Abstract
Metazoans rapidly respond to changes in oxygen availability by regulating gene expression. The transcription factor hypoxia-inducible-factor (HIF), which controls the expression of several genes, ‘senses' the oxygen concentration indirectly through the hydroxylation of two proline residues that earmarks the HIF-α subunits for proteasomal degradation. We review the expression, regulation and function of the HIF prolyl hydroxylases or prolyl hydroxylases domain proteins, which are genuine oxygen sensors.
Keywords: cell signalling, hypoxia inducible factor, oxygen sensing, prolyl hydroxylase domain protein
Introduction
All organisms respond to changes in their environment by activating networks of signal transduction pathways that are regulated by post-translational modifications and that culminate in changes in gene expression. Studies of the adaptation to low O2 availability have now identified the hypoxia-inducible-factor (HIF) transcription factor, and a further mechanism of signal transduction involving a ‘new' post-translational modification that is inherently O2-dependent: hydroxylation on proline residues. This review summarizes our current knowledge of the prolyl hydroxylase domain-containing proteins (PHDs) that catalyse HIF prolyl hydroxylation.
The hypoxia-inducible-factor transcriptional complex
HIF is central to O2 homeostasis during embryonic development and postnatal life in both physiological and pathophysiological processes such as tumour growth, ischaemia and tissue repair (Semenza, 1998). Indeed, HIF regulates the transcription of many genes involved in cellular and systemic responses to hypoxia. HIF consists of one of the three O2-regulated HIF-α subunits (HIF-1α, HIF-2α and HIF-3α) and the constitutively expressed HIF-1β subunit. The most extensively studied isoform is the ubiquitous HIF-1α.
HIF activation is a multistep process involving HIF-α stabilization, nuclear translocation, hetero-dimerization, transcriptional activation and interaction with other proteins. Several steps in this activation process are independently regulated by O2. However, O2-dependent regulation of the proteasomal degradation of HIF-α is the most crucial step (Huang et al, 1996; Salceda & Caro, 1997).
Looking for the oxygen sensor
In well-oxygenated cells, HIF-α is an exceptionally short-lived protein (half-life less than 5 min at 21% O2) and steady-state levels are very low. By contrast, reduced O2 availability induces HIF-α accumulation by relaxing its ubiquitin-proteasome degradation. A central oxygen-dependent degradation domain (ODDD) mediates HIF-α proteolytic degradation. The mechanism by which O2 deprivation increases HIF-α stability remained obscure until Ratcliffe's group showed that ubiquitylation and proteasomal degradation of HIF-α requires pVHL, the product of the von Hippel–Lindau tumour suppressor gene (Maxwell et al, 1999). pVHL is the HIF-α recognition component of a multiprotein complex, which functions as a ubiquitin E3 ligase (Kim & Kaelin, 2003). Subsequent studies have shown that stabilization of HIF-α is due to a disruption of the pVHL/HIF-α interaction under hypoxic conditions. Two laboratories simultaneously reported that O2-dependent hydroxylation of two conserved proline residues (Pro402 and Pro564 in human HIF-1α) in the ODDD triggers pVHL binding and thus HIF-1α proteasome targeting (Ivan et al, 2001; Jaakkola et al, 2001). Within months of these initial reports, the PHDs—the enzymes catalysing the hydroxylation reaction—were identified (see Fig 1; Bruick & McKnight, 2001; Epstein et al, 2001).
Figure 1.
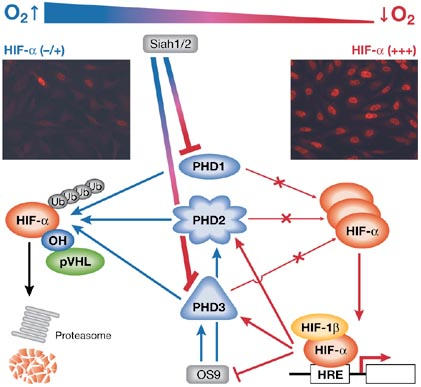
Prolyl-hydroxylase-domain proteins regulate hypoxia inducible factor-α in response to O2 availability. Under aerobic conditions (blue arrows), prolyl-hydroxylase-domain proteins (PHDs) hydroxylate hypoxia inducible factor-α (HIF-α), which allows the von Hippel–Lindau protein (pVHL) to bind and thus target HIF-α to the proteasome. Likewise, by binding to PHD2 and PHD3, OS9 promotes HIF-α hydroxylation. A decrease in O2 availability (red arrows) inhibits the PHDs; HIF-α accumulates and induces the expression of target genes. In addition, Siah 1 and 2 trigger PHD1 and PHD3 degradation under hypoxic conditions. Interestingly, hypoxia controls PHD2, PHD3, OS9 and Siah 1 and 2 expression by a feedback loop mechanism. Immunofluorescence inserts show expression of HIF-1α in HeLa cells at 20% O2 (left) and 1–2% O2 (right). HRE, hypoxia response element.
