Abstract
A central function of telomeres is to prevent chromosome ends from being recognized as DNA double-strand breaks (DSBs). Several proteins involved in processing DSBs associate with telomeres, but the roles of these factors at telomeres are largely unknown. To investigate whether the Mre11/Rad50/Nbs1 (MRN) complex is involved in the generation of proper 3′ G-overhangs at human telomere ends, we used RNA interference to decrease expression of MRN and analysed their effects. Reduction of MRN resulted in a transient shortening of G-overhang length in telomerase-positive cells. The terminal nucleotides of both C- and G-rich strands remain unaltered in Mre11-diminished cells, indicating that MRN is not responsible for specifying the final end-processing event. The reduction in overhang length was not seen in telomerase-negative cells, but was observed after the expression of exogenous telomerase, which suggested that the MRN complex might be involved in the recruitment or action of telomerase.
Keywords: telomere, 3′ G-overhang, Mre11/Rad50/Nbs1, DNA double-strand break repair, telomere end processing
Introduction
The physical ends of human chromosomes, telomeres, can form special structures, t-loops, by inserting the 3′ single-stranded G-rich overhang into duplex telomeric DNA. T-loops presumably help prevent chromosome ends from being recognized as damaged DNA and preserve genome stability (de Lange, 2002; d'Adda di Fagagna et al, 2004; Ferreira et al, 2004; Maser & DePinho, 2004). Human cells possess long G-overhangs ranging from <50 nt to several hundred nucleotides (nt) (Makarov et al, 1997; McElligott & Wellinger, 1997; Stewart et al, 2003; Chai et al, 2005). They are thought to be generated by incomplete DNA replication at linear chromosome ends (Olovnikov, 1973; Lingner et al, 1995) and further processing by unidentified nucleases and/or helicases (Wellinger et al, 1996).
Telomeres shorten progressively during the proliferation of normal cells partly owing to these reasons. Cells senesce when telomeres become critically short. Most cancer cells express a specialized reverse transcriptase, telomerase, to prevent telomere shortening by adding TTAGGG repeats using the G-overhang as a template. However, telomeres are not extended by telomerase indefinitely, with the length of telomeric repeats being generally kept within 3–15 kb. Telomere length homeostasis is the result of a balance between telomere shortening and telomere lengthening activities (Smogorzewska & de Lange, 2004). Understanding the mechanism for telomere end processing and its communication with telomerase-mediated telomere lengthening may help in developing methods to drive the equilibrium towards telomere shortening and facilitate treatment of cancer.
The identities of the enzymes that are involved in human telomere end processing remain unknown. Increasing evidence suggests that at least some DNA damage repair factors are involved. Deficiency in DNA-dependent protein kinase catalytic subunit causes chromosome end fusions involving only leading strands (Bailey et al, 2001). The ERCC1/XPF endonuclease that is involved in processing homologous recombination intermediates is responsible for cleavage of G-overhangs after TRF2 inhibition (Zhu et al, 2003).
The Mre11/Rad50/Nbs1 (MRN) complex is recruited to double-strand breaks (DSBs) at the very early stage of DNA repair to initiate a series of downstream repair events (Jackson, 2002; Valerie & Povirk, 2003). In vitro, Mre11 exhibits 3′ → 5′ exonuclease and endonuclease activity (Paull & Gellert, 1998), and it promotes the formation of 3′ overhangs at DSBs through unclear molecular mechanisms (Jackson, 2002; Valerie & Povirk, 2003; Lavin, 2004; Sancar et al, 2004). The MRN complex is also required for proper maintenance of telomere integrity in mammals, insects, plants and yeast (Kironmai & Muniyappa, 1997; Boulton & Jackson, 1998; Gallego & White, 2001; Ranganathan et al, 2001; Bi et al, 2004; Ciapponi et al, 2004). The human MRN complex has been shown to associate with human telomeres through interaction with TRF2, particularly during the S phase (Lombard & Guarente, 2000; Zhu et al, 2000). Telomere DNA ends resemble DSBs immediately after replication and the MRN complex has been suggested to promote C-strand resection. Studies in yeast and flies have demonstrated the role of the MRN complex in telomere end processing (Tomita et al, 2003; Bi et al, 2004; Ciapponi et al, 2004; Larrivee et al, 2004; Takata et al, 2005). In this study, we investigated whether the MRN complex is involved in the proper generation of G-overhangs in human cells. Reduction of the MRN complex in telomerase-positive but not telomerase-negative cells resulted in G-overhang shortening, and did not alter the terminal nucleotides at either G- or C-rich strands. The MRN complex thus influences overhang length in a telomerase-dependent manner, but is not responsible for specifying the terminal nucleotides at human telomere ends.
