Summary
Search engines are increasingly important tools for browsing the vast spaces of the Internet. How good are they for searching through scientific literature?
Unless you read EMBO reports cover to cover, or were alerted by this issue's table of contents, chances are that you found this article by using a search engine. So, what were you looking for? Information on search engines? Google? PubMed? Literature searches? Web services, SOAP, WSDL, text retrieval or text mining? Systems biology? If so, any of these keywords or phrases was probably enough for a search engine to put this article high on its ‘hit list'.
If this article is not exactly what you were after, you will have to start again, using a new selection of search terms that may give you a better fit to your expectations. In either case, you will probably not look beyond the first page of results, considering it easier and faster to reiterate this trial-and-error process than to sift carefully through hundreds, if not thousands, of other hits.
So how smart is your favourite search engine? Can you usually rely on its results? Given the enormous reach of the worldwide web, do we need better search engines? Do you really care? Most scientists quite sensibly expect that search engines, similar to many other ‘under the hood' technologies, should work quickly, efficiently, accurately and above all, unobtrusively—not an unreasonable demand in today's research environment where you may feel that you need to outrun Lewis Carroll's Red Queen simply to stay where you are.
Nevertheless, in the same way that the quality of reagents is crucial to the success or failure of an experiment, the quality and limitations of a search engine, as well as the skill with which it is used, largely determine the success or failure of a web search. I still recall the early days when literature searching was almost the exclusive domain of the trained librarian, who needed to be fully briefed before embarking on a series of detailed searches that often lasted for several days. Today, most life scientists will not bother their librarian, but will instead use the National Library of Medicine's PubMed service at one stage or another during their literature search. However, an increasing proportion of young researchers prefer Google or Google Scholar as, at least, a starting point and, more worryingly, also as an endpoint. Furthermore, they expect it to produce significant results from only a few relevant keywords. The average query length in Google is approximately 2.2 words (O'Reilly, 2004) and most users seem to think that this is enough to find the most recent and most notable literature within the top ten search results. Imagine what the result would be if the same two-word question was fired at a trained librarian.
Behind their diverse web fronts, all search engines share very similar algorithms at various stages of their operation. As a first step, they break query text down into individual words, ignoring those that are too common to be of predictive value. The remaining words are then stemmed—reduced to a common root by removing suffixes—and stored in one or more indexes together with their frequency of occurrence in any given document. This has a widening effect on the search request, because the deletion of suffixes removes nuances in the original query. Stemming can therefore generate unexpected results through the introduction of ambiguities. Words with different meanings reduce to the same stem—such as secrete, secretion and secretive, which all reduce to secret. By the same process, acronyms and gene symbols are reduced to generic forms that have different meanings from their use in the original query text, for example, FACS (fluorescence activated cell sorting) becomes FAC, one of the symbols for the Fanconi anaemia gene, FANCC.
…in the same way that the quality of reagents is crucial to the success or failure of an experiment, the quality and limitations of a search engine … largely determine the success or failure of a web search
Differences begin to emerge when we look at how various search engines handle queries. Most will allow the use of Boolean operators (AND, OR, NOT) to combine and/or exclude specific keywords. Most will also allow the user to define phrases by including them in quotation marks. Some search engines offer so-called proximity operators to search for the co-occurrence of words or phrases within a specified distance. The results of Boolean queries are usually sharply defined: each document is considered in isolation, search terms are equally weighted, and they either co-occur or not. This straightforward approach, although logical, limits the power of the search engine to find related documents that do not contain all keywords or phrases. Some specialist engines therefore allow terms to be differentially weighted, and others implement multi-word frequency comparisons using vector–cosine or similar algorithms (Salton & Buckley, 1991), allowing ‘more like this' queries to return further documents related to the initial query. The Vivisimo metasearch engine is one example of such an algorithm and both its general web version (www.clusty.com/) and the PubMed-based scientific variant (http://demos.vivisimo.com/projects/medline) present the outcome of searches as informative, hierarchically arranged folders, in which related links or documents are grouped together.
In the text above I occasionally used the term ‘keyword'. For most of us, a keyword is no more than one of the most important words that we are looking for in a document and that a search engine indexes. A query using this keyword will retrieve all documents that contain it wherever it occurs, irrespective of its importance or relevance to the topic it describes. As an example, take the word ‘occasionally' from the beginning of this paragraph: as a search term, it is obviously not very useful—as smart as search engines aim to be, they are not very good at guessing what the user really wants to find.
