Abstract
Regulated gene expression, achieved through the coordinated assembly of transcription factors, co-regulators and the basal transcription machinery on promoters, is an initial step in accomplishing cell specificity and homeostasis. Traditional models of transcriptional regulation tend to be static, although gene expression profiles change with time to adapt to developmental and environmental cues. Furthermore, biochemical and structural studies have determined that initiation of transcription progresses through a series of ordered events. By integrating time into the analysis of transcription, chromatin immunoprecipitation assays and live-cell imaging techniques have revealed the dynamic, cooperative, functionally redundant and cyclical nature of gene expression. In this review, we present a dynamic model of gene transcription that integrates data obtained by these two techniques.
Keywords: nuclear receptors, transcriptional cycling, ChIP, FRAP model
Introduction
The phenotypic diversity of cells, and their response and adaptation to their environment, are achieved through the regulation of gene transcription. Understanding how transcription is modulated is vital for describing the generation of cell-specific transcriptome and proteome profiles. Current models of transcription initiation tend to be static and centre on promoter elements that provide a platform for the assembly of intermediate complexes and the basal transcription machinery. The discoveries that transcription takes place in a repressive environment and that chromatin structure influences transcription further expanded these models (Ahmad & Henikoff, 2002; Brown, 1999). In parallel, a massively increasing number of co-regulators and protein complexes involved in transcription were identified (Belandia & Parker, 2003; McKenna & O'Malley, 2002; Narlikar et al, 2002), provoking the realization that functional redundancy is a generally applicable feature of transcriptional attainment. Together, these insights increased the complexity of transcriptional modulation and implied that the dynamics of recruitment are significant in gene expression. Moreover, the three-dimensional structure of transcription factors also has a significant impact on events (Asturias, 2004). Collectively, achieving transcription requires the integration of five variables: cis-acting factors (DNA and chromatin), trans-acting factors (transcriptional activators and associated complexes), the basal transcription machinery (including RNA polymerase II (Pol II) and TATA-binding protein (TBP)), three-dimensional structures and time. Additional hierarchical parameters, such as nuclear organization, also have an impact on these local events (van Driel et al, 2003).
Kinetic descriptions of transcriptional activation have been generated during the past five years by laboratories using nuclear receptor (NR)-driven gene expression as a model system. NRs are a subfamily of transcription factors, and include ligand-dependent transcription factors, such as receptors for oestrogens (ER-α and ER-β), androgens (ARs), glucocorticoids (GRs), progesterone (PR), thyroid hormones (TRs), vitamin D (VDR) and retinoids/rexinoids (RARs/RXRs; Robinson-Rechavi et al, 2003). Chromatin immunoprecipitation (ChIP) assays and fluorescence recovery after photobleaching (FRAP) have been used to evaluate transcriptional processes kinetically, albeit on different time-scales; ChIP has a time resolution of several minutes, whereas FRAP resolves events in the sub-second range. Essentially, each technique provides a different view of transcription, with ChIP indicating that, on average, transcriptional processes take tens of minutes and FRAP demonstrating a rapid exchange between transcription factors and their target promoters. In this review, we present a model that reconciles both data sets.
Chromatin and transcription
The organization of DNA into chromatin in vivo generates regulatory constraints that have central roles in many cellular processes (Khorasanizadeh, 2004). The basic organization of chromatin as a succession of nucleosomes separated by linker DNA is often likened to beads on a string. A nucleosome consists of 146 bp of DNA wrapped around histone octamers made of dimers of each of the core histones H2A, H2B, H3 and H4 (Luger et al, 1997). Chromatin structure has a dual influence on transcription: it organizes genomic information in three dimensions, which is important for the coordinated regulation of genome expression (Perkins et al, 2004), and it restricts the access of promoter sequences to the transcriptional machinery (Dillon & Festenstein, 2002). This structural restriction of chromatin on gene expression is overcome by multi-subunit protein complexes that have three main activities. First, reversible post-translational modifications (such as phosphorylation, acetylation, methylation, ubiquitylation and sumoylation) of the amino-terminal tails of the histones on lysine (K), arginine (R), serine (S) and threonine (T) residues modify chromatin structure. These alterations are directed by enzymes (for example, kinases, phosphatases, histone acetyl transferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), ubiquitin and SUMO ligases) that associate with sequence-specific transcription factors binding directly to DNA (Gill, 2004; Narlikar et al, 2002). Specific sets of histone modifications are associated with genes that are actively transcribed and with those that are repressed. This defines the ‘histone code' ( Jenuwein & Allis, 2001), in which specific histone modifications imposed by one factor provoke the sequential recruitment of subsequent transcriptional factors. This adds further combinatorial and dynamic aspects to transcriptional regulation and increases the complexity of the information contained in chromatin: it is not only the sum of the charges on nucleosome tails that is important, but also their spatial combination and the order of their development. Second, plasticity of chromatin is induced by ATP-dependent remodelling complexes, which rearrange the organization of the nucleosomes in the chromatin fibre (Sif, 2004). Third, coupling CpG methylation by DNA methyltransferases (DNMTs) with deacetylation and methylation of histone lysines induces profound gene silencing and is typical in the organization of heterochromatin (Hermann et al, 2004). Finally, besides these enzymatic and energy-dependent processes, dynamic and competitive interactions of histone H1 and variants also modulate the local structure of the chromatin fibre (Catez et al, 2004; Bustin et al, 2005).