Results and Discussion
End processing in telomerase-positive cells
RNA interference (RNAi) targeting Mre11 in HeLa cells resulted in >90% of reduction of Mre11 expression (Fig 1A), and led to marked reduction of G-overhang length, as determined by the recently developed telomere overhang protection assay (Chai et al, 2005; Fig 1B,C). Coexpression of Mre11 short hairpin RNA (shRNA) and untargeted Mre11 restored the overhang length, indicating that the RNAi was specific (supplementary Fig 1A,B online). The abundance of the single-stranded G-overhangs was reduced by about 50% as well, as determined by the nondenaturing in-gel hybridization assay (supplementary Fig 1C online). We also observed the effect of Mre11 RNAi on G-overhang shortening in normal human fibroblasts (BJ) expressing an exogenous hTERT (supplementary Fig 2 online) and in A549 lung cancer cells (supplementary Fig 3 online). Thus, the requirement of Mre11 in proper G-overhang generation is not cell line specific.
Figure 1.
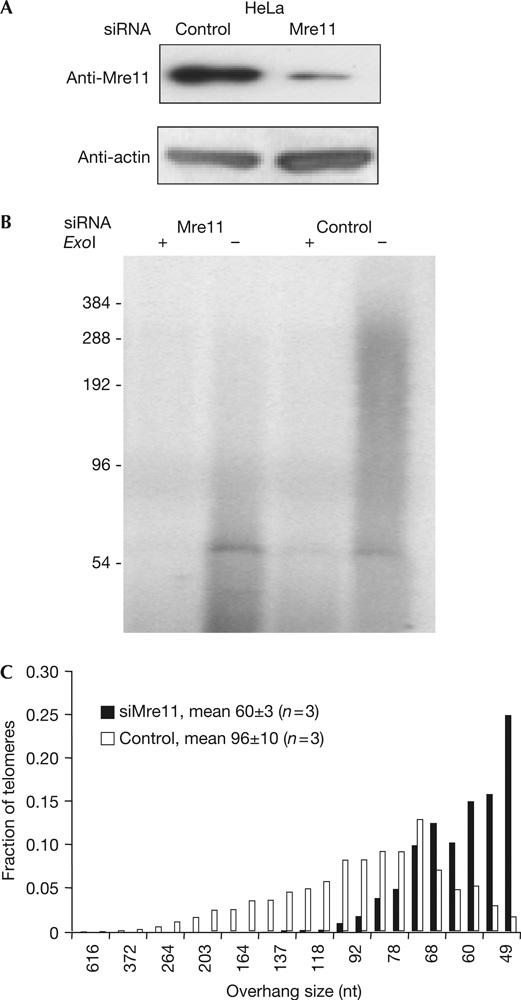
Reducing Mre11 expression in HeLa cells induces overhang shortening. (A) Western blot showing small interfering RNAs (siRNA) targeting Mre11 in HeLa cells. Cells were collected 72 h after being transfected with siRNA. Proteins were extracted for immunoblotting and DNA was isolated at the same time for the telomere overhang protection assay. (B) Telomere overhang protection assay using HeLa cells with reduced Mre11 expression. The smears represent the heterogeneity of overhang sizes. ExoI specifically digests 3′ overhangs and the ExoI plus (+) lanes show the background. The 54 nt band has been shown to be nonspecific (Chai et al, 2005). (C) Overhang size distribution in Mre11-RNAi HeLa cells. Gels from the overhang protection assay were analysed by ImageQuant as described (Chai et al, 2005). The signal from each size region was divided by its size in nucleotides to compensate for the increase in probe hybridization with size. The resulting value was then expressed as a fraction of the total from all the regions spanning the entire lane above 45 nt. Mean overhang sizes were measured from three independent experiments. Errors represent 1 s.d. The fraction of short overhangs was significantly increased in Mre11-RNAi cells.
