Abstract
Recent analyses have shown that the activity of the yeast nuclear exosome is stimulated by the Trf4p–Air1/2p–Mtr4p polyadenylation (TRAMP) complex. Here, we report that strains lacking the Rrp6p component of the nuclear exosome accumulate polyadenylated forms of many different ribosomal RNA precursors (pre-rRNAs). This polyadenylation is reduced in strains lacking either the poly(A) polymerase Trf4p or its close homologue Trf5p. In contrast, polyadenylation is enhanced by overexpression of Trf5p. Polyadenylation is also markedly increased in strains lacking the RNA helicase Mtr4p, indicating that it is required to couple poly(A) polymerase activity to degradation. Tandem affinity purification-tagged purified Trf5p showed polyadenylation activity in vitro, which was abolished by a double point mutation in the predicted catalytic site. Trf5p co-purified with Mtr4p and Air1p, indicating that it forms a complex, designated TRAMP5, that has functions that partially overlap with the TRAMP complex.
Keywords: polyadenylation, RNA degradation, RNA surveillance, TRAMP complex, yeast
Introduction
Related exosome complexes of 3′–5′ exonucleases are found in the nucleus and cytoplasm of all eukaryotes examined, and a similar complex is found in archaea. The nuclear exosome is implicated in the maturation and degradation of many different RNA species. The exosome purified from Saccharomyces cerevisiae shows limited activity in vitro (Mitchell et al, 1997), but can be activated by the Trf4p–Air1/2p–Mtr4p polyadenylation (TRAMP) complex, which contains the poly(A) polymerase Trf4p, one of two functionally redundant zinc-knuckle proteins Air1p or Air2p and the putative RNA helicase Mtr4p/Dob1p (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005).
Trf4p shows 56% similarity (48% identity) to Trf5p (Castano et al, 1996), but purified Trf5p did not initially show in vitro poly(A) polymerase activity, which suggests that it might have a different function (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). This was unexpected, as Trf5p shows clear homology to the CID/gld-2 class of poly(A) polymerases (Wang et al, 2000, 2002; Saitoh et al, 2002; Kwak et al, 2004). Moreover, Trf5p shows at least partial functional redundancy with Trf4p, as strains lacking both proteins are inviable (Castano et al, 1996) and the depletion of Trf5p enhanced RNA maturation defects seen in the trf4Δ strain (LaCava et al, 2005).
We show that Trf5p functions in vivo and in vitro as a poly(A) polymerase in a TRAMP-like complex (TRAMP5). We previously reported that Trf5p lacks polymerase activity. However, owing to an error in the genomic TRF5 (DNA topoisomerase I-related function) sequence, previously analysed tandem affinity purification (TAP)-tagged Trf5p constructs had an incorrect carboxy-terminal region.
Results
Trf5p shows poly(A) polymerase activity in vivo
Previous analyses showed that rrp6Δ strains accumulate polyadenylated ribosomal RNA precursors, and implicated Trf4p in this activity (Kuai et al, 2004; LaCava et al, 2005; Wyers et al, 2005). However, we observed that, although polyadenylation of the 23S RNA was reduced in rrp6Δ, trf4Δ strains relative to the rrp6Δ single mutants, it was not eliminated (LaCava et al, 2005). This strongly suggested the participation of another poly(A) polymerase.
To assess whether Trf5p contributes to pre-rRNA polyadenylation, we compared total and poly(A)+ selected RNA isolated from rrp6Δ strains that also lacked either Trf4p or Trf5p (Fig 1). In rrp6Δ single mutant strains, polyadenylated forms were detected for all tested pre-rRNAs (35S, 32S, 27S, 23S, 21S, 20S, 17S′, 7S and 5.8S+30), as well as for mature 25S and 18S rRNAs (Fig 1A–J; see supplementary Fig S1 online for descriptions of the RNA species observed; TSA1 messenger RNA was used as a control for loading and poly(A)+ RNA recovery). For all these species, the fraction that was polyadenylated was decreased in the rrp6Δ strain lacking Trf5p. Polyadenylation of these RNAs was also reduced by loss of Trf4p, but, for most species, greater reduction in polyadenylation was seen in the rrp6Δ, trf5Δ strain than in the rrp6Δ, trf4Δ strain (see Fig 1N for quantification of data). These reductions were seen for both normal processing intermediates and aberrant species detected only in rrp6Δ strains, such as 5.8S+30 and 17S′.
