were immunostained with TIP47 antibodies (red). Tgl3p–GFP fluorescence is in green. Two separate microscopy fields are shown in the upper and lower panels.
In conclusion, our results show that ATGL regulates adiposome size and has a significant role in their degradation in mammalian cells. We show that ATGL regulates LD size in non-adipocyte cells, which complements the previously published work indicating its role in LD triglyceride degradation in adipocytes (Jenkins et al, 2004; Villena et al, 2004; Zimmermann et al, 2004). ATGL is a member of the superfamily of patatin domain lipases that also includes Tgl3p, the distantly related but functionally similar yeast triglyceride lipase. Tgl3p has been localized to adiposomes and shown to mobilize triglycerides from them when overexpressed. On the basis of these results, we propose a minor alteration in name for ATGL from ‘adipose triglyceride lipase' to ‘adiposome triglyceride lipase'. In both HeLa and COS-7 cells, overexpression of the ATGL lipase, but not of a catalytically inactive mutant form of ATGL, leads to a striking decrease in the size of adiposomes. In HeLa cells, depletion of ATGL by RNA interference results in a significant increase in the size of adiposomes. Hence, ATGL causes adiposome degradation, not only in fat-storage cells, but also in other eukaryotic cells.
Methods
Antibodies and plasmid constructs. See the supplementary information online.
Cell culture and immunofluorescence microscopy. HeLa and COS-7 cells were cultured on glass coverslips in DMEM supplemented with 10% FBS and allowed to adhere overnight. Cells were transfected using FuGENE 6 (Roche, Indianapolis, IN, USA) transfection reagent, according to the manufacturer's protocol. To increase triacylglycerol synthesis and storage in cells, 62.5–400 μM oleic acid complexed to albumin (OA/BSA) was added to the medium and incubated overnight. To visualize LDs using dyes, Nile Red or BODIPY 493/503 dye (Molecular Probes, Eugene, OR, USA) was added to the medium 15 min before fixation and then, additionally, during incubation with antibodies. Cells were fixed with 3.7% formaldehyde and mounted on glass microscope slides using ProLong mounting media (Molecular Probes). Images were captured using a Perkin-Elmer UltraView confocal microscope. For TIP47 displacement quantification, see the supplementary information online. For LD size quantifications, additional images were taken using Zeiss 510 laser-scanning confocal microscope and analysed using the 510 Image Analyzer software.
RNA interference. For ATGL expression silencing, siGENOME:TTS-2.2 SMARTpool reagent was used (Dharmacon, Lafayette, CO, USA). As a control, a SMARTpool reagent against an unrelated mRNA was used. Two consecutive transfections of HeLa cells were carried out using 50 nM siRNA and Lipofectamine™ 2000 reagent (Invitrogen, Carlsbad, CA, USA) at 24 h intervals and cells were collected 72 h after the first transfection. 400 μM OA/BSA was added for 22 h before fixation, where indicated.
Phylogenetic analysis See the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400559-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Dr P. Shaw (University of Nottingham, Nottingham, UK) for providing the ATGL complementary DNA.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G (2003) YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J Biol Chem 278: 23317–23323 [DOI] [PubMed] [Google Scholar]
- Brown DA (2001) Lipid droplets: proteins floating on a pool of fat. Curr Biol 11: R446–R449 [DOI] [PubMed] [Google Scholar]
- Haemmerle G, Zimmermann R, Zechner R (2003) Letting lipids go: hormone-sensitive lipase. Curr Opin Lipidol 14: 289–297 [DOI] [PubMed] [Google Scholar]
- Hickenbottom SJ, Kimmel AR, Londos C, Hurley JH (2004) Structure of a lipid droplet protein; the PAT family member TIP47. Structure (Camb) 12: 1199–1207 [DOI] [PubMed] [Google Scholar]
- Huang Z, Payette P, Abdullah K, Cromlish WA, Kennedy BP (1996) Functional identification of the active-site nucleophile of the human 85-kDa cytosolic phospholipase A2. Biochemistry 35: 3712–3721 [DOI] [PubMed] [Google Scholar]
- Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW (2004) Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 279: 48968–48975 [DOI] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG (2004) Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem 279: 3787–3792 [DOI] [PubMed] [Google Scholar]
- Londos C, Sztalryd C, Tansey JT, Kimmel AR (2005) Role of PAT proteins in lipid metabolism. Biochimie 87: 45–49 [DOI] [PubMed] [Google Scholar]
- Lush MJ, Li Y, Read DJ, Willis AC, Glynn P (1998) Neuropathy target esterase and a homologous Drosophila neurodegeneration-associated mutant protein contain a novel domain conserved from bacteria to man. Biochem J 332: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner H, Daum G (2004) Dynamics of neutral lipid storage in yeast. Acta Biochim Pol 51: 323–347 [PubMed] [Google Scholar]
- Raben DM, Baldassare JJ (2005) A new lipase in regulating lipid mobilization: hormone-sensitive lipase is not alone. Trends Endocrinol Metab 16: 35–36 [DOI] [PubMed] [Google Scholar]
- Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R (2004) Association of stomatin with lipid bodies. J Biol Chem 279: 23699–23709 [DOI] [PubMed] [Google Scholar]
- Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS (2004) Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem 279: 47066–47075 [DOI] [PubMed] [Google Scholar]
- Wolins NE, Rubin B, Brasaemle DL (2001) TIP47 associates with lipid droplets. J Biol Chem 276: 5101–5108 [DOI] [PubMed] [Google Scholar]
- Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE (2005) S3-12, adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem 280: 19146–19155 [DOI] [PubMed] [Google Scholar]
- Yeaman SJ (2004) Hormone-sensitive lipase—new roles for an old enzyme. Biochem J 379: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Strauss JG, Haemmerle G, Lass A, Zimmermann R (2005) Lipolysis: pathway under construction. Curr Opin Lipidol 16: 333–340 [DOI] [PubMed] [Google Scholar]
- Zimmermann R et al. (2004) Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306: 1383–1386 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