The family of HIF prolyl hydroxylases
Epstein and colleagues identified egg-laying abnormal-9 (EGL-9) as the HIF prolyl hydroxylase in Caenorhabditis elegans and they described a family of three human and mouse phd genes that are homologous to egl-9 (Epstein et al, 2001). Unfortunately, several acronyms were coined to describe these genes. PHD1, PHD2 and PHD3 (used in the initial paper and in this review) have also been called EGL nine homologue 2 (EGLN2), EGLN1 and EGLN3, or HIF prolyl hydroxylase 3 (HPH3), HPH2 and HPH1, respectively. Each of these homologues has a conserved gene structure, which suggests duplication in the lineage leading to vertebrates (Taylor, 2001).
The PHDs belong to the superfamily of iron- and 2-oxoglutarate-dependent dioxygenases. These enzymes need O2 as a co-substrate, which provides the molecular basis for their O2-sensing function. In the hydroxylation reaction, one oxygen atom is added to a peptidyl proline to form hydroxyproline, whereas the other is used in a coupled decarboxylation reaction that converts 2-oxoglutarate to succinate; thus, PHDs also require 2-oxoglutarate as a co-substrate. Likewise, PHDs use Fe2+ and ascorbate as co-factors. Fe2+ is crucial for activating O2 and as a template for the orderly binding of reactants. Ascorbate seems to act by reducing Fe3+, which binds to the active site of the enzyme after the decarboxylation reaction, and is therefore necessary for its re-activation (Fig 2).
Figure 2.
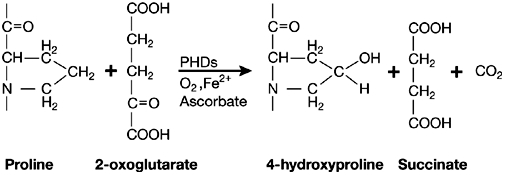
Catalytic function of the prolyl-hydroxylase-domain proteins. These enzymes need O2 and 2-oxoglutarate as co-substrates, and Fe2+ and ascorbate as co-factors. The hydroxylation reaction forms hydroxyproline and succinate. PHDs, prolyl-hydroxylase-domain proteins.
Substrate specificity. PHDs hydroxylate two proline residues in a conserved LxxLAP sequence motif. In vitro studies have assigned their relative activities as PHD2>>PHD3>PHD1 (Huang et al, 2002). However, these results are different from those of another report, which showed similar specific activities for PHD2 and PHD3, and a lower activity for PHD1 (Tuckerman et al, 2004).
The two HIF-1α prolines are differentially hydroxylated by the PHDs (Epstein et al, 2001; Chan et al, 2005). All three PHDs can hydroxylate HIF-1α Pro564, but only PHD1 and PHD2 are able to hydroxylate HIF-1α Pro402. Furthermore, the PHD1 and PHD2 Km values for a Pro402-containing peptide were about 20–50 times higher than those for a Pro564-containing peptide, which suggests important differences between Pro402 and Pro564 (Huang et al, 2002).
Despite the in vitro analysis, the existence of three highly conserved PHDs in mammalian cells raised important questions as to whether and in what way each PHD contributes to HIF regulation in vivo. It is now well established that all three PHD proteins regulate HIF-α (Appelhoff et al, 2004). However, the contribution of each PHD is dependent on its relative abundance, which supports the theory that HIF-α hydroxylation is a non-equilibrium reaction. We have shown that PHD2 has a dominant role, as it is the rate-limiting enzyme that sets the low steady-state levels of HIF-1α in normoxia (Berra et al, 2003). Carmeliet's group have also shown that the knockdown of phd2 is lethal in mice, whereas phd1 or phd3 knockout mice have no important defects (P. Carmeliet, personal communication). Conversely, we have detected a role for PHD1 and PHD3 in HIF-1α degradation during long-term hypoxic stress (A.G., J.P. and E.B., unpublished data).