Figure 1.

Both Trf4p and Trf5p participate in the polyadenylation of ribosomal RNA precursors. Otherwise isogenic strains were grown at 25°C. Strains with GAL-regulated alleles were grown on 2% galactose and shifted to 2% glucose for 24 h before analysis. A 5 μg portion of total RNA and poly(A)+ RNA recovered from 50 μg total RNA was loaded per lane on 1.2% agarose/glyoxal (A–F) or 8% polyacrylamide/urea gels (G–M). (A–M) Northern hybridization with probes to pre-rRNA, rRNA and small nucleolar RNA species or the loading control TSA1, a highly expressed messenger RNA, the level of which is unaffected by exosome mutations. Probes used were (A) 003, (B) 020, (C) 004, (D) 007, (E) 008, (F) 499, (G) 020, (H) 015, (I) 041, (J) 499, (K) 202, (L) 214 and (M) 499. See supplementary Fig S1 online for descriptions of the pre-rRNA species and Supplementary Table S2 online for oligonucleotide probe sequences. RNA species marked with an asterisk are likely to represent as yet uncharacterized pre-rRNA degradation intermediates. (N) Graphs showing the fraction of each (pre-)rRNA species that is polyadenylated, calculated relative to the TSA1 mRNA control. The data are an average of three biological replicates; bars represent standard errors. Polyadenylated forms resolved on the polyacrylamide gels are indicated with pA. U24 is excised from the intron of the BEL1 pre-mRNA (Qu et al, 1995; Bousquet-Antonelli et al, 2000) and the band marked U24-int is likely to correspond to the species extended to the 5′ end of the intron (Qu et al, 1995; Bousquet-Antonelli et al, 2000).
For low-molecular-weight RNAs separated on polyacrylamide gels (Fig 1G–M), polyadenylated RNA species (marked pA) are resolved from the background of non-polyadenylated RNAs, showing a characteristic pattern, with a cutoff ∼15 nt above the non-adenylated species. This represents the minimal poly(A) tract that is retained on the oligo(dT) beads. A low level of nonspecific background recovery on the oligo(dT) column probably accounts for the 1%–2% signal observed for the mature rRNAs in the wild-type strain. The absence of Trf4p reduced ribosome synthesis in an otherwise wild-type background (Fig 1A–F), whereas the absence of only Trf5p induced no obvious defects in rRNA synthesis. This may reflect the mild growth impairment seen in trf4Δ strains, as ribosome synthesis rates are tightly linked to the growth rate.
These data indicate that Trf5p is largely responsible for pre-rRNA polyadenylation in strains lacking Rrp6p. This is not, however, the case for all stable RNA precursors. 3′-extended and polyadenylated forms of the U14 and U24 small nucleolar RNAs (snoRNAs) accumulate in rrp6Δ strains (Allmang et al, 1999; van Hoof et al, 2000; LaCava et al, 2005; Wyers et al, 2005). A comparison of the double mutant strains indicates that Trf4p is largely responsible for polyadenylation of U14 (Fig 1K) and deletion of either polymerase leads to a similar reduction in polyadenylation of U24 (Fig 1L).
Growth of a trf4Δ, GAL-trf5 strain on galactose medium caused overexpression of the TRF5 mRNA (Fig 2C), and presumably also results in overexpression of Trf5p. This suppressed most of the defects caused by the lack of Trf4p, consistent with functional redundancy between Trf4p and Trf5p. However, simultaneous depletion of Trf4p and Trf5p by growth of the trf4Δ, GAL∷trf5 strain on glucose medium did not clearly alter pre-rRNA levels compared with the trf4Δ single mutant, except for the previously reported accumulation of the aberrant 23S RNA (LaCava et al, 2005). As expected in a strain lacking both putative pre-rRNA poly(A) polymerases, this 23S RNA was not detectably polyadenylated (Fig 1).
Figure 2.