Next, we used RNAi to reduce expression of the other two components of the MRN complex, Rad50 and Nbs1 (Fig 2). The overhang length was greatly reduced in HeLa cells transiently expressing Nbs1 small interfering RNAs (siRNAs) (Fig 2B). Stable expression of Rad50 shRNA in H1299 lung cancer cells (Fig 2C) also showed a reduction in the length of G-overhangs (Fig 2D). Taken together, our data provide strong evidence that the MRN complex may be involved in proper G-overhang length generation. Alternatively, the reduced G-overhang length could be due to overhang degradation resulting from an unknown mechanism influenced by MRN deficiency.
Figure 2.
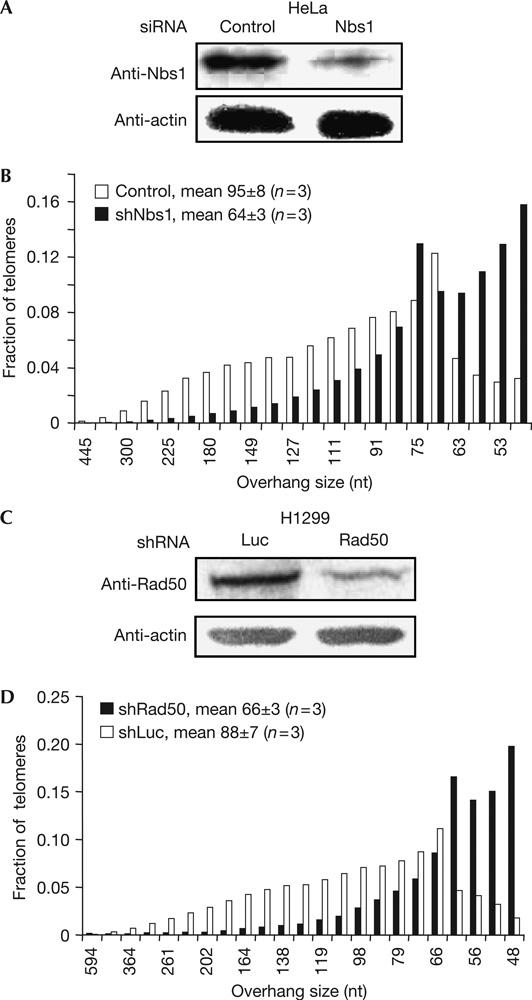
Reducing Nbs1 or Rad50 expression induces overhang shortening. (A) Western blot showing small interfering RNAs (siRNA) targeting Nbs1 in HeLa. Cells were collected 72 h after being transfected with siRNA. Proteins were extracted for immunoblotting and DNA was isolated at the same time for the telomere overhang protection assay. (B) Overhang size distribution with weighted mean overhang sizes of G-overhangs from Nbs1-RNAi HeLa cells. Mean overhang sizes were measured from three independent experiments. The gel from which this distribution was derived is shown in supplementary Fig 5A online. The fraction of short overhangs was significantly increased in Nbs1-RNAi cells. (C) Western blot showing reduced Rad50 expression in H1299 cells after stably expressing small hairpin RNA (shRNA) targeting Rad50. Cells were collected after 7 days of selection. (D) Overhang size distribution with weighted mean overhang sizes of G-overhangs from Rad50-RNAi H1299 cells. Mean overhang sizes were measured from three independent experiments. The gel from which this distribution was derived is shown in supplementary Fig 5B online. The fraction of short overhangs was significantly increased in Rad50-RNAi cells.
G-overhang shortening induced by MRN reduction seemed to be transient. In spite of persistent Mre11 knockdown, the decreased overhangs seen in A549 cells 12 days after infection returned to normal 8 days later (supplementary Fig 3 online). Similar transient decreases in overhang length were also seen in BJ/hTERT cells (data not shown), indicating that compensation by other factors may substitute for Mre11. Reduction of overhang size did not alter the telomere length or cause loss of bulk telomeres (supplementary Fig 4 online), presumably owing to the transient nature of the effect. No increase in telomere-associated DNA damage was observed, as measured by γ-H2AX staining (data not shown), suggesting that telomere structure was not disrupted by MRN reduction.