…an increasing proportion of young researchers prefer Google or Google Scholar as, at least, a starting point and, more worryingly, also as an endpoint
However, keywords have special meanings to librarians and information professionals. Any combination of these form the ‘metadata' for a particular document: they hold information that someone—an author, librarian or database curator—considers to be both relevant and useful for understanding and describing a given text. Metadata in scientific articles usually include at least the author and affiliation, journal title, volume and page numbers, and date of publication. Furthermore, the metadata record may contain keywords from a controlled vocabulary assigned to the article by an expert, so that the document can be classified, searched and retrieved systematically and in a reproducible way.
PubMed's metadata includes gene names or symbols and Medical Subject Headings (MeSH; www.nlm.nih.gov/mesh/), which is a hierarchically branched tree of internationally agreed terms and their synonyms in the biomedical sciences. Each item of the metadata is then stored in a separate index. Thus, unlike Google and other general search engines, PubMed allows the user to specify the index, such as author name or journal, through which to search. Furthermore, unless specifically instructed otherwise, it automatically carries out query–term translation, a process that speeds up searches by matching words in the search box against translation tables for MeSH, and author and journal title indexes. In the case of a match with a MeSH term, the query is expanded to include related entries and their aliases, thereby widening the scope of the search. This is valuable in instances where several terms describe different aspects of a particular topic. For instance a PubMed search for ‘haematopoiesis' without query–term translation generates about 21,300 hits. The same search with query–term translation returns almost 31,000 results, because ‘haematopoiesis' has been expanded to include the more specific processes of erythro-, leuko- and thrombopoiesis.
Along similar lines, a small group of search engines are concept-based. That is, they try to determine what the user means rather than what he or she ‘says'. They achieve this by distilling meanings from both specific combinations of words and the context in which these are used in individual documents and document collections. One example of this type of approach, the Collexis-engine-based E-BioSci service (http://e-biosci.embo.org), maps the words in user queries and in the documents searched to terms in one or more branches of an internal thesaurus. At present, this consists of a modified MeSH thesaurus, but could easily expand to use the CAB Thesaurus, Gene Ontology or some other controlled vocabulary. Other engines use sophisticated computational methods, such as latent semantic indexing (LSI; Yu et al, 2002), to relate documents both to each other and to the query.
Concept-based systems work best with long queries, which are rich in concepts and allow semantically equivalent queries to be made to several information resources. In principle, these systems facilitate streamlined navigation and interconnectivity between, for example, a literature article, a sequence or disease database entry, and a patent record. In practice, it is difficult to keep hand-curated thesauri or ontologies up-to-date in rapidly developing research fields, which limit the broad application of concept-based systems. However, for areas such as systems biology, in which full semantic interlinkage of information is essential for modelling, this approach—in combination with LSI-based methods for automated thesaurus or ontology construction—needs to be pursued further.
So much for features common to search engines in general. But what about Google and other search engines that are becoming increasingly popular for searching full-text scientific literature? Can these engines be relied on to provide results that are equivalent to more dedicated literature services? The answers to these questions require a quick survey of three important topics: content, metadata and page-ranking algorithms.
Web-based search engines use automatic programmes—known as robots, spiders or crawlers—to visit web pages and collect their content. This information is sent to a central indexing engine, while the robot itself continues a semi-random walk through cyberspace by following any links it discovers in the pages it collects. This already highlights the first problem with web searches: web pages that do not have an incoming link are unlikely to be visited unless the robot is specifically sent to them. The same holds true for information in databases—content that is usually referred to as the ‘deep web'. Estimates of the amount of high-quality information in the deep web vary widely, but it could be anything up to 40-fold larger than the part of the worldwide web that is now indexed by robots. An increasing number of publishers and database curators allow robots to access their content to make its existence more widely known. Yet, any information on a web page or database that is not accessible to these robots is invisible to web-based search engines and therefore to their users.
Unlike scientific articles, web pages do not contain conventional metadata. Unless the author explicitly defines keywords for a particular page, the search engine must hazard a guess as to what these may be. It does this by applying a set of rules, for instance if the author has specified keywords in the web page's source code. Otherwise the engine will give greater weighting to words in the page's title, its headings or in its initial sections, or to words that simply occur more frequently throughout the page.
In the early days of the worldwide web, simple ranking of metadata and content was usually sufficient for relevant documents to surface in response to a corresponding query. Now, with more than 8 billion web pages and an escalating war of attrition between search-engine programmers, search-engine result ‘optimizers' and web spammers—the last two of which try to manipulate the results of web searches—engines no longer fully trust web pages to provide an accurate description of their own content. As early as 2002, keyword metadata was regarded as untrustworthy (Sullivan, 2002) and is now avoided by all of the main search engines. The Dublin Core Metadata Initiative, dedicated to promoting the widespread adoption of interoperable metadata standards, has also confirmed this. Thus, search engines now use indirect, but less spam-sensitive, measures to rank hits emerging from a search.