Overexpression of Trf5p causes hyperadenylation. Trf5p was overexpressed in the absence of Trf4p and Rrp6p by growth of the GAL∷trf5, trf4Δ, rrp6Δ strain in galactose medium. (A,B) Polyadenylation of 5.8S+30 pre-ribosomal RNA. (C): Overexpression of TRF5 messenger RNA on galactose. Samples were processed as described in the legend to Fig 1. Probes used were (A) 020, (B) 499, (C) random primed probe to TRF5 open reading frame and (D) 403. (C,D) Poly(A)+ RNA only; PGK1 is an mRNA loading control.
The excised 5′-external transcribed spacer (5′-ETS) region of pre-rRNA is strongly accumulated in strains with defects in the exosome or Mtr4p (de la Cruz et al, 1998). However, no clear accumulation of 5′-ETS was seen in trf4Δ or trf5Δ mutants, or in the trf4Δ, GAL∷trf5 strain, in glucose medium (data not shown). These strains also failed to show the defects in 7S pre-rRNA maturation that are seen in exosome and Mtr4p mutants (de la Cruz et al, 1998; Allmang et al, 1999).
To analyse an rrp6Δ strain lacking both Trf4p and Trf5p, we attempted to create the triple mutant strain rrp6Δ, trf4Δ, GAL∷trf5. However, whereas the resulting strain showed overexpression of TRF5 mRNA on galactose medium, its depletion was not seen on glucose. Further analysis showed that a rearrangement had taken place during transformation, giving two copies of the TRF5 gene, only one of which was under GAL control. Repeated attempts to construct a strain of rrp6Δ, trf4Δ, GAL∷trf5 genotype that does not have such a rearrangement were unsuccessful. Analysis of the strain, however, provided further evidence that Trf5p acts as a poly(A) polymerase in vivo. Overexpresson of Trf5p in the rrp6Δ strain by growth on galactose medium (Fig 2C) resulted in greatly increased polyadenylation of the 5.8S+30 pre-rRNA (Fig 2A). A similar degree of hyperadenylation was seen for the U14 snoRNA (data not shown). As this strain lacks Trf4p, it is likely that the observed hyperadenylation is the result of Trf5p activity. Some hyperadenylation of U14 and U24 was also seen in the trf4Δ, GAL∷trf5 strain on galactose in the presence of functional Rrp6p (Fig 1K,L, lane 12).
Trf4p and Trf5p associate with the putative RNA helicase Mtr4p (Kadaba et al, 2004; LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). However, strains depleted of Mtr4p show pre-rRNA defects that were not seen in the trf4Δ or trf5Δ mutants (de la Cruz et al, 1998). We speculated that Mtr4p might link polyadenylation by Trf4p and Trf5p to RNA degradation by the exosome. This model predicts that Mtr4p-depleted cells would show accumulation of pre-rRNAs that had been polyadenylated by Trf4p and/or Trf5p, but inefficiently degraded because of the absence of exosome recruitment. As shown in Fig 3, this proved to be the case. Particularly notable was the large accumulation of 23S RNA and 20S pre-rRNA, of which 30–40% was polyadenylated. Accumulation of polyadenylated forms of the 35S, 32S, 21S and 7S pre-rRNAs and the excised 5′-ETS region was also seen in this strain (Fig 3). The accumulation of most polyadenylated pre-rRNAs was greater in strains depleted of Mtr4p or Rrp6p than in strains depleted of the core exosome components Rrp41p and Rrp44p (Fig 3). It may be that following recruitment by Mtr4p, deadenylation is preferentially carried out by Rrp6p, with subsequent degradation by the core exosome.
Figure 3.

Mtr4p connects polyadenylation to degradation. Depletion of Mtr4p by growth of the GAL∷mtr4 strains on glucose medium results in RNA hyperadenylation. Strains were grown and analysed as described in Fig 1. Probes used were (A) 003, (B) 020, (C) 004, (D) 007, (E) 008, (F) 499, (G) 020, (H) 033, (I) 015, (J) 041, (K) 499 and (L) Phosphorlmager quantification of data from panel C.
Trf5p shows polyadenylation activity in vitro
The in vivo analyses strongly indicated that Trf5p functions as a poly(A) polymerase, and we therefore reassessed its in vitro activity. While cloning TRF5 to make the necessary tagged constructs, we noticed that our clones consistently differed from the TRF5 sequence given in the Saccharomyces Genome Database (www.yeastgenome.org; Castano et al, 1996). Our sequence has a single inserted cytosine, located within a run of cytosine residues, leading to a frameshift relative to the published sequence that alters the last 30 amino acids of the protein. Alignment of this sequence with Trf5p homologues, predicted from the genomic sequences of other yeast species, shows that this region is highly conserved (Fig 4A) and therefore likely to be functionally important. This conservation also makes it likely that our revised sequence is correct.