Tissue distribution and alternative splicing. PHD1, PHD2 and PHD3 are expressed in all tissues albeit at different levels (Cioffi et al, 2003; Lieb et al, 2002). PHD2 and PHD3 messenger RNAs are subjected to alternative splicing (Hirsila et al, 2003), although no enzyme activity has been detected for any of the alternatively spliced forms. Therefore, changes in the splicing pattern can be expected to influence the production and activity of the enzymes.
Localization. Intracellular localization of the PHDs has been reported by using chimeric proteins fused to the green fluorescent protein (Metzen et al, 2003). PHD1 is present exclusively in the nucleus, PHD2 is located mainly in the cytoplasm and PHD3 is distributed homogeneously in the cytoplasm and nucleus. Despite its mostly cytoplasmic localization, PHD2 is able to shuttle between the cytoplasm and the nucleus. Indeed, our data indicate that PHD2 accumulates in the nucleus after 4 h of incubation with leptomycin B, which inhibits nuclear export (A.G., D. Roux, J.P. and E.B., unpublished data). We have previously shown that HIF-1α can be degraded in the nucleus and cytoplasm (Berra et al, 2001b); therefore, PHD2 has all the attributes to target HIF-1α degradation in both cellular compartments.
Regulation of PHD expression. PHD expression is regulated in three ways. First, there is hypoxia/HIF-dependent regulation as the expression of PHD2 and PHD3 is transcriptionally induced by hypoxia, which promotes a negative feedback loop (Berra et al, 2001a; Berra et al, 2003; Epstein et al, 2001). Indeed, hypoxic induction of PHDs is HIF-1-dependent (del Peso et al, 2003). Silencing of HIF-1α or HIF-2α results in decreased hypoxia-induced PHD3 expression. By contrast, PHD2 is not affected by HIF-2α (Aprelikova et al, 2004). Second, there is hypoxia-dependent/HIF-independent regulation. Similar to HIF-α, PHD1 and PHD3 are also degraded by the proteasome (Nakayama et al, 2004). Degradation of PHDs by Siah 1 and 2, their specific E3 ligases, is enhanced by hypoxia and Siah 1 and 2 are transcriptionally upregulated during hypoxia in a HIF-1-independent manner. Third, physiological stimuli other than hypoxia can also regulate the expression of PHDs. Indeed, PHD1 mRNA is upregulated by estrogens in the ZR75-1 breast cancer cell line (Seth et al, 2002). SM20, the rat orthologue of human PHD3, is induced in response to p53 activation (Madden et al, 1996). Nerve growth factor withdrawal also induces SM20 in PC-12 cells, as do platelet-derived growth factor and angiotensin II in smooth muscle cells (Lipscomb et al, 2001). Likewise, overexpression of the Drosophila melanogaster cyclin-dependent protein kinase complex CycD/Cdk4 increases the expression of Fatiga (the Drosophila homologue of PHD) at the post-transcriptional level (Frei & Edgar, 2004).
Regulation of PHD activity. PHDs are effective O2 sensors. In keeping with their Km values for O2 (230–250 μM), which are slightly above the atmospheric concentration, the activity of PHDs is tightly regulated by the full range of O2 concentrations from normoxia (21% O2) to the lowest (<0.1% O2) hypoxic level (Hirsila et al, 2003). Nevertheless, as mentioned above, prolonged hypoxia can unexpectedly enhance PHD activity. Whether PHD accumulation during hypoxia and/or extra mechanisms are responsible for this re-activation is unknown. Whatever the explanation, it is noteworthy that prolonged hypoxia would limit HIF-1 activation, which supports the regulatory feedback loop we proposed previously (Berra et al, 2001a).
Although the availability of O2 serves as a general determinant of PHD activity, several further parameters regulate this activity in a more complex manner to subtly adapt HIF function in a variety of dynamic microenvironments. Depletion of intracellular ascorbate and competitive inhibitors of 2-oxoglutarate lead to PHD inhibition. Similarly, normoxic stabilization of HIF-1α by some oncogenes is mediated by the inhibition of prolyl hydroxylation—such as in RasVal12 and v-Src activation (Chan et al, 2002). Mutations in the tumour suppressor succinate dehydrogenase (SDH) have also been linked to PHDs (Selak et al, 2005; Lee et al, 2005); succinate, which accumulates as a result of SDH mutations, inhibits PHD activity. Thus, succinate transmits an oncogenic signal from mitochondria to the cytosol. Interestingly, an independent study that links mitochondria to PHD activity was reported simultaneously: HIF-1 activity is repressed by mRpL12, the D. melanogaster homologue of the mammalian mitochondrial ribosomal protein MRPL12a (Frei et al, 2005).