Regulation of transcription by nuclear receptors
NRs are transcription factors that bind as dimers to cognate response elements (PuGGTCA or PuG(G/A)ACA) that are organized in palindromic or direct repeats, or as monomers (Claessens & Gewirth, 2004). After binding to a promoter, NRs modulate transcription by recruiting transcriptional co-regulators and components of the basal transcription machinery. In the absence of ligand or, in the case of ER, when bound to partial antagonists such as tamoxifen (Lavinsky et al, 1998), NRs recruit repressive complexes to target promoters; these include HDACs, ATP-dependent remodelling complexes and corepressors such as SMRT and NCoR. These complexes generate a local chromatin environment that actively restricts transcription (Bowen et al, 2004). An exception to this are the ‘classical' steroid receptors, such as GR, which, in the absence of ligand, reside in the cytoplasm. Binding of agonistic ligands induces the exchange of corepressors for coactivators through a structural rearrangement of the NR (Glass & Rosenfeld, 2000). So far, more than 100 cofactors of NRs have been identified, including ATP-dependent remodelling complexes such as SWI/SNF, complexes with HAT activities (such as the SRC/p160 family, CBP/p300 proteins and ADA complexes), and proteins with HMT activities (including CARM1 and PRMT1; Klinge, 2000; Lonard & O'Malley, 2005; McKenna & O'Malley, 2002). Another class of coactivators, known as TRAP–DRIP–mediator complexes, has also been identified (Fondell et al, 1996); these interact with NRs at the identical surface to the p160/HAT proteins (Ren et al, 2000). Conceptually, recruitment of p160 and TRAP–DRIP complexes are mutually exclusive, as they cannot bind simultaneously to the same surface, which implies that they interact consecutively with the NR. Specific recruitment of the repressor of oestrogen receptor activity (REA), receptor-interacting protein 140 (RIP140) and ligand-dependent nuclear receptor corepressor (LCoR) by agonist-bound NR also displaces coactivators (Martini & Katzenellenbogen, 2003; White et al, 2004). Allosteric changes induced within interacting partners, such as ER-α and TBP (Warnmark et al, 2001), or NRs and HATs (Demarest et al, 2002) also define ordered and sequential interactions. Furthermore, HATs and HMTs modify target proteins other than histones (Wang C et al, 2001), with reciprocal post-translational modifications provoking sequential recruitment of protein complexes (Chen et al, 1999). Additional allosteric effects integrate chromatin remodelling with transcriptional regulation by NRs. For instance, binding of CBP to promoters is alleviated by H3 methylation (Wang H et al, 2001), and acetylation of H4 stabilizes recruitment of SWI/SNF (Hassan et al, 2001). Collectively, these data indicate that sequential, highly ordered processes define transcriptional events.
ChIP analysis of ER-α-mediated transcription
The most detailed ChIP-based analysis of the dynamic mechanisms involved in transcriptional initiation has been obtained for ER-α-mediated gene expression. The kinetics of association of ER-α and Pol II on four promoters—pS2/TFF1, cyclin D1, cathepsin D (CATD) and c-Myc—show a periodicity of 40–60 min (Shang et al, 2000; Reid et al, 2003; Liu & Bagchi, 2004; Park et al, 2005). By using α-amanitin to synchronize responsive promoters, a non-productive, cyclical interaction between the pS2 promoter and unliganded ER-α was shown (Reid et al, 2003; Métivier et al, 2003). Furthermore, three types of cycling occur in the presence of oestradiol (E2), namely an initial unproductive cycle that prepares the pS2 promoter for subsequent transcription, followed by two alternating, transcriptionally productive cycles (Fig 1; Métivier et al, 2003). Importantly, sequential immunoprecipitations of chromatin (Re-ChIP), which detect the simultaneous presence of two proteins on the same pool of promoters, have identified six ER-α-containing complexes in the presence of E2 (Métivier et al, 2003). In the cycles, a given enzymatic function (for example, HAT, HMT or HDAC activity) is provided by one of the alternative proteins in these complexes, such that combinations of functionally redundant enzyme complexes accomplish transcription by different routes.