Figure 4.
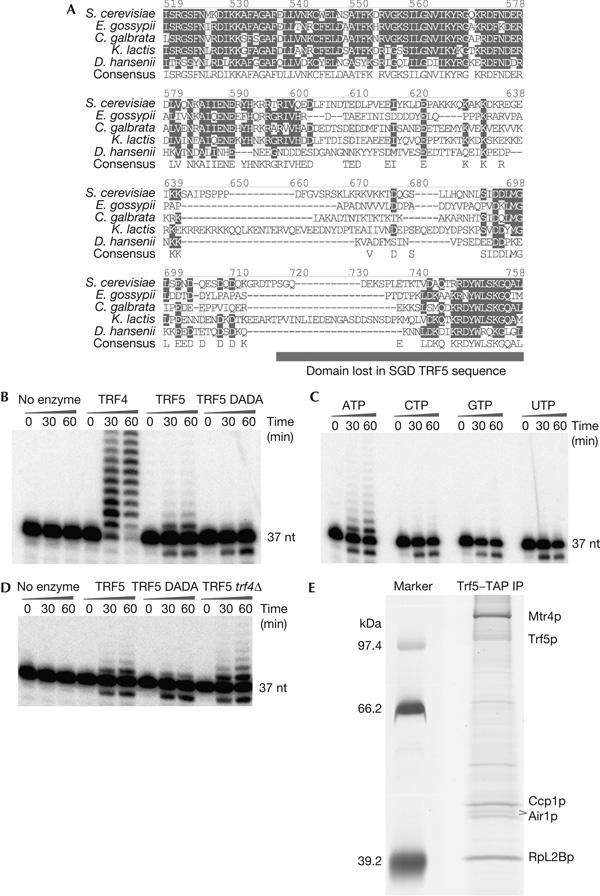
The TRAMP5 complex shows Trf5p-dependent polyadenylation activity. (A) The predicted sequence of the carboxy-terminal region of Saccharomyces cerevisiae Trf5p was compared with the predicted open reading frames from the genomes of related yeasts. (B–D) In vitro poly(A) polymerase assays. (B) Comparison of the activity of purified Trf4p, Trf5p and the catalytically inactive Trf5p DADA mutant. (C) Comparison of polymerase activity with different nucleotide triphosphates. (D) Purified Trf5p retains poly(A) polymerase activity when purified from a strain lacking Trf4p. Proteins were isolated by means of C-terminal tandem affinity purification (TAP) tags from yeast grown to OD600 nm 1.0 at 25°C in YPD. Trf4–TAP and Trf5–TAP concentrations were normalized by western blotting of the immunoprecipitated (IP) fractions using an anti-TAP antibody that recognizes the calmodulin binding peptide region of the tag. The substrate is a 5′-labelled 37-nt transcript derived from the pBS linker. Samples were taken at the indicated time points and resolved on a 12% polyacrylamide/urea gel. (E) SDS gel analysis of proteins co-precipitated with Trf5p. Marker, molecular weight markers; Trf5–TAP IP, precipitated proteins. Species identified by mass spectrometry are labelled. Rpl2Bp and Ccp1p (cytochrome c peroxidase) are abundant proteins that are likely to be contaminants.
Our previous analysis of Trf5p (LaCava et al, 2005) used a C-terminal TAP-tagged strain obtained from Open Biosystems, in which the position of the tag was based on the published sequence. Sequencing of TRF5 in this strain showed that the TAP tag is in an incorrect position. The tag is expressed because the construct contains another frameshift mutation 6 nt upstream of the site of the TAP fusion, which brings the tag back into frame.