The role of reactive oxygen species (ROS) in the control of HIF-1α stability is also important. This role remains controversial despite the identification of PHDs as the oxygen sensors. This is mainly owing to the fact that hypoxia concomitantly inhibits PHD activity and induces the production of ROS by mitochondria. An interesting explanation for this duality of action arose from the work of Metcha-Gregoriou and colleagues, who showed that the accumulation of ROS in JunD−/− cells decreases the availability of Fe2+, which reduces the activity of PHDs (Gerald et al, 2004). Therefore, any stress capable of inducing a persistent boost of free radicals should affect the stability of HIF-1α by indirectly inhibiting PHDs. Another source of controversy arose from blocking respiration through the inhibition of mitochondria, which prevents HIF-1α stabilization in moderate hypoxia. One school of thought, which we favour, points to the redistribution of O2 in the cell, whereas another proposes the abrogation of ROS (for a discussion see Hagen et al, 2003; Doege et al, 2005; Kaelin, 2005).
In addition, an increasing number of proteins such as OS9 seem to bind to PHDs and to regulate their activity (Baek et al, 2005). Several second messengers also modify PHD activity (Appelhoff et al, 2004; Hirota et al, 2004; Temes et al, 2005).
Functions of PHDs. PHDs are pivotal components of the signalling pathways that are elicited to assure O2 homeostasis. So far, more than 70 HIF target genes have been identified, including genes involved in the development and function of the vascular system, erythropoiesis, energy metabolism, pH regulation, cell proliferation and migration. This represents an elegantly adaptive mechanism for survival, but these target genes are also implicated in the pathogenesis of several serious diseases including myocardial ischaemia, stroke, pulmonary hypertension, pre-eclampsia, and almost every type of cancer. Therefore, PHDs, by modulating HIF-α stability and thus HIF activity, are at the heart of these pathophysiological processes.
Apart from their known function in the cellular response to low O2 availability, some data suggest that PHDs have a role in several other physiological and pathophysiological processes, such as the control of cell size. Indeed, loss of function in Fatiga suppresses the growth but not the proliferation function of CycD/Cdk4 in D. melanogaster (Frei & Edgar, 2004). Likewise, C. elegans lacking egl-9 show an egg-laying defect and are resistant to an otherwise paralytic Pseudomona aeruginosa toxin. Further analysis is necessary to explain definitively whether PHDs drive these functions through a HIF-dependent mechanism or through hydroxylation of new PHD substrates. We have shown the regulatory mechanisms and function of PHDs in Fig 3.
Figure 3.
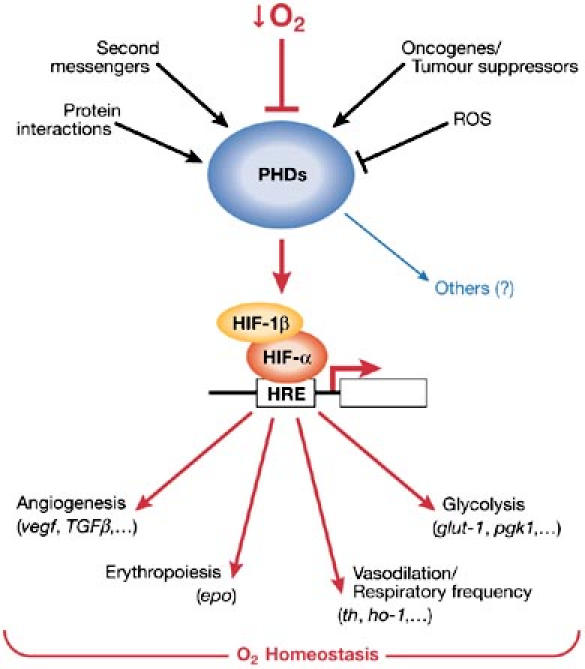
Schematic model of prolyl-hydroxylase domain regulation and function. Several physiological stimuli, in addition to O2 availability, regulate prolyl hydroxylase domain (PHD) activity, which promotes fine-tuning adaptation to the microenvironment. PHDs, by modulating hypoxia-inducible factor-α (HIF-α) stability and thus HIF activity, are pivotal in O2 homeostasis. Nevertheless, PHDs might have a role in other physiological and pathological processes. We still do not know whether PHDs drive these functions through a HIF-dependent mechanism or through hydroxylation of new PHD substrates. HRE, hypoxia response element; ROS, reactive oxygen species.