Figure 1.
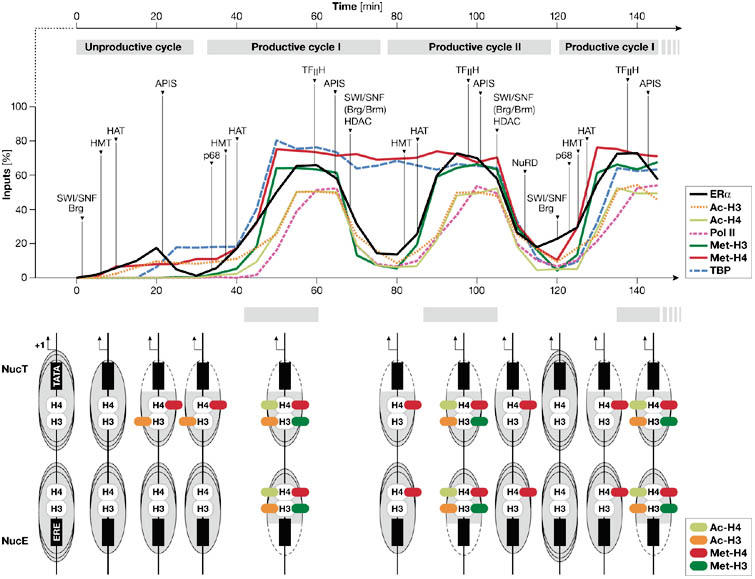
Cyclical recruitment of transcription factors to the pS2 promoter. The recruitment of cofactors (top) and the dynamics of the nucleosome (bottom) mediated by oestrogen receptor-α (ER-α) on the pS2 promoter in MCF-7 cells in the presence of oestrogen. The periodic association of HATs, HDACs, HMTs and SWI/SNF (Brg/Brm), as well as other important complexes that contribute to ER-α dynamics and promoter clearance are shown with arrows. The association phase of each productive cycle is shown by grey bars. Location of the modified histones in nucleosome E (NucE) and nucleosome T (NucT) are shown, with increased accessibility of either the TATA box or the ERE shown by dashed lines. Schemes are based on Métivier et al (2003) and Reid et al (2003) and our unpublished data. Specific recruitment of NuRD at the end of the second transcriptionally productive cycle corresponds to NucT remodelling, displacement of TBP and demethylation of dimethylated H4 R3 (either complete or with only one CH3 group). This step, which provokes the promoter to return to the basal state, delineates the two transcriptionally productive cycles. Ac-H3, acetylated histone 3 (K14); Ac-H4, acetylated histone 4 (K16); APIS, AAA ATPase proteins independent of 20S; ERE, oestrogen response element; HAT, histone acetyltransferase; HDAC, histone deacetylase; HMT, histone methyltransferase; Met-H3, dimethylated histone 3 (R17); Met-H4, dimethylated histone 4 (R3); NucE, nucleosome including the ERE; NucT, nucleosome including the TATA box; NuRD, nucleosome remodelling and deacetylating complex; p68, p68 RNA helicase; TBP, TATA-binding protein.
Achieving transcription
The initial cycle in the presence of ligand and cycling of unliganded ER-α are similar in character. Both generate a chromatin environment that is permissive for transcription without attaining transcription itself (Reid et al, 2003). ER-α initiates the association of chromatin remodelling complexes, with SWI/SNF recruited by liganded ER-α, whereas, in the absence of E2, the complex responsible for the initial remodelling has not been identified. This relocates the nucleosome associated with the TATA box of the pS2 promoter, such that the TATA box lies outside the DNA occluded by the histone core. The additional recruitment of complexes that have HMT and HAT activities then defines a transcriptionally permissive promoter. The achievement of transcription during successive cycles is initiated by ER-α, which induces the sequential recruitment of intermediate transcription factors, then the basal transcription machinery, which in turn recruits and activates Pol II. After initiation, the sequential and ordered recruitment of factors defines the direction of cycling; transcriptional attainment is thus achieved through a transcriptional ratchet that ensures expression of the pS2 gene. Unexpectedly, the presence of certain factors and post-translational modifications, such as the association of TBP and the dimethylation of arginine 3 (R3) in H4, persist over two cycles. Additionally, rearrangement of nucleosome phasing changes at the completion of every double cycle (Fig 1). These events reflect a sequential difference in the clearance phase of alternating transcriptionally productive cycles, in which complete resetting of chromatin organization correlates with removal of TBP. Physiologically, transcriptional cycling achieves the continuous sampling of oestradiol exposure and ensures an appropriate limitation to responsiveness.
Limiting transcription
Periodic limitation of transcription is generated by events that clear the pS2 promoter of transcription factors and induce a restrictive chromatin environment (Métivier et al, 2003). The proteins involved in resetting the pS2 promoter are generally implicated in transcriptional repression, but recent data have questioned whether this is their exclusive role (Ma, 2005). Activation of Pol II induces recruitment of ‘repressive' complexes such as HDACs that direct the termination of each cycle. Three complexes act on the promoter to limit transcription (Fig 1). At the end of all productive cycles, HDACs, in association with the SWI/SNF complex, remodel local chromatin structure such that histone deacetylation restricts transcriptional engagement and the oestrogen response element (ERE) becomes associated with the nucleosome core. At the end of the second cycle, another ATP-dependent remodelling complex, NuRD, repositions the nucleosome associated with the TATA box, resulting in its occlusion and the exclusion of factors such as TBP. Illustrating the concept of the histone code, a transcriptionally engaged pS2 promoter can be defined by the presence of dimethylated H4 R3 and acetylated H3 K14. So far, the enzymes that achieve demethylation of H4 and H3 residues during transcription cycles of the pS2 promoter have not been identified. Candidates are PADI1/PAD4 and LSD1, which deiminate or demethylate the dimethylated residues, respectively (Shi et al, 2004; Wang et al, 2004). Rapid histone replacement through re-deposition, as shown during transcription elongation in yeast (Schwabish & Struhl, 2004), is an as-yet uninvestigated, alternative possibility.
The kinetically appropriate recruitment of E3 ligases and proteasomal components indicates that the degradation of the assembled transcriptionally active complex is also involved in the clearance phase of the pS2 promoter. Moreover, ubiquitylated proteins are found on the pS2 promoter, and inhibition of proteasome degradation abrogates transcription (Reid et al, 2003). Although ubiquitylated ER-α has never been detected on the pS2 promoter, and ER-α degradation and transcriptional activity can be dissociated in certain circumstances (Valley et al, 2005), proteasome function and transcription are inherently linked processes.
Transcription dynamics evaluated by live-cell imaging
Real-time, single live-cell imaging of transcription factors tagged with fluorescent proteins has also illustrated the dynamic nature of transcriptional activation, by showing that NRs are highly mobile in the nucleus (Hager et al, 2004; Maruvada et al, 2003; Rayasam et al, 2005; Schaaf & Cidlowski, 2003; Reid et al, 2003). Imaging of NR-directed transcription has been greatly facilitated by the use of tandem arrays of responsive promoters, which generate a high local concentration of responsive elements that are visible as a discrete locus when associated with labelled proteins (McNally et al, 2000; Tsukamoto et al, 2000). Fluorescence recovery after photobleaching (FRAP) analyses have shown that NRs and interacting cofactors rapidly exchange on these arrays (Becker et al, 2002; Stenoien et al, 2001a). This continuous sampling of responsive promoters suggests that transcription activation is achieved through stochastic mechanisms, generally known as ‘hit-and-run' (McNally et al, 2000); consequently, stochastic, sequential initiation and limitation of transcription are predicted to result from the high mobility of transcription factors and other chromatin-associated proteins (Phair et al, 2004).
Reconciliating ChIP and FRAP data: a model
The apparent discrepancy between rapid events seen with live-cell imaging and longer cycling times determined by ChIP assays arises in part from the different time-scales examined in these experiments. This notwithstanding, the data obtained by ChIP and FRAP are robust. Whereas FRAP experiments mainly detect the bulk, rapid and potentially transient binding of factors, ChIP assays only detect productive associations of promoter sequences with specific transcription factors. Taking into account these limitations, we propose a model (Fig 2) that integrates the high mobility of NR as observed by FRAP with the longer cycle times determined by ChIP.