In telomerase activity-negative BJ cells, stable expression of Mre11 shRNA did not alter overhang length (Fig 3B), indicating that overhang shortening induced by MRN reduction is dependent on telomerase expression. We and others have observed that telomerase produces longer overhangs when expressed in normal diploid cells (Stewart et al, 2003; Chai et al, 2005). Our results are consistent with the possibility that MRN deficiency may transiently disturb telomerase elongation of telomere overhangs, causing them to have the shorter overhangs of telomerase-negative cells. The shorter overhangs that we observed in telomerase-positive cells would thus not be a direct effect of the MRN complex on overhang generation, but an indirect consequence of altering telomerase recruitment or action. This is consistent with the work in Saccharomyces cerevisiae implicating the MRX complex in the recruitment of telomerase (Nugent et al, 1998; Diede & Gottschling, 2001; Takata et al, 2005). We also found that reduction of Mre11 in BJ cells induced growth arrest about 10–15 population doublings after selection (Fig 3C), with no effects on telomere shortening (data not shown), possibly owing to the non-telomeric functions of Mre11. Analysis of DNA contents showed an increased sub-G1 population, which was indicative of apoptosis, and a disappearance of G2 population in BJ cells stably expressing Mre11 shRNA (Fig 3D).
Figure 3.
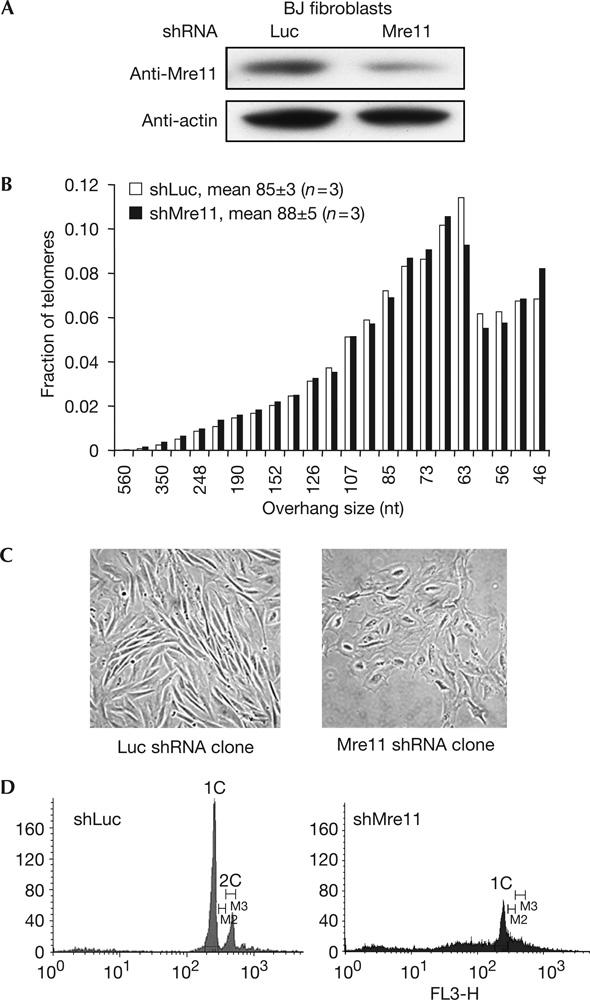
MRN reduction does not alter overhang sizes in telomerase-negative BJ fibroblasts. (A) Western blot showing reduced Mre11 expression in BJ cells after stably expressing small hairpin RNA (shRNA) targeting Mre11. Cells were collected 7 days after selection. (B) Overhang size distribution with weighted mean averages of BJ cells with reduced Mre11 expression. Results are representative of three independent experiments. The gel from which this distribution was derived is shown in supplementary Fig 5C online. Reduced Mre11 expression in BJ cells does not change overhang size distribution. (C) Reduction of Mre11 induces growth arrest of BJ cells. Clones were isolated after stable expression of Mre11 shRNA and luciferase control shRNA. Mre11-RNAi cell growth arrested 24 days after selection, showing enlarged and flattened morphology compared with the control. A total of 20 clones were isolated from each group and 19 of the Mre11 shRNA clones showed such morphology alteration, whereas none of the luciferase control shRNA clones showed morphology or growth change. One representative clone from each group is shown. (D) Fluorescence-activated cell sorter analysis of DNA contents of population of BJ cells expressing Mre11 shRNA and luciferase control shRNA at 10 days after selection.