Protein hydroxylation
Besides prolyl hydroxylation, HIF-1α is hydroxylated on an asparagine residue (Asn803)—in its carboxy-terminal transactivation domain—by factor inhibiting HIF (FIH-1; Lando et al, 2002). FIH-1, which could be subjected to the same regulatory mechanisms as the PHDs, suppresses HIF transcriptional activity under normoxic conditions by blocking its association with the coactivator p300/CBP.
In addition to HIF-α, prolyl hydroxylation has long been known to be important for collagen biosynthesis through stabilization of the triple-helical conformation of collagen fibres (Uitto & Lichtenstein, 1976). Furthermore, recent reports implicate the LxxLAP motif in the pVHL-dependent ubiquitylation of subunit 1 of RNA polymerase II (Hanson et al, 2003), and the hydroxylation of iron regulatory protein 2 (IRP2) in its interaction with the ubiquitin machinery (Kuznetsova et al, 2003). With database predictions of new iron- and 2-oxoglutarate-dependent dioxygenases such as AlkB (Welford et al, 2003), these data hint that protein hydroxylation extends beyond the HIF system and might be widely involved in cell signalling.

Acknowledgments
We thank all the laboratory members for their help and support and C. Brahimi-Horn for reading the manuscript. We apologize to the many research groups whose work was omitted owing to space constraints. Financial support was from the Centre National de la Recherche Scientifique, Ministère de l'Education, de la Recherche et de la Technologie, Ligue Nationale Contre le Cancer (Equipe labellisée), and the GIP HMR (contract No. 1/9743B-A3).
References
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM (2004) Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem 279: 38458–38465 [DOI] [PubMed] [Google Scholar]
- Aprelikova O, Chandramouli GV, Wood M, Vasselli JR, Riss J, Maranchie JK, Linehan WM, Barrett JC (2004) Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J Cell Biochem 92: 491–501 [DOI] [PubMed] [Google Scholar]
- Baek JH, Mahon PC, Oh J, Kelly B, Krishnamachary B, Pearson M, Chan DA, Giaccia AJ, Semenza GL (2005) OS-9 interacts with hypoxia-inducible factor 1α and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1α. Mol Cell 17: 503–512 [DOI] [PubMed] [Google Scholar]
- Berra E, Richard DE, Gothie E, Pouyssegur J (2001a) HIF-1-dependent transcriptional activity is required for oxygen-mediated HIF-1α degradation. FEBS Lett 491: 85–90 [DOI] [PubMed] [Google Scholar]
- Berra E, Roux D, Richard DE, Pouyssegur J (2001b) Hypoxia-inducible factor-1α (HIF-1α) escapes O2-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep 2: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J (2003) HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J 22: 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Denko NC, Giaccia AJ (2002) Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1α. J Biol Chem 277: 40112–40117 [DOI] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Yen SE, Giaccia AJ (2005) Coordiante regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor-1α. Mol Cell Biol 25: 6415–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi CL, Liu XQ, Kosinski PA, Garay M, Bowen BR (2003) Differential regulation of HIF-1α prolyl-4-hydroxylase genes by hypoxia in human cardiovascular cells. Biochem Biophys Res Commun 303: 947–953 [DOI] [PubMed] [Google Scholar]
- del Peso L, Castellanos MC, Temes E, Martin-Puig S, Cuevas Y, Olmos G, Landazuri MO (2003) The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J Biol Chem 278: 48690–48695 [DOI] [PubMed] [Google Scholar]
- Doege K, Heine S, Jensen I, Jelkmann W, Metzen E (2005) Inhibition of mitochondrial respiration elevates oxygen concentration, but leaves regulation of hypoxia-inducible factor (HIF) intact. Blood 106: 2311–2317 [DOI] [PubMed] [Google Scholar]
- Epstein AC et al. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Frei C, Edgar BA (2004) Drosophila cyclin D/Cdk4 requires Hif-1 prolyl hydroxylase to drive cell growth. Dev Cell 6: 241–251 [DOI] [PubMed] [Google Scholar]
- Frei C, Galloni M, Hafen E, Edgar BA (2005) The Drosophila mitochondrial ribosomal protein mRpL12 is required for Cyclin D/Cdk4-driven growth. EMBO J 24: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, Mechta-Grigoriou F (2004) JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell 118: 781–794 [DOI] [PubMed] [Google Scholar]