The details of these methods are, for obvious reasons, closely guarded trade secrets, but the general principles are usually the same. In addition to calculating a web page's relevance by matching words in the query to the text in the page, search engines use some measure of authority or popularity, as shown by the number and wording of hyperlinks that come from other sites. If these in turn have large numbers of incoming links, they too are regarded as authoritative sources. This approach (Fig 1) was pioneered by the founders of Google (Page et al, 1998), but has since been adopted by most other search engines, often in combination with other measures. Smart though this system may be, it is still not safe from abuse. Take, for example, Google's top-ranking response to the query ‘miserable failure': a link to the biography of US President George W. Bush on the official White House homepage. A quick search will confirm that this page lacks both of the query words. The result is generated simply by creating appropriately worded hyperlinks to the White House site, with probably as few as 32 websites being necessary to achieve the effect (BBC News, 2003). Interestingly, the second result of this query is the homepage of film-maker Michael Moore, which shows that Bush supporters have also learned how to play the game.
Figure 1.
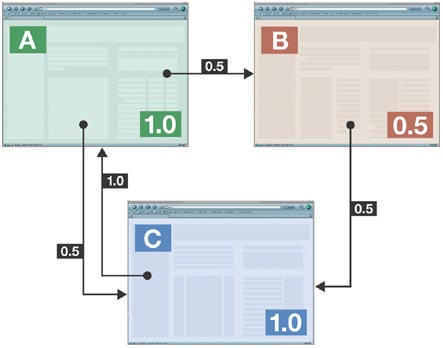
A simplified example of page ranking by link analysis of three web pages, A, B and C, adapted from Page et al, 1998. Each page on the worldwide web is initially assigned a raw page rank value of 1. About 85% of this value is passed on to pages that this page points to, divided roughly equally across all links made. A page's final raw rank value can therefore only be calculated by a series of iterations because links can be made back and forth between any number of pages. The result is the sum of the initial page value plus anything that gets assigned to it from incoming links. Obviously, the more links that lead to a web page, the higher its score. In the last step of the calculation, raw rank values are converted to PageRank scores by use of an algorithm known only to the search engine company that applies it. This value constantly changes as new challenges from web spammers and results ‘optimizers' are met. Those who depend on their Google ranking for their economic survival can be either pleasantly or unpleasantly surprised by the outcome (Battelle, 2005).
Of course, none of this should be relevant to a PubMed record that in principle contains only the metadata of a published article together with its abstract. I also assume—I hope not naively—that even in today's highly competitive research world, there is nothing to be gained from link-targeting or web-spamming particular articles, as described above. Nonetheless, those who use a standard Google query to find relevant PubMed records are clearly prepared to spend—or rather waste—valuable time separating the valid hits from the unrelated results that surface high on hit lists as a result of page-rank optimization efforts by advertisers and lobby groups. However, even with services such as Google Scholar (http://scholar.google.com/) or Elsevier's Scirus (www.scirus.com), which focus more specifically on scientific literature, the user should be aware that hits are not ranked solely on relevance to the original query. Aside from the full text of each article, Google Scholar uses “the author, the publication in which the article appeared, and how often the piece has been cited in other scholarly literature”, according to their website. Elsevier's Scirus uses a combination of term frequency and position, with link analysis. It also examines the length of the page's address, or Universal Resource Locator (URL), assuming for some reason that a short URL is more authoritative than a long one (Scirus, 2004). How the link analysis works is not entirely clear, but it is striking that most search queries will return results in which at least one of Elsevier's ScienceDirect publications occupy positions high on the list.
…with more than 8 billion web pages and an escalating war of attrition between search-engine programmers, search-engine result ‘optimizers' and web spammers…engines no longer fully trust web pages to provide an accurate description of their own content
In short, web engines are great for finding out where to buy the cheapest iPod, to download elusive music clips or to learn more about Madonna. At first sight, they also provide a seemingly simple and attractive entry point into the vast spaces of the scientific literature. Nevertheless, users should not be lulled into the illusion that search engines produce complete results or list them by their relevance.
In EMBO reports in 2002, I discussed the need for new tools to facilitate the processes of discovery and analysis of information in the scientific literature (Grivell, 2002). Since then, the content of PubMed, which covers just the biomedical disciplines, has grown by almost 40% from 11 million to more than 15 million records. In that time there has also been a growing demand for systems-based approaches, in which interconnectivity of biological information—contained in databases or published articles—has a key role.