MRN and terminal telomeric nucleotides
The 5′ end of human telomeres is very specific, with more than 80% of human telomeres ending at CCAATC-5′ at the C-strand and more varied terminal nucleotides at the G-strand (Sfeir et al, 2005). Reduction of the telomere-binding protein Pot1 leads to randomization of the terminal nucleotides in HeLa cells (Hockemeyer et al, 2005). We used the recently developed ligation-mediated PCR assay (C- and G-STELA) (Sfeir et al, 2005) to determine whether Mre11 reduction altered the final nucleotides of the C- and G-strands. The same preference for ATC-5′ terminating at the C-strand (Fig 4B) and the same pattern of G-terminal nucleotides (Fig 5B) was found in Mre11-diminished and control cells. Thus, Mre11 is not involved in the determination of the correct terminal nucleotides.
Figure 4.

Mre11 RNA interference does not alter C-rich strand terminal nucleotides. (A) Schematic representation of C-STELA (Sfeir et al, 2005). Six individual C-telorettes are ligated in separate reactions to the same amount of DNA and then amplified using a forward Xp/Yp chromosome-specific subtelomeric primer and a reverse Teltail primer. The six C-telorettes and the 5′ ends to which they can ligate are shown. Only the C-telorette annealing adjacent to the last base of the C-strand will be ligated to the telomere end and can produce a PCR product. Most of the telomeres in human cells end at ATC-5′ (Sfeir et al, 2005). (B) PCR products of C-STELA using DNA from HeLa cells with control short interfering RNA (siRNA) and Mre11 siRNA. Six individual ligation reactions used different C-telorettes at 10−5 μM concentration. PCR products were run in separate lanes. Reduction of Mre11 by RNAi did not alter the terminal nucleotide of the C-strand. The same Mre11 siRNA-treated HeLa cell DNA characterized in Fig 1, showing reduced overhang length, was used for this analysis.
Figure 5.
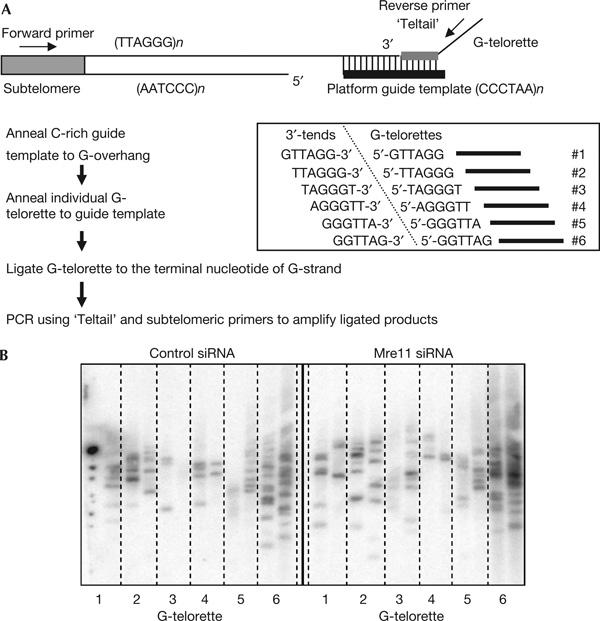
Mre11 RNA interference does not alter G-rich strand terminal nucleotides. (A) Schematic representation of G-STELA (Sfeir et al, 2005). A C-rich guide template is first annealed to G-overhangs to provide a platform for subsequently annealing G-telorettes to the 3′ end of the G-strand. Six individual G-telorettes are ligated in separate reactions to the same amount of DNA and then amplified using a forward Xp/Yp-chromosome-specific subtelomeric primer and a reverse Teltail primer. The six G-telorettes and the 5′ ends to which they can ligate are shown. Only the G-telorette annealing adjacent to the last base of the G-strand will be ligated to the telomere end and can produce a PCR product. The terminal nucleotide at G-strands is less precise than that at C-strands (Sfeir et al, 2005). (B) PCR products of G-STELA using DNA from HeLa cells with control small interfering RNA (siRNA) and Mre11 siRNA. Six individual ligation reactions using different G-telorettes at 10−5 μM concentration are shown. PCR products were run in separate lanes. Results are representative of three independent experiments. Reduction of Mre11 by RNAi did not alter the terminal nucleotide of the G-strand. The same Mre11 siRNA-treated HeLa cell DNA characterized in Fig 1, showing reduced overhang length, was used for this analysis. Insufficient numbers of bands were analysed to determine whether the slight shift in favoir of the specific TAG-3′ end, previously observed in telomerase-positive versus -negative cells (Sfeir et al, 2005), was affected by Mre11.