As it seemed likely that these mutations might have contributed to the lack of in vitro activity previously observed, we constructed strains expressing full-length Trf5–TAP based on our new sequence. We additionally constructed an otherwise identical strain carrying two point mutations, D233A and D235A, in the putative catalytic site of Trf5p, which are predicted to abolish any catalytic activity (DADA mutation). TAP purifications of the Trf5p fusions and Trf4p were carried out and polymerase activities were assayed, using equivalent amounts of each protein, on a 5′-labelled 37-nt RNA substrate (Fig 4B). Trf5–TAP reproducibly showed poly(A) polymerase activity. This was abolished by the DADA catalytic site mutation, showing that the activity stems from Trf5p rather than from a co-precipitating contaminant. In contrast, no activity was observed using the truncated Trf5p–TAP construct from Open Biosystems, isolated under identical conditions (data not shown). The polymerase activity of Trf5–TAP was also assessed using ATP, GTP, CTP or UTP as substrates, and polymerization was observed only with ATP (Fig 4C). We conclude that Trf5p is a functional poly(A) polymerase. The in vitro activity of purified Trf5–TAP was substantially lower than that of an equivalent quantity of Trf4–TAP. This may reflect intrinsic differences, but it is also possible that the activity of Trf5p is adversely affected by the C-terminal TAP tag, even in the context of the full-length open reading frame (ORF).
To confirm that Trf5p acts independently of Trf4p, we deleted trf4 in the TRF5–TAP strain and then isolated Trf5p and assessed the polyadenylation activity as above (Fig 4D). The activity of TRAMP5 was not abolished in the absence of Trf4p and was mildly enhanced. This shows that the TRAMP5 complex does not require Trf4p.
In previous analyses, no association of Trf4p with Trf5p was detected, showing them to be in distinct complexes (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). However, whereas Trf4p was found to be associated with Mtr4p plus either Air1p or Air2p in the TRAMP complex, the truncated Trf5–TAP construct was previously found to be associated with Mtr4p but not with Air1p or Air2p (LaCava et al, 2005). To determine whether full-length Trf5p is present in a TRAMP-like complex, we analysed proteins that co-precipitated with the corrected Trf5–TAP construct (Fig 4E). Both Mtr4p and Air1p were identified by mass spectrometry as bands that showed approximate stoichiometry with Trf5–TAP.
We conclude that the in vivo poly(A) polymerase activity of Trf5p is likely to be carried out by the complex, which we designate TRAMP5, consisting of Trf5p, Air1p and Mtr4p.
Discussion
We have shown that budding yeast contains two related nuclear poly(A) polymerase complexes, which include either Trf4p or Trf5p. We previously reported that purified Trf5p lacked poly(A) polymerase activity and failed to co-precipitate with Air1p or Air2p (LaCava et al, 2005). However, it now seems that this was related to the finding reported here that the TRF5 sequences in the Saccharomyces genomic database contain a single nucleotide deletion (www.yeastgenome.org; Castano et al, 1996). The sequence that we are using is likely to be correct, as it generates a predicted ORF with a C-terminal region that is highly conserved in other Ascomycete genomes. The region of Trf5p that is absent from previously used constructs lacks clear sequence motifs, but its high evolutionary conservation indicates that it has some important function. It is unclear whether the absence of poly(A) polymerase activity observed with the truncated Trf5p was because of an intrinsic defect in its catalytic activity or lack of association with Air1p.
The Trf5p complex we isolated added poly(A) tails of only ∼1–7 nt in vitro. It is possible that, under normal circumstances, the TRAMP5 complex adds only short poly(A) tails in vivo, which would not readily be detected by poly(A) selection or RNase H/oligo(dT) cleavage. However, the poly(A) tails seen in rrp6Δ strains lacking Trf4p, as well as the hyperadenylated RNAs detected in strains overexpressing Trf5p, are considerably longer. It is therefore possible that the short tails seen in vitro are a consequence of the presence of the TAP tag, suboptimal conditions or the use of a non-physiological substrate.
The relative effects of the loss of Trf4p or Trf5p from the rrp6Δ strain were variable for different RNA species. In some cases, such as the 23S pre-rRNA and U14 snoRNA, polyadenylation was greatly reduced in the absence of only Trf4p. Although polyadenylation of most pre-rRNA species was affected more strongly by the absence of Trf5p, polyadenylation of the U24 snoRNA seemed to be carried out almost equally by both proteins. These differences strongly indicate that the TRAMP and TRAMP5 complexes do not have identical specificities. The factors that determine the functional differences between the Trf4p and Trf5p versions of the TRAMP complex are unclear. Trf5p was reported to be enriched in the nucleolus (Huh et al, 2003), and this may be related to its effects on pre-rRNA polyadenylation. Our analyses of the function of Trf5p have largely used strains lacking Rrp6p, and the RNA species affected might not fully reflect the relative roles of Trf4p and Trf5p in wild-type cells. However, it seems clear that Trf5p interacts functionally with Rrp6p and therefore with the nuclear exosome complex.
Overexpression of Trf5p led to hyperadenylation of pre-rRNAs, whereas overexpression of Trf4p led to hyperadenylation of an aberrant pre-tRNA (Kadaba et al, 2004), indicating that they are limiting in normal cells. Depletion of Mtr4p resulted in substantial accumulation of polyadenylated pre-rRNA, which was not replicated in a strain depleted of a core exosome component Rrp44p. Together, these observations support the model that Mtr4p links the polyadenylation activities of Trf4p and Trf5p to deadenylation and degradation by the exosome.
In conclusion, budding yeast contains two nuclear poly(A) polymerase complexes that are likely to function preferentially on different substrates. Our data provide no clear evidence that the TRAMP or TRAMP5 complexes are directly required for ribosome synthesis or other RNA maturation pathways. It is, however, clear that Trf4p and Trf5p are required to polyadenylate ribosomal and other RNA precursors that would normally be degraded by Rrp6p. This is likely to form part of a surveillance pathway that removes RNA molecules that have failed to undergo correct processing and/or assembly with proteins. Such defects apparently occur at significant levels in all wild-type cells, but can be detected only when the surveillance system is impaired by inactivation of TRAMP or exosome components.
Methods
Strains and media. Strains were grown in YPD medium (2% peptone, 1% yeast extract, 2% glucose) or YPGal medium (2% peptone, 1% yeast extract, 2% galactose) at 25°C. Transformation was carried out as described (Gietz et al, 1992). Strains of S. cerevisiae used in this study are listed in supplementary Table S1 online.
In vivo analyses. Yeast RNA extraction and northern analysis were carried out as described (Tollervey, 1987). Oligonucleotide probes used are listed in supplementary Table S2 online. Polyadenylated RNAs were purified from 200 μg of total RNAs using the ‘PolyA Tract mRNA Isolation System IV' (Promega, Madison, WI, USA) modified as described (LaCava et al, 2005). The ratio of the total RNA input to the recovered poly(A)+ fraction that was loaded on the gels was 1:10. Quantification was carried out using a Storm PhosphorImager.
In vitro assays. Polyadenylation assays were carried out as described (LaCava et al, 2005). Proteins were isolated using C-terminal TAP tags from yeast grown to OD600 nm 1.0 at 25°C in YPD medium, as previously described (Rigaut et al, 1999; LaCava et al, 2005). Enzyme concentrations were normalized by western blotting of the immunoprecipitated fractions using ‘anti-TAP' antibody (Open Biosystems, Huntsville, AL, USA) at 1:5,000 dilution. Equivalent amounts of each protein were incubated with a 5′-labelled 37-nt substrate transcribed from the pBS linker. Samples were resolved on a 12% polyacrylamide/8 M urea gel.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400612-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank C. Schneider, C. Dez, K. Kotovic and J. LaCava for reagents, helpful advice and critical reading of the manuscript. This work was supported by the Wellcome Trust and EU Grant QLG2-CT-2001-01554.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Castano IB, Heath-Pagliuso S, Sadoff BU, Fitzhugh DJ, Christman MF (1996) A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res 24: 2404–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L, Fang F, Butler JS, Sherman F (2004) Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101: 8581–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M (2004) Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA 101: 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 21: 713–724 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome; a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonuclease activities. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Qu LH, Henry Y, Nicoloso M, Michot B, Azum MC, Renalier MH, Caizergues-Ferrer M, Bachellerie JP (1995) U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res 23: 2669–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P (2002) Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109: 563–573 [DOI] [PubMed] [Google Scholar]
- Tollervey D (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J 6: 4169–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316 [DOI] [PubMed] [Google Scholar]
- Wang SW, Toda T, MacCallum R, Harris AL, Norbury C (2000) Cid1, a fission yeast protein required for S–M checkpoint control when DNA polymerase δ or ɛ is inactivated. Mol Cell Biol 20: 3234–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F et al. (2005) Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