And yet, during that time, several surveys (Tenopir, 2003) show that a growing number of younger scientists are using generic web search engines as their preferred and sometimes only search tool for literature or databases. In a nationwide survey of students and academics in the Netherlands, most respondents reported that they were self-taught web searchers who relied on a trial-and-error strategy to find information (Voorbij, 1999). Nearly two-thirds believed that searching the web was important or very important, and most thought that their web searches yielded enough information. This is a worrying trend, not only for the reasons discussed above, but also because the exclusive use of such systems precludes any further analysis of the information returned, unless the user is prepared to painstakingly repeat the query process on each of the original information resources.
However, the past couple of years have seen various promising developments of web services for molecular biological resources, still largely unnoticed by most people outside the realm of bioinformatics and computer science. The principle of these services, of which E-BioSci was possibly one of the earliest, is that they use a common language (WSDL, the Web Services Description Language; Christensen et al, 2001) and protocol (usually SOAP, the Simple Object Access Protocol; Gudgin et al, 2003) to describe the structure of a molecular or bibliographic database and to inform the user—which may be another web-based information resource—how queries are made and what kind of information is returned as a result. For the human user, most of these underlying interactions and negotiations between different web services are invisible, but the outcome is worthwhile: physically, geographically and structurally distinct information resources can be queried either simultaneously or sequentially, without the user needing to know anything about the inner workings of any of them. The information is returned in XML (Extensible Markup Language), a format that offers huge potential for further analysis, storage in a local database, re-formatting at different levels of detail, and so forth.
…users should not be lulled into the illusion that search engines produce complete results or list them by their relevance
Alongside these developments, there is progress being made to construct web service ‘pipelines', in which the output given in response to an initial query is automatically used as input to a subsequent service, which in turn passes its results on to another service for analysis. An example of such a pipeline application is the Taverna workbench (Oinn et al, 2004), developed as part of the MyGrid project (www.mygrid.org.uk). Taverna and the underlying web service infrastructures, such as the BioMoby project (Wilkinson et al, 2005), are still in their infancy, but we should all make the effort to appreciate this technology, rather than simply leaving it to Google and comparable search engines to dictate how the information we have so laboriously gathered and curated is organized and made accessible to us.

References
- Battelle J (2005) The Search: How Google and Its Rivals Rewrote the Rules of Business and Transformed Our Culture. Boston, MA, USA: Nicholas Brearley [Google Scholar]
- BBC News (2003) ‘Miserable failure' links to Bush. 7 Dec. http://news.bbc.co.uk/2/hi/americas/3298443.stm
- Christensen E, Curbera F, Meredith G, Weerawarana S (2001) Web Services Description Language (WSDL) 1.1. W3C Note, 15 Mar. www.w3.org/TR/wsdl [Google Scholar]
- Grivell L (2002) Mining the bibliome: searching for a needle in a haystack? EMBO Rep 3: 200–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudgin M, Hadley M, Mendelsohn N, Moreau J -J, Nielsen HF (2003) SOAP Version 1.2 Part 1: Messaging Framework. W3C Recommendation, 24 Jun. www.w3.org/TR/soap12/ [Google Scholar]
- Oinn T et al. (2004) Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics 20: 3045–3054 [DOI] [PubMed] [Google Scholar]
- O'Reilly D (2004) Web-user satisfaction on the upswing. PC World, 7 May. http://pcworld.about.com/news/May072004id116060.htm [Google Scholar]
- Page L, Brin S, Motwani R, Winograd T (1998) The PageRank Citation Ranking: Bringing Order to the Web. CiteSeerIST. http://citeseer.ist.psu.edu/page98pagerank.html [Google Scholar]
- Salton G, Buckley C (1991) Global text matching for information retrieval. Science 253: 1012–1015 [DOI] [PubMed] [Google Scholar]
- Scirus (2004) How Scirus Works. Amsterdam, Netherlands: Elsevier. www.scirus.com/press/pdf/WhitePaper_Scirus.pdf [Google Scholar]
- Sullivan D (2002) Death of a Meta Tag. Search Engine Watch, 1 Oct. http://searchenginewatch.com/sereport/02/10-meta.html [Google Scholar]
- Tenopir C (2003) Use and Users of Electronic Library Resources: An Overview and Analysis of Recent Research Studies. Washington, DC, USA: Council on Library and Information Resources [Google Scholar]
- Voorbij HJ (1999) Searching Scientific Information on the Internet: A Dutch Academic User Survey. J Am Soc Inf Sci 50: 598–615 [Google Scholar]
- Wilkinson M, Schoof H, Ernst R, Haase D (2005) BioMOBY successfully integrates distributed heterogeneous bioinformatics Web Services. The PlaNet exemplar case. Plant Physiol 138: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Cuadrado J, Ceglowski M, Payne JS (2002) Patterns in Unstructured Data: Discovery, Aggregation and Visualization. http://research.nitle.org/lsi/ [Google Scholar]