Figure 2.
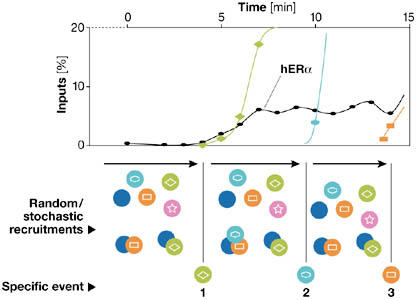
Proposed allosteric, stochastic and dynamic model. This model integrates the general concepts that have emerged from detecting oestrogen receptor-α (ER-α) in live-cell imaging experiments such as fluorescence recovery after photobleaching (FRAP) and those from chromatin immunoprecipitation (ChIP) assays. This model incorporates both stochastic and deterministic concepts into transcriptional attainment. We postulate that transcriptionally productive complexes, which have slower mobility, are rarely formed on promoters. Transcription initiation requires a specific sequence of events to occur, defining a transcriptional ratchet that orientates progression through the cycles. Before that one deterministic event takes place, many rapid stochastic and transient associations of factors occur that are unproductive. It is only when a specific required factor is recruited at the appropriate time that progress is made. Allostery is instrumental in these transitions, as functional, three-dimensional changes are anticipated to occur on all participating partners (namely, proteins, DNA and RNA). Whereas FRAP experiments mainly detect the rapid, unproductive binding of factors, ChIP assays allow the determination of the precise kinetics of the productive associations and of the time required for the transition from one step to another.
We postulate that transcriptionally productive complexes have a slower mobility than transcription factors not engaged on a promoter. In general, and as determined by ChIP assays, initiation of transcription requires specific sequences of events to occur; these are ordered, kinetic and directional. These processes define a transcriptional ratchet that orientates progression through the cycles and is dependent on productive events that occur infrequently from many rapid, stochastic, transient and unproductive associations of factors. On average, many factors rapidly but non-productively associate with a promoter before a deterministic event takes place. Such continuous scanning is essential for transcription and is mirrored in the high mobility seen by FRAP. It is only when a specific and required factor becomes recruited at an appropriate time that progress is made. Allostery is instrumental in these transitions, as functional, three-dimensional changes are anticipated to occur on all participating partners (that is, proteins, DNA and RNA). Specific events that modulate chromatin also orientate the sequence of recruitment and act as a transcriptional ratchet that determines the direction of cycling. After promoter synchronization, kinetic ChIP evaluations determine the average time required for transition from one phase of transcription to another.
This model proposes that transcriptionally productive complexes, with slow mobility, are rarely formed on promoters. In accordance with these principles, functional engagement of a protein results in a restriction of mobility. By inference, the overall high mobility of NRs reflects a low probability that association with a promoter element is functionally productive. Interestingly, there are at least two kinetic components in the rate of recovery after photobleaching. Although some mathematical models indicate that, in some instances, a single-molecule population can generate biphasic FRAP curves (Sprague & McNally, 2005), other interpretations suggest that these components might reflect the existence of a rapid fraction, probably consisting of non-productive, freely diffusing and scanning NR, and a slower component that perhaps reflects productively engaged NR (Phair et al, 2004). In addition, this model implies that some events do not follow a stochastic binding process. For instance, histone modifications define a given promoter state; this acts to ensure the direction and progress between phases of the transcriptional cycle.
In accordance with this model, the interaction of GR with structural chromatin components, such as high mobility group box 1 (HMGB1), slows down the mobility of GR (Agresti et al, 2005). In each period of residence, a bound factor has to recruit another available and required protein. If this partner is not recruited, then the factor dissociates. If it is recruited, then the resulting complex becomes stabilized on the promoter, thereby advancing the cycle and precipitating the next event. Therefore, it should be possible to define the complexes that are present at each step of the cycle by their association and dissociation kinetics. Interestingly, this probabilistic, deterministic model is in accordance with data from Dundr and colleagues (Dundr et al, 2002), who showed that the assembly of the RNA polymerase I transcription complex proceeds in a sequential manner through metastable intermediates created through random ‘collisions' (Vermeulen & Houtsmuller, 2002). Another inference of this model is the distinction of two ‘clearance' mechanisms: one that is inherent to the stochastic recruitment and stimulation of the transcriptional machinery; and a second, cyclical ‘active clearance' inherent to a promoter that has cyclical activity. This distinction is a kinetic distinction not related to the relative energy dependency of each of these mechanisms, as rapid cycling of at least some transcription factors requires ATP (Karpova et al, 2004; Stenoien et al, 2001b).
How is transcriptional cycling initiated? Potentially, transient dissociation of nucleosomes is sufficient to expose an NR-binding site and to provoke subsequent transcriptional regulation. However, it is difficult to envisage how spontaneous dissociation from nucleosomes can create short windows of binding opportunity to allow promoters from a cell population to act synchronously after their release from transcriptional blockade. Alternatively, active clearance allows synchrony, with each ChIP peak representing a mean of stochastic, asynchronous states. The kinetics seen by ChIP therefore reflects the delay required between each state. If spontaneous dissociation of nucleosomes is vital to initiate the system, this also questions the mechanisms of DNA-binding specificity and sequence recognition: are highly mobile NRs continuously scanning the entire genome for adequate binding sequences? ChIPs performed on specific arrays found that ER was associated with large regions of chromosomes 21 and 22, with some located outside E2-dependent gene sequences (Caroll et al, 2005; Laganière et al, 2005). This may be in accordance with a scanning process.
A model for NR-mediated transcriptional activation?
How general is the phenomenon of promoter cycling? In addition to ER-α, detailed kinetic ChIP analysis of the association of transcription factors with a cognate promoter have been reported for AR, TR and VDR (Kang et al, 2002; Sharma & Fondell, 2002; Vaisanen et al, 2005). In each case, and without promoter synchronization using α-amanitin treatment, cyclical recruitment of transcriptionally competent complexes has been observed with a periodicity of 50–80 min. In contrast to AR and ER-α, TR persists on responsive promoters (Sharma & Fondell, 2002) and represses target promoters in the absence of ligand, perhaps indicating that TR-dependent cycles are generated by sequential changes in the transactivation capacity of TR. Other specific mechanistic details of NR-mediated cyclical transcription also exist. For instance, inhibition of the proteasome stimulates GR activity, which is in direct contrast to ER, although both NRs become immobilized on proteasome inhibition (Reid et al, 2003; Wallace & Cidlowski, 2001). Furthermore, on the mouse mammary tumour virus promoter, proteasome activity is required for PR clearance, but it is chaperones that are involved in GR clearance (Freeman & Yamamoto, 2002; Dennis et al, 2005).
Although ER-α-mediated transcriptional cycles have been observed on four gene promoters, a more complete analysis evaluating different types of oestrogen-responsive promoters would address potential correlations between promoter structure, cycling periodicity and cofactor engagement. For example, the presence of many EREs affects ER-α transactivation capacity (Hall et al, 2002). TRAP/Mediator and p160 proteins associate simultaneously on the CATD promoter (Shang et al, 2000), in contrast to the situation found with the pS2 promoter (Métivier et al, 2003), which suggests a diversity in how individual promoters achieve transcription. Additional cognate binding sites for transcription factors on promoters have an impact on the sequence of recruitment. For instance, on the pS2 promoter, Sp proteins and AP1 influence ER-α activity (Barkhem et al, 2002; Sun et al, 2005). It is also probable that the association of the general transcription machinery on TATA− promoters might generate kinetics of association that are different to those depicted for the TATA+ pS2 promoter. It is likely that additional specificity exists at the level of the recruitment of general transcription factors, depending on the architecture of the core promoter (Smale & Kadonaga, 2003).
Concluding remarks
It is evident that, for a limited proportion of promoters at least, transcription is attained by a cyclical progression of generating transcriptional competence, achieving transcription, then limiting this process through removal of the transcriptional machinery and resetting the histone code. Inherent to this progression is the concept of a transcriptional ratchet, in which the general use of post-translational modifications, acting as directional markers in time, orientates and progresses movement through the cycle. These new insights into transcriptional attainment and regulation provide new opportunities in understanding and influencing gene expression. They also offer new challenges, such as studying the kinetic interconnection between transcription initiation and splicing, and RNA maturation, which are processes regulated by NRs (Auboeuf et al, 2002). Finally, an outstanding issue will be to understand how to reconcile these highly dynamic models with other hierarchical regulatory elements of transcription, such as nucleus organization and recently identified transcription factories (Osborne et al, 2004).

References
- Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME (2005) GR and HMGB1 interact only within chromatin and influence each other's residence time. Mol Cell 18: 109–121 [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S (2002) Epigenetic consequences of nucleosome dynamics. Cell 111: 281–284 [DOI] [PubMed] [Google Scholar]
- Asturias FJ (2004) RNA polymerase II structure, and organization of the preinitiation complex. Curr Opin Struct Biol 14: 121–129 [DOI] [PubMed] [Google Scholar]
- Auboeuf D, Honig A, Berget SM, O'Malley BW (2002) Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298: 416–419 [DOI] [PubMed] [Google Scholar]
- Barkhem T, Haldosen LA, Gustafsson JA, Nilsson S (2002) pS2 gene expression in HepG2 cells: complex regulation through crosstalk between the estrogen receptor α, an estrogen-responsive element, and the activator protein 1 response element. Mol Pharmacol 61: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Becker M, Baumann C, John S, Walker DA, Vigneron M, McNally JG, Hager GL (2002) Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep 3: 1188–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B, Parker MG (2003) Nuclear receptors: a rendezvous for chromatin remodelling factors. Cell 114: 277–280 [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA (2004) Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta 1677: 52–57 [DOI] [PubMed] [Google Scholar]
- Brown K (1999) Nuclear structure, gene expression and development. Crit Rev Eukaryot Gene Expr 9: 203–212 [DOI] [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim JH (2005) The dynamics of histone H1 function in chromatin. Mol Cell 17: 617–620 [DOI] [PubMed] [Google Scholar]
- Caroll JS et al. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M (2004) Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol 24: 4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98: 675–686 [DOI] [PubMed] [Google Scholar]
- Claessens F, Gewirth DT (2004) DNA recognition by nuclear receptors. Essays Biochem 40: 59–72 [DOI] [PubMed] [Google Scholar]
- Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE (2002) Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415: 549–553 [DOI] [PubMed] [Google Scholar]
- Dennis AP, Lonard DM, Nawaz Z, O'Malley BW (2005) Inhibition of the 26S proteasome blocks progesterone receptor-dependent transcription through failed recruitment of RNA polymerase II. J Steroid Biochem Mol Biol 94: 337–346 [DOI] [PubMed] [Google Scholar]
- Dillon N, Festenstein R (2002) Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet 18: 252–258 [DOI] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T (2002) A kinetic framework for a mammalian RNA polymerase in vivo. Science 298: 1623–1626 [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA 93: 8329–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR (2002) Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296: 2232–2235 [DOI] [PubMed] [Google Scholar]
- Gill G (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18: 2046–2059 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Hager GL, Nagaich AK, Johnson TA, Walker DA, John S (2004) Dynamics of nuclear receptor movement and transcription. Biochim Biophys Acta 1677: 46–51 [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP, Korach KS (2002) Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol 16: 469–486 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL (2001) Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Hermann A, Gowher H, Jeltsch A (2004) Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci 61: 2571–2587 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kang Z, Pirskanen A, Janne OA, Palvimo JJ (2002) Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J Biol Chem 277: 48366–48371 [DOI] [PubMed] [Google Scholar]
- Karpova TS, Chen TY, Sprague BL, McNally JG (2004) Dynamic interactions of a transcription factor with DNA are accelerated by a chromatin remodeller. EMBO Rep 5: 1064–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorasanizadeh S (2004) The nucleosome: from genomic organization to genomic regulation. Cell 116: 259–272 [DOI] [PubMed] [Google Scholar]
- Klinge CM (2000) Estrogen receptor interaction with coactivators and corepressors. Steroids 65: 227–251 [DOI] [PubMed] [Google Scholar]
- Laganière J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguère V (2005) Location analysis of estrogen receptor a target promoters reveals that FoxA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA 102: 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky RM et al. (1998) Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA 95: 2920–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Bagchi MK (2004) Recruitment of distinct chromatin-modifying complexes by tamoxifen-complexed estrogen receptor at natural target gene promoters in vivo. J Biol Chem 279: 15050–15058 [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW (2005) Expanding functional diversity of the coactivators. Trends Biochem Sci 30: 126–132 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Ma J (2005) Crossing the line between activation and repression. Trends Genet 21: 54–59 [DOI] [PubMed] [Google Scholar]
- Martini PG, Katzenellenbogen BS (2003) Modulation of estrogen receptor activity by selective coregulators. J Steroid Biochem Mol Biol 85: 117–122 [DOI] [PubMed] [Google Scholar]
- Maruvada P, Baumann CT, Hager GL, Yen PM (2003) Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. J Biol Chem 278: 12425–12432 [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BWO (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108: 465–474 [DOI] [PubMed] [Google Scholar]
- McNally JG, Muller WG, Walker D, Wolford R, Hager GL (2000) The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F (2003) Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115: 751–763 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Osborne CS et al. (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Park KJ, Krishnan V, O'Malley BW, Yamamoto Y, Gaynor RB (2005) Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol Cell 18: 71–82 [DOI] [PubMed] [Google Scholar]
- Perkins TJ, Hallett M, Glass L (2004) Inferring models of gene expression dynamics. J Theor Biol 230: 289–299 [DOI] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T (2004) Global nature of dynamic protein–chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol 24: 6393–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam GV, Elbi C, Walker DA, Wolford R, Fletcher TM, Edwards DP, Hager GL (2005) Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodelling in vitro. Mol Cell Biol 25: 2406–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Hübner MR, Métivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F (2003) Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell 11: 695–707 [DOI] [PubMed] [Google Scholar]
- Ren Y, Behre E, Ren Z, Zhang J, Wang Q, Fondell JD (2000) Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol Cell Biol 20: 5433–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Escriva Garcia H, Laudet V (2003) The nuclear receptor superfamily. J Cell Sci 116: 585–586 [DOI] [PubMed] [Google Scholar]
- Schaaf MJ, Cidlowski JA (2003) Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol Cell Biol 23: 1922–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K (2004) Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843–852 [DOI] [PubMed] [Google Scholar]
- Sharma D, Fondell JD (2002) Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci USA 99: 7934–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Sif S (2004) ATP-dependent nucleosome remodelling complexes: enzymes tailored to deal with chromatin. J Cell Biochem 91: 1087–1098 [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT (2003) The RNA polymerase II core promoter. Annu Rev Biochem 72: 449–459 [DOI] [PubMed] [Google Scholar]
- Sprague BL, McNally JG (2005) FRAP analysis of binding: proper and fitting. Trends Cell Biol 15: 84–91 [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Nye AC, Mancini MG, Patel K, Dutertre M, O'Malley BW, Smith CL, Belmont AS, Mancini MA (2001a) Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor α–coactivator complexes in living cells. Mol Cell Biol 21: 4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O'Malley BW, Mancini MA (2001b) FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat Cell Biol 3: 15–23 [DOI] [PubMed] [Google Scholar]
- Sun JM, Spencer VA, Li L, Yu Chen H, Yu J, Davie JR (2005) Estrogen regulation of trefoil factor 1 expression by estrogen receptor α and Sp proteins. Exp Cell Res 302: 96–107 [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL (2000) Visualization of gene activity in living cells. Nat Cell Biol 2: 871–878 [DOI] [PubMed] [Google Scholar]
- Vaisanen S, Dunlop TW, Sinkkonen L, Frank C, Carlberg C (2005) Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1α,25-dihydroxyvitamin D3. J Mol Biol 350: 65–77 [DOI] [PubMed] [Google Scholar]
- Valley CC, Métivier R, Solodin NM, Fowler AM, Mashek MT, Hill L, Alarid ET (2005) Differential regulation of estrogen-inducible proteolysis and transcription by the estrogen receptor α N terminus. Mol Cell Biol 13: 5417–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel R, Fransz PF, Verschure PJ (2003) The eukaryotic genome: a system regulated at different hierarchical levels. J Cell Sci 116: 4067–4075 [DOI] [PubMed] [Google Scholar]
- Vermeulen W, Houtsmuller AB (2002) The transcription cycle in vivo. A blind watchmaker at work. Mol Cell 10: 1264–1266 [DOI] [PubMed] [Google Scholar]
- Wallace AD, Cidlowski JA (2001) Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem 276: 42714–42721 [DOI] [PubMed] [Google Scholar]
- Wang C et al. (2001) Direct acetylation of the estrogen receptor α hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem 276: 18375–18383 [DOI] [PubMed] [Google Scholar]
- Wang H et al. (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293: 853–857 [DOI] [PubMed] [Google Scholar]
- Wang Y et al. (2004) Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306: 279–283 [DOI] [PubMed] [Google Scholar]
- Warnmark A, Wikstrom A, Wright AP, Gustafsson JA, Hard T (2001) The N-terminal regions of estrogen receptor α and β are unstructured in vitro and show different TBP binding properties. J Biol Chem 276: 45939–45944 [DOI] [PubMed] [Google Scholar]
- White JH, Fernandes I, Mader S, Yang XJ (2004) Corepressor recruitment by agonist-bound nuclear receptors. Vitam Horm 68: 123–143 [DOI] [PubMed] [Google Scholar]