In S. cerevisiae, deletion of Mre11 results in shorter overhangs and telomere shortening (Boulton & Jackson, 1998; Ritchie & Petes, 2000; Larrivee et al, 2004; Takata et al, 2005). MRX deficiency in S. cerevisiae reduces the association of Cdc13 and Est1 to telomeres in the late S phase (Diede & Gottschling, 2001; Takata et al, 2005), leading to reduced recruitment of telomerase to telomeres and telomere shortening. It has been proposed that passage of the replication fork at telomeres allows transient access of MRX to telomere ends, which then processes telomere ends as DSBs (Takata et al, 2005). Mre11 levels are higher in knockdowns (human cells) than in knockouts (yeast), and this may contribute to some differences. Our results showing that MRN reduction leads only to a transient overhang shortening and has no effect on telomere length nonetheless suggest potential differences in end-processing steps between yeast and human telomeres.
Our findings that overhang length is rapidly restored in Mre11-diminished cells may partly explain the observation that G-overhang length is unchanged 7 PD after Mre11 disruption in chicken cells (Wei et al, 2002), and further suggest that other factors may substitute for Mre11 in MRN-deficient cells. Many nucleases and helicases have been identified in mammalian cells (Shevelev & Hubscher, 2002). Determining which nuclease and/or helicases are involved will be important to understand the molecular mechanism for telomere end processing.
Methods
Cells and antibodies. Cells were cultured at 37°C under 5% CO2 in a 4:1 mixture of Dulbecco's modified Eagle's medium and medium 199 supplemented with 10% cosmic calf serum (HyClone, South Logan, UT, USA). The following primary antibodies were used: monoclonal anti-Mre11 (GeneTex, San Antonio, TX, USA), anti-Nbs1 (BD Biosciences, San Jose, CA, USA), anti-Rad50 (CalBiochem, La Jolla, CA, USA) and goat anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Horseradish peroxidase-conjugated anti-mouse or anti-goat IgG were used as secondary antibodies.
RNA interference. siRNAs were designed and synthesized for the target sequence of Mre11 (ACAGGAGAAGAGATCAACT), Rad50 (GGAGAAGGAAATACCAGAA) and Nbs1 (CAGGAGGAAGATGTCAATG). The control siRNA targets luciferase and its sequence is available from BD Biosciences. HeLa cells were transfected with siRNA with Oligofectamine (Invitrogen, Carlsbad, CA, USA). Cells were collected 72 h after transfection to isolate DNA for overhang measurement and to isolate protein for immunoblotting. For stable expression of shRNA, the corresponding double-strand DNA sequences were cloned into pSIREN-retroQ retroviral vector (BD Biosciences), and cells were infected and selected for stable puromycin-resistant colonies.
Overhang assays. Telomere overhang protection assay and non-denaturing in-gel hybridization assay were carried out as described (Chai et al, 2005). The relative abundance of overhangs from non-denaturing gel assays was calculated by normalizing signals obtained from hybridization to an 18-mer C-rich probe under non-denaturing conditions (representing overhang signals) by signals obtained from hybridization of the same gel to the Alu-repeat probe under denaturing conditions (representing total genomic DNA).