- Hagen T, Taylor CT, Lam F, Moncada S (2003) Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1α. Science 302: 1975–1978 [DOI] [PubMed] [Google Scholar]
- Hanson ES, Rawlins ML, Leibold EA (2003) Oxygen and iron regulation of iron regulatory protein 2. J Biol Chem 278: 40337–40342 [DOI] [PubMed] [Google Scholar]
- Hirota K, Fukuda R, Takabuchi S, Kizaka-Kondoh S, Adachi T, Fukuda K, Semenza GL (2004) Induction of hypoxia-inducible factor 1 activity by muscarinic acetylcholine receptor signaling. J Biol Chem 279: 41521–41528 [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J (2003) Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem 278: 30772–30780 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao Q, Mooney SM, Lee FS (2002) Sequence determinants in hypoxia-inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J Biol Chem 277: 39792–39800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, Bunn HF (1996) Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its α subunit. J Biol Chem 271: 32253–32259 [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Jaakkola P et al. (2001) Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Kaelin WG (2005) ROS: really involved in oxygen sensing. Cell Metab 1: 357–358 [DOI] [PubMed] [Google Scholar]
- Kim W, Kaelin WG (2003) The von Hippel–Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr Opin Genet Dev 13: 55–60 [DOI] [PubMed] [Google Scholar]
- Kuznetsova AV, Meller J, Schnell PO, Nash JA, Ignacak ML, Sanchez Y, Conaway JW, Conaway RC, Czyzyk-Krzeska MF (2003) von Hippel–Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci USA 100: 2706–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 16: 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S et al. (2005) Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromacytoma genes: developmental culling and cancer. Cancer Cell 8: 155–167 [DOI] [PubMed] [Google Scholar]
- Lieb ME, Menzies K, Moschella MC, Ni R, Taubman MB (2002) Mammalian EGLN genes have distinct patterns of mRNA expression and regulation. Biochem Cell Biol 80: 421–426 [DOI] [PubMed] [Google Scholar]
- Lipscomb EA, Sarmiere PD, Freeman RS (2001) SM-20 is a novel mitochondrial protein that causes caspase-dependent cell death in nerve growth factor-dependent neurons. J Biol Chem 276: 5085–5092 [DOI] [PubMed] [Google Scholar]
- Madden SL, Galella EA, Riley D, Bertelsen AH, Beaudry GA (1996) Induction of cell growth regulatory genes by p53. Cancer Res 56: 5384–5390 [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275 [DOI] [PubMed] [Google Scholar]
- Metzen E et al. (2003) Intracellular localisation of human HIF-1α hydroxylases: implications for oxygen sensing. J Cell Sci 116: 1319–1326 [DOI] [PubMed] [Google Scholar]
- Nakayama K et al. (2004) Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell 117: 941–952 [DOI] [PubMed] [Google Scholar]
- Salceda S, Caro J (1997) Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem 272: 22642–22647 [DOI] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7: 77–85 [DOI] [PubMed] [Google Scholar]
- Semenza GL (1998) Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev 8: 588–594 [DOI] [PubMed] [Google Scholar]
- Seth P, Krop I, Porter D, Polyak K (2002) Novel estrogen and tamoxifen induced genes identified by SAGE (Serial Analysis of Gene Expression). Oncogene 21: 836–843 [DOI] [PubMed] [Google Scholar]
- Taylor MS (2001) Characterization and comparative analysis of the EGLN gene family. Gene 275: 125–132 [DOI] [PubMed] [Google Scholar]
- Temes E, Martin-Puig S, Acosta-Iborra B, Castellanos MC, Feijoo-Cuaresma M, Olmos G, Aragones J, Landazuri MO (2005) Activation of HIF-prolyl hydroxylases by R59949, an inhibitor of the diacylglycerol kinase. J Biol Chem 280: 24238–24244 [DOI] [PubMed] [Google Scholar]
- Tuckerman JR, Zhao Y, Hewitson KS, Tian YM, Pugh CW, Ratcliffe PJ, Mole DR (2004) Determination and comparison of specific activity of the HIF-prolyl hydroxylases. FEBS Lett 576: 145–150 [DOI] [PubMed] [Google Scholar]
- Uitto J, Lichtenstein JR (1976) Defects in the biochemistry of collagen in diseases of connective tissue. J Invest Dermatol 66: 59–79 [DOI] [PubMed] [Google Scholar]
- Welford RW, Schlemminger I, McNeill LA, Hewitson KS, Schofield CJ (2003) The selectivity and inhibition of AlkB. J Biol Chem 278: 10157–10161 [DOI] [PubMed] [Google Scholar]