C- and G-strand STELA. Determination of the terminal nucleotides on C- and G-strands was carried out as described previously (Sfeir et al, 2005).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400600-s1.pdf).
Supplementary Material
Supplemental Materials
Acknowledgments
We thank members of Shay/Wright lab for technical assistance. This work was supported by Ruth L. Kirschstein NRSA Individual Fellowship (W.C.) and NIH AG01228. W.E.W. is an Ellison Medical Foundation Senior Scholar.
References
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH (2001) Strand-specific postreplicative processing of mammalian telomeres. Science 293: 2462–2465 [DOI] [PubMed] [Google Scholar]
- Bi X, Wei SC, Rong YS (2004) Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr Biol 14: 1348–1353 [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J 17: 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Shay JW, Wright EW (2005) Human telomeres maintain their overhang length at senescence. Mol Cell Biol 25: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, Gorski MM, Engels WR, Gatti M (2004) The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol 14: 1360–1366 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Teo SH, Jackson SP (2004) Functional links between telomeres and proteins of the DNA-damage response. Genes Dev 18: 1781–1799 [DOI] [PubMed] [Google Scholar]
- de Lange T (2002) Protection of mammalian telomeres. Oncogene 21: 532–540 [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE (2001) Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol 11: 1336–1340 [DOI] [PubMed] [Google Scholar]
- Ferreira MG, Miller KM, Cooper JP (2004) Indecent exposure: when telomeres become uncapped. Mol Cell 13: 7–18 [DOI] [PubMed] [Google Scholar]
- Gallego ME, White CI (2001) RAD50 function is essential for telomere maintenance in Arabidopsis. Proc Natl Acad Sci USA 98: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T (2005) POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 24: 2667–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23: 687–696 [DOI] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa K (1997) Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2: 443–455 [DOI] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ (2004) The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF (2004) The Mre11 complex and ATM: a two-way functional interaction in recognising and signaling DNA double strand breaks. DNA Repair (Amsterdam) 3: 1515–1520 [DOI] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR (1995) Telomerase and DNA end replication: no longer a lagging strand problem? Science 269: 1533–1534 [DOI] [PubMed] [Google Scholar]
- Lombard DB, Guarente L (2000) Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res 60: 2331–2334 [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88: 657–666 [DOI] [PubMed] [Google Scholar]
- Maser RS, DePinho RA (2004) Telomeres and the DNA damage response: why the fox is guarding the henhouse. DNA Repair (Amsterdam) 3: 979–988 [DOI] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ (1997) The terminal DNA structure of mammalian chromosomes. EMBO J 16: 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol 8: 657–660 [DOI] [PubMed] [Google Scholar]
- Olovnikov AM (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41: 181–190 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M (1998) The 3′–5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979 [DOI] [PubMed] [Google Scholar]
- Ranganathan V et al. (2001) Rescue of a telomere length defect of Nijmegen breakage syndrome cells requires NBS and telomerase catalytic subunit. Curr Biol 11: 962–966 [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD (2000) The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE (2005) Telomere end processing: the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138 [DOI] [PubMed] [Google Scholar]
- Shevelev IV, Hubscher U (2002) The 3′–5′ exonucleases. Nat Rev Mol Cell Biol 3: 364–376 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Stewart SA, Ben-Porath I, Carey VJ, O'Connor BF, Hahn WC, Weinberg RA (2003) Erosion of the telomeric single-strand overhang at replicative senescence. Nat Genet 33: 492–496 [DOI] [PubMed] [Google Scholar]
- Takata H, Tanaka Y, Matsuura A (2005) Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol Cell 17: 573–583 [DOI] [PubMed] [Google Scholar]
- Tomita K et al. (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie K, Povirk LF (2003) Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22: 5792–5812 [DOI] [PubMed] [Google Scholar]
- Wei C, Skopp R, Takata M, Takeda S, Price CM (2002) Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res 30: 2862–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA (1996) Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet 25: 347–352 [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12: 1489–1498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials


