Abstract
Two key features of RNA viruses are their compacted genomes and their high mutation rate. Accordingly, deleterious mutations are common and have an enormous impact on viral fitness. In their multicellular hosts, robustness can be achieved by genomic redundancy, including gene duplication, diploidy, alternative metabolic pathways and biochemical buffering mechanisms. However, here we review evidence suggesting that during RNA virus evolution, alternative robustness mechanisms may have been selected. After briefly describing how genetic robustness can be quantified, we discuss mechanisms of intrinsic robustness arising as consequences of RNA-genome architecture, replication peculiarities and quasi-species population dynamics. These intrinsic robustness mechanisms operate efficiently at the population level, despite the mutational sensitivity shown by individual genomes. Finally, we discuss the possibility that viruses might exploit cellular buffering mechanisms for their own benefit, producing a sort of extrinsic robustness.
Keywords: fitness, deleterious mutations, quasi-species, genetic robustness, virus evolution
Introduction
RNA viruses have the highest mutation rate among living species (that is, between 10−3 and 10−5 errors per nucleotide and replication cycle), very small and compacted genomes, short generation times and extremely large populations (Domingo & Holland, 1997). This might be beneficial in the long-term, as it allows viral populations to quickly explore genotypic space and find beneficial mutations. However, it is clearly detrimental in the short-term as most mutations have deleterious fitness effects. The balance between the continuous generation of mutants and the action of selection leads to a dynamic population structure, known as ‘quasi-species' (Domingo & Holland, 1997).
In recent years, the interest of evolutionary biologists in the mechanisms, consequences and evolution of genetic robustness has been revitalized by new and powerful techniques that allow the tracking and manipulation of genotypes (de Visser et al, 2003). Robustness is defined as a reduced sensitivity to perturbations affecting phenotypic expression. If perturbations are inheritable, then we talk about genetic robustness; if they are not (for example, changes in physical and chemical parameters, or developmental noise), then we talk about environmental robustness. Robustness should occur when there are several copies of a single gene, when several genes contribute to the same function or through biochemical buffering mechanisms. This includes gene duplication, polyploidy, alternative metabolic pathways or chaperone proteins. As illustrated in Fig 1A, a lack of robustness is expected in haploid genomes that have no duplications, overlapping gene functions, repair systems and arepleiotropic. A small number of mutations can produce a strong effect, but as mutations accumulate, they affect the same function with increasing probability and, thus, their marginal contribution to fitness diminishes. Hence, the observed fitness is above the expected multiplicative value or, in other words, epistasis is antagonistic (Wolf et al, 2000). By contrast, in the presence of redundancy and buffering mechanisms, the fitness of genomes is only mildly affected; however, as the mutation load increases, these mechanisms ultimately collapse. Fitness will therefore be lower than the expected multiplicative value, which means that there will be synergistic epistasis (Fig 1B).
Figure 1.
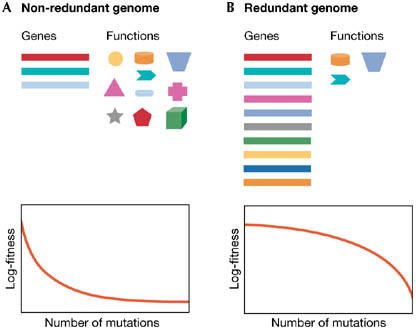
Hypothetical genetic systems. (A) Non-robust genomes should have fewer genes than functions; single mutations have strong fitness effects and positive epistasis. (B) Robust genomes should have more genes than functions; single mutations have mild fitness effects and show negative epistasis.
In principle, genetic robustness might evolve for one of the following reasons. First, as long as robustness has a heritable basis, shows variability among individuals and affects the probability of survival, it can be a target for selection and evolutionary optimization (Wilke & Adami, 2003). The selection pressure for increasing robustness depends on the occurrence of mutations. The more frequent mutations are, the more efficient selection will be at promoting the evolution of robustness. Second, it might evolve because buffering is a necessary consequence of character adaptation; that is, robustness is a side-effect of stabilizing selection acting on different traits (Meiklejohn & Hartl, 2002). Third, given that environmental fluctuations often have a strong impact on fitness, selection would efficiently favour mechanisms of environmental robustness. On the basis of theoretical arguments and RNA folding simulations, some authors have predicted that genetic robustness should be intrinsically correlated to environmental robustness and, thus, that the former could evolve as a correlated response to selection favouring the latter (Ancel & Fontana, 2000; Wagner et al, 1997). This is an appealing hypothesis because, during their life cycle, RNA viruses must cope not only with the deleterious effect of mutations but also with dramatic and fast fluctuations in their environments such as alternating among host species, tissue- and organ-specific microenvironments or the presence of antiviral agents.
Measuring genetic robustness
Although it might be interesting to measure mutational robustness in specific biochemical pathways, gene expression patterns or developmental plans, it is more informative, from an evolutionary standpoint, to determine the effects of mutations on fitness. To quantify the fitness effects of a given mutation, i, population geneticists make use of the selection coefficient si = Wi − 1, in which Wi is the relative fitness of the mutant; that is, the number of descendants per wild-type descendant. Most mutations are deleterious and thus the average selection coefficient, s̄ is negative. Whenever information about the average fitness effects of mutations is available, s̄ can be seen as a measure of mutational sensitivity and, henceforth, as an inverse approximation to robustness.
A simple experimental approach to estimate s̄ is the Bateman–Mukai mutation-accumulation (MA) method (Mukai, 1964). Such experiments consist of the parallel accumulation of spontaneous mutations in which the effect of natural selection is minimized—for example, by population bottlenecks or by endogamic crosses. The main limitation of this kind of experiment is that highly unviable or lethal mutations cannot accumulate, and therefore the average effects of mutations tend to be underestimated. This drawback can be minimized by a mutagenesis approach, using radiation, chemical treatments, transposons, proofreading-defective polymerases or site-directed mutagenesis.
RNA virus hypersensitivity to deleterious mutations
During the past decade, virologists have established that RNA viruses are very sensitive to the effect of deleterious mutations. In a pioneering study, Chao (1990) performed an MA experiment by imposing dramatic population bottlenecks on the phage φ6. After the bottlenecks took effect, the average fitness of φ6 was reduced by 22%. This experiment was followed by others under similar demographic circumstances: vesicular stomatitis (VSV), foot and mouth disease (FMDV) and human immunodeficiency type 1 (HIV-1) viruses suffered fitness losses of 38%, 35% and 82%, respectively (Duarte et al, 1992; Escarmís et al, 1996; Yuste et al, 1999), and bacteriophage MS2 lost as much as 16% fitness in a single bottleneck event (de la Peña et al, 2000). The rate of log-fitness decay equals U × s̄ (where U is the genomic mutation rate) and, therefore, high fitness losses can be explained by a high U-value. However, when Escarmís et al (1996) obtained consensus sequences for the bottlenecked FMDV populations, only a few mutations were fixed in each lineage, suggesting that each one imposed a strong fitness burden. A similar conclusion was obtained for HIV-1 (Yuste et al, 2000). Indeed, there are several strategies for disentangling the contributions of U and s̄ to the rate of fitness decline, such as computing the among-lines variance in MA experiments, sequencing the MA lines, or undertaking a mutagenesis experiment. Using site-directed mutagenesis, Sanjuán et al (2004a) showed that up to 40% of random single-nucleotide substitutions in VSV were lethal and that, for non-lethal mutations, fitness reductions were as high as 46% (supplementary Table S1 online).
After an extensive review of the literature on mutational effects (supplementary Table S1 online), the median selection coefficient against single mutations across different RNA viruses has been calculated to ∼10.8%. This high sensitivity to mutation contrasts sharply with the estimates obtained for DNA organisms, in which many mutations are believed to be neutral (Conant & Wagner, 2004; Davies et al, 1999; Steinmetz et al, 2002) and a review of available data on mutational fitness effects in these organisms yields a median deleterious mutational effect across species of ∼1.7% per generation (supplementary Table S2 online). Therefore, the data support the idea that RNA viruses are genetically unprotected against mutation, possibly due to the compactness of their genomes, with frequently overlapping functional regions and few alternative pathways.
Although s̄ provides important information about genetic robustness, it might not reflect the complete picture because when several mutations are present simultaneously in the same genotype, they might interact to determine fitness. As we hypothesized above, synergistic epistasis would be diagnostic for robustness mechanisms, whereas antagonistic epistasis would be characteristic of hypersensitive genomes. Although reports of synergistic epistasis in eukaryotes exist (de Visser et al, 1996; Mukai, 1969; Whitlock & Bourguet, 2000), others failed to detect a general trend in prokaryotes and eukaryotes (de Visser & Hoekstra, 1998; Elena & Lenski, 1997). Among RNA viruses, no evidence for epistasis was obtained from MA experiments with phage MS2 (de la Peña et al, 2000), FMDV (Elena, 1999) and poliovirus (Burch & Chao, 2004), although in these studies the number and type of the mutations accumulated was unknown and the statistical power was probably too low. However, more recent and exhaustive MA and site-directed mutagenesis experiments carried out with φ6 (Burch & Chao, 2004), VSV (Sanjuán et al, 2004b) and HIV-1 (Bonhoeffer et al, 2004) have revealed a tendency towards antagonistic epistasis.
For loci to be epistatic, they must be involved in the expression of the same trait. Whether this produces antagonistic or synergistic epistasis depends on the way these loci participate in the expression of the character. If the contribution of several loci is necessary, then epistasis will be antagonistic, as the effect of a mutation at one locus is the same as mutations at several loci; this is the case when the loci involved belong to the same gene. Antagonistic epistasis should therefore be characteristic of small and compact genomes. Finally, in RNA genomes, there is a strong selection pressure for maintaining secondary structures, and compensatory mutations can occur relatively often (Wilke et al, 2003). Both features might explain the tendency of RNA viruses to display antagonistic epistasis.
Population mechanisms of intrinsic viral robustness
RNA viruses show hypersensitivity to mutational effects, but the question remains: how do they face deleterious mutations? Although it might sound paradoxical, individual mutational hypersensitivity produces robustness at the population level. The efficiency of natural selection to purge deleterious alleles from a population, or more generally, the relative contribution of selection and genetic drift to evolutionary change, depends on the product N × s̄ (where N is the population size). The efficiency of natural selection increases at high N provided that ecological resources are limited. In this scenario, a form of soft selection operates through the differential survival and associated competition among individuals. This is likely to be the case in RNA viruses. Usually, enormous values of N are reached even in a single infected host and many more viral particles are produced than available cells. The contribution of large N acts in positive synergy with the above-mentioned large s̄, making selection remarkably efficient. Indeed, Krakauer & Plotkin (2002) showed that lineages existing as very large N-values might become more robust at the population level, whereas individual sensitivity to mutation is increased. This anti-redundant strategy relies on the efficient elimination of mutated genotypes and the preservation of the non-mutated genotype.
Population genetics theory predicts that the optimal evolutionary mutation rates are largely determined by s̄ (Johnson & Barton, 2002; Orr, 2000) and thus, the mutational hypersensitivity shown by RNA viruses might allow them to tolerate particularly high mutation rates. This interpretation provides an evolutionary basis for the characteristically high mutation rates observed in RNA viruses but apparently fails to account for the concomitantly high variability of these systems. However, in good agreement with the anti-redundancy strategy, increasing evidence suggests that much of this variation is rapidly purged from populations and that only a few beneficial changes are responsible for adaptation (Novella et al, 1999; Rodríguez et al, 2000).
Opposite to the anti-redundancy strategy is the idea of ‘survival of the flattest', originally postulated by Schuster & Swetina (1988). Standard population genetics theory has shown that, under some simplifying assumptions, the average fitness of a population equals exp(−U) (Kimura & Maruyama, 1966). However, when neutral and back mutations are considered, the population average equilibrium fitness also depends on the geometry of the fitness landscape, which, at a first instance, can be described by s̄ (Wilke & Adami, 2003). This being the case, another selective pressure comes into play at high values of U that pushes populations towards regions of the landscape where the density of neutral mutations is higher (Wilke, 2001). As a consequence, the whole population evolves increased robustness against mutations. This phenomenon of selection for robustness at high values of U can be understood as a pressure for populations to occupy either highly connected areas on a neutral network or broad rather than narrow fitness peaks; populations occupying high but narrow peaks are easily mutated to have low fitness, whereas populations living in low but broad peaks are less susceptible to mutations (Fig 2). Despite the fact that the effect of survival of the flattest has been observed in silico (Wilke et al, 2001), and has been supported by some in vitro observations (Burch & Chao, 2000), definitive experimental proof for its relevance is still missing.
Figure 2.
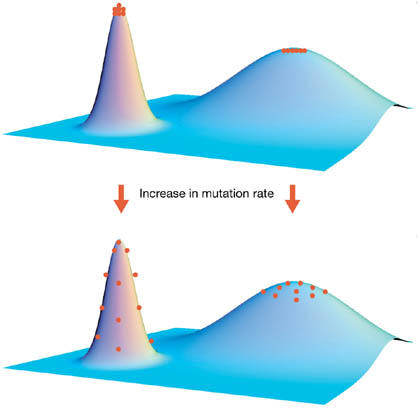
Schematic representation of a landscape characterized by a peak of high fitness but low robustness and another one of low fitness but high neutrality (that is, robustness). At a high mutation rate, populations at the high peak are pushed down, whereas those at the low peak remain unchanged. Adapted from Wilke & Adami (2003) with permission from Elsevier.
A common misconception is that most RNA viruses are haploid. However, it is more convenient to consider viruses as n-ploid systems, n being variable during the infectious cycle. At initial stages, the multiplicity of infection (infectious particles per available cell) is low and, consequently, viruses are effectively haploid. However, as the host is invaded, high multiplicities of infection ensure frequent co-infection events and increasing ploidy. An immediate consequence of polyploidy is genetic complementation. If all co-existing genomes in an infected cell contribute to the same protein pool, and provided that viral proteins diffuse freely in the cytoplasm, mutant genomes can use proteins in which they are deficient as long as they are produced by a co-existing genome. In agreement with this idea, the maintenance of lethal mutations by complementation has been a common observation in animal, plant and bacterial viruses (Cicin-Sain et al, 2005; Moreno et al, 1997; Roux et al, 1991). For example, recent experiments have shown that whenever the probability of co-infection is maximized, low-fitness mutant VSV genotypes can co-exist with the wild-type virus in a frequency-dependent manner (Wilke et al, 2004).
Another population robustness mechanism that might be important for RNA viruses is sex, as it results in not only recombination between homologous molecules, but also the segregation of segments in a multipartite genome. Sex recreates mutation-free genotypes and helps to keep the average population fitness high (Otto & Lenormand, 2002). Both forms of sex are common among RNA viruses (Lai, 1992). The fitness advantages associated with recombination and segregation are amply illustrated. Fernández-Cuartero et al (1994) found that a recombinant RNA 3 created from cucumber mosaic and tomato aspermy viruses outcompeted both parental species. Chao et al (1997) found that populations of the tripartite φ6, which were previously subjected to MA (Chao, 1990), recovered fitness through compensatory mutations. However, hybrids produced by crossing the populations recovered fitness to a greater extent, illustrating the positive effect of segregation.
Mechanisms of extrinsic robustness: cellular chaperones
Heat-shock proteins (HSPs) are a set of highly conserved proteins, the expression of which is induced by a variety of cellular stresses; however, most of them also have essential functions under normal physiological conditions. Chaperones are a specific subset of the HSPs (Hartl et al, 1994). They belong to structurally unrelated protein families that share the ability to recognize and bind to aberrant protein conformations, such as solvent-exposed hydrophobic domains. Under normal physiological conditions, chaperones function in different ways: by preventing nascent polypeptide chains from forming aberrant associations before the remainder of the protein has emerged from the ribosome; by assisting the entrance of newly synthesized polypeptides into organelles; or by masking the exposed surfaces of oligomeric proteins before they assemble with their partners. Under stressful conditions, chaperones prevent misfolding and aggregation, or can even restore the proper conformation of proteins and actively disaggregate damaged ones. Mutations may lead to the destabilization of proteins and increase their tendency to misfold and aggregate. Chaperones can keep mutationally altered proteins functional and thereby buffer detrimental mutations (Fares et al, 2002).
It is well known that viral infections induce the cellular stress response. Indeed, increases in Hsp70 chaperone levels after viral infection have been widely observed (Mayer, 2005). Furthermore, during viral infection of animals, host chaperones that bind to viral particles can act as flags on the cellular surface, triggering the immune response (Oglesbee et al, 2002). However, is it possible that viruses also use chaperones for their own benefit to buffer mutational effects? It has been shown that most viruses need cellular chaperones during their life cycle both to solve their own protein-folding problems as well as to interfere with cellular processes such as signal transduction (Mayer, 2005). Table 1 provides examples of the interaction between cellular chaperones and viral factors at different stages of the infectious cycle. The fact that mutant misfolded viral proteins elicit the expression of chaperones has been illustrated by Jockusch et al (2001). They showed that the presence of tobacco mosaic virus mutant misfolded coat proteins (CPs) triggers the overexpression of Hsp70, with the intensity of the response proportional to the number of mutant CPs. By contrast, the presence of wild-type CPs does not elicit such a response. Moreover, perhaps the most convincing evidence of the importance of chaperones during the virus life cycle is the existence of virus-encoded HSP-like proteins (Table 1, beet yellow closterovirus Hsp70h). Taken together, these observations indicate that viruses might benefit from cellular chaperones as an extrinsic robustness mechanism.
Table 1.
Phases of the viral infectious cycle in which chaperones have a role
| Phase of the infectious cycle | Examples | HSP involved | Viral factor | Reference |
|---|---|---|---|---|
| Cell entry | Rotavirus | Hsc70 | VP5 coat protein | Guerrero et al (2002) |
| Human T lymphotropic type 1 | Hsc70 | gp46 | Fang et al (1999) | |
| Uncoating | Adenovirus | Hsp70 | Hexon (major coat protein) | Niewiarowska et al (1992) |
| Hsc70 + Bag3 | Penton (fibre protein) | |||
| Replication | Human hepatitis B | Hsp60 | Polymerase | Park et al (2003) |
| λ-phage | DnaK, DnaJ, GrpE | λP, λO, oriλ | Alfano & McMacken (1989) | |
| Human papillomavirus 11 | Hsp70, Hdj1, Hdj2 | DNA helicase E1 | Liu et al (1998) | |
| Gene expression | Flock house | Hsp60 | RNA-replication complex assembly | Kampmueller & Miller (2005) |
| Tobacco mosaic | Hsp101 | 5′ leader W sequence | Gallie (2002) | |
| Encapsidation | Citrus tristeza | Virus-encoded Hsp70h | Major and minor coat proteins, p61 protein | Satyanarayana et al (2004) |
| Polyomavirus | Hsc70 | VP1, VP2, VP3 | Cripe et al (1995) | |
| Cytopathic effect and release | Measles | Hsp72 | Nucleocapside | Vasconcelos et al (1998) |
| Cell-to-cell movement | Potato X, barley stripe mosaic | Beet yellow closterovirus Hsp70h | Movement protein | Agranovsky et al (1998) |
Hsc, heat-shock cognate; Hsp, heat-shock protein; VP, viral protein.
Concluding remarks
Far from being passive victims of their error-prone replication, RNA viruses cope with the deleterious effects associated with an excess of mutations. In sharp contrast to their cellular hosts, these buffering mechanisms are generally not encoded in their genomes, but are rather a consequence of their population dynamics and parasitic lifestyle. Unravelling the mechanisms of robustness in RNA viruses is a promising research avenue for understanding their survival and evolution.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material

Acknowledgments
S.F.E. is supported by grants from the Spanish Ministerio de Educación y Ciencia (MEC-FEDER), the Generalitat Valenciana, and the EMBO Young Investigator Program. J.-A.D. is supported by grants from MEC-FEDER and the Generalitat Valenciana. R.S. is contracted under the Consejo Superior de Investigaciones Científicas I3P postdoctoral programme.
References
- Agranovsky AA, Folimonov AS, Folimonova SY, Morozov SY, Schiemann J, Lesemann D, Atabekov JG (1998) Beet yellows closterovirus HSP70-like protein mediates the cell-to-cell movement of a potexvirus transport-deficient mutant and a hordeivirus-based chimeric virus. J Gen Virol 79: 889–895 [DOI] [PubMed] [Google Scholar]
- Alfano C, McMacken R (1989) Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of phage lambda DNA replication. J Biol Chem 264: 10709–10718 [PubMed] [Google Scholar]
- Ancel LW, Fontana W (2000) Plasticity, evolvability, and modularity in RNA. J Exp Zool 288: 242–283 [DOI] [PubMed] [Google Scholar]
- Bonhoeffer S, Chappey C, Parkin NT, Whitcomb JM, Petropoulos CJ (2004) Evidence for positive epistasis in HIV-1. Science 306: 1547–1550 [DOI] [PubMed] [Google Scholar]
- Burch CL, Chao L (2000) Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature 406: 625–628 [DOI] [PubMed] [Google Scholar]
- Burch CL, Chao L (2004) Epistasis and its relationship to canalization in the RNA virus φ6. Genetics 167: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L (1990) Fitness of RNA virus decreased by Muller's ratchet. Nature 348: 454–455 [DOI] [PubMed] [Google Scholar]
- Chao L, Tran TT, Tran TT (1997) The advantage of sex in the RNA virus φ6. Genetics 147: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicin-Sain L, Podlech J, Messerle M, Reddehase MJ, Koszinowski UH (2005) Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected hosts. J Virol 79: 9492–9502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wagner A (2004) Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc R Soc Lond B 271: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripe TP, Delos SE, Estes PA, Garcea RL (1995) In vivo and in vitro association of hsc70 with polyomavirus capsid proteins. J Virol 69: 7807–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies EK, Peters AD, Keightley PD (1999) High frequency of cryptic deleterious mutations in Caenorhabditis elegans. Science 285: 1748–1751 [DOI] [PubMed] [Google Scholar]
- de la Peña M, Elena SF, Moya A (2000) Effect of deleterious mutation-accumulation on the fitness of RNA phage MS2. Evolution 54: 686–691 [DOI] [PubMed] [Google Scholar]
- de Visser JAGM, Hoekstra RF (1998) Synergistic epistasis between loci affecting fitness: evidence in plants and fungi. Genet Res Camb 71: 39–49 [Google Scholar]
- de Visser JAGM, Hoekstra RF, van den Ende H (1996) The effect of sex and deleterious mutations on fitness in Chlamydomonas. Proc R Soc Lond B 263: 193–200 [Google Scholar]
- de Visser JAGM et al. (2003) Evolution and detection of genetic robustness. Evolution 57: 1959–1972 [DOI] [PubMed] [Google Scholar]
- Domingo E, Holland JJ (1997) RNA virus mutations and fitness for survival. Annu Rev Microbiol 51: 151–178 [DOI] [PubMed] [Google Scholar]
- Duarte EA, Clarke DK, Moya A, Domingo E, Holland JJ (1992) Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc Natl Acad Sci USA 89: 6015–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF (1999) Little evidence for synergism among deleterious mutations in a nonsegmented RNA virus. J Mol Evol 49: 703–707 [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE (1997) Test of synergistic interactions among deleterious mutations in bacteria. Nature 390: 395–398 [DOI] [PubMed] [Google Scholar]
- Escarmís C, Dávila M, Charpentier N, Bracho MA, Moya A, Domingo E (1996) Genetic lesions associated with Muller's ratchet in an RNA virus. J Mol Biol 264: 255–267 [DOI] [PubMed] [Google Scholar]
- Fang D, Haraguchi Y, Jinno A, Soda Y, Shimizu N, Hoshino H (1999) Heat shock cognate protein 70 is a cell fusion-enhancing factor but not an entry factor for human T-cell lymphotrophic virus type I. Biochem Biophys Res Commun 261: 357–363 [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E (2002) Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature 417: 398. [DOI] [PubMed] [Google Scholar]
- Fernández-Cuartero B, Burgyán J, Aranda MA, Salánki K, Moriones E, García-Arenal F (1994) Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology 203: 373–377 [DOI] [PubMed] [Google Scholar]
- Gallie DR (2002) The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res 30: 3401–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero CA, Bouyssounade D, Zarate S, Isa P, López T, Espinosa R, Romero P, Méndez E, López S, Aras CF (2002) Heat shock cognate protein 70 is involved in rotavirus cell entry. J Virol 76: 4096–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hlodan R, Langer T (1994) Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem Sci 19: 20–25 [DOI] [PubMed] [Google Scholar]
- Jockusch H, Wiegand C, Mersch B, Rajes D (2001) Mutants of tobacco mosaic virus with temperature-sensitive coat proteins induces heat shock response in tobacco leaves. Mol Plant Microbe Interact 14: 914–917 [DOI] [PubMed] [Google Scholar]
- Johnson T, Barton NH (2002) The effect of deleterious alleles on adaptation in asexual populations. Genetics 162: 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmueller KM, Miller DJ (2005) The cellular chaperone heat shock protein 90 facilitates flock house virus RNA replication in Drosophila cells. J Virol 79: 6827–6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Maruyama T (1966) The mutational load with epistatic gene interactions in fitness. Genetics 54: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer DC, Plotkin JB (2002) Redundancy, antiredundancy, and the robustness of genomes. Proc Natl Acad Sci USA 99: 1405–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MMC (1992) RNA recombination in animal and plant viruses. Microbiol Rev 56: 61–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, Makhov AM, Cyr DM, Griffith JD, Broker TR, Chow LT (1998) Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem 273: 30704–30710 [DOI] [PubMed] [Google Scholar]
- Mayer MP (2005) Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol 153: 1–46 [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL (2002) A single mode of canalization. Trends Ecol Evol 17: 468–473 [Google Scholar]
- Moreno IM, Malpica JM, Rodríguez-Cerezo E, García-Arenal F (1997) A mutation in tomato aspermy cucumovirus that abolishes cell-to-cell movement is maintained to high levels in the viral population by complementation. J Virol 71: 9157–9162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T (1964) The genetic structure of natural populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 50: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T (1969) The genetic structure of natural populations of Drosophila melanogaster. VII. Synergistic interaction of spontaneous mutant polygenes controlling viability. Genetics 61: 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiarowska J, D'Halluin JC, Belin MT (1992) Adenovirus capsid proteins interact with HSP70 proteins after penetration in human or rodent cells. Exp Cell Res 201: 408–416 [DOI] [PubMed] [Google Scholar]
- Novella IS, Hershey CL, Escarmis C, Domingo E, Holland JJ (1999) Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J Mol Biol 287: 459–465 [DOI] [PubMed] [Google Scholar]
- Oglesbee MJ, Pratt M, Carsillo T (2002) Role for heat shock proteins in the immune response to measles virus infection. Viral Immunol 15: 399–416 [DOI] [PubMed] [Google Scholar]
- Orr HA (2000) The rate of adaptation in asexuals. Genetics 155: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Lenormand T (2002) Resolving the paradox of sex and recombination. Nat Rev Genet 3: 252–261 [DOI] [PubMed] [Google Scholar]
- Park SG, Lee SM, Jung G (2003) Antisense oligodeoxynucleotides targeted against molecular chaperonin Hsp60 block human hepatitis B virus replication. J Biol Chem 278: 39851–39857 [DOI] [PubMed] [Google Scholar]
- Rodríguez LL, Bunch TA, Fraire M, Llewellyn ZN (2000) Re-emergence of vesicular stomatitis in the western United States is associated with distinct viral genetic lineages. Virology 271: 171–181 [DOI] [PubMed] [Google Scholar]
- Roux L, Simon AE, Holland JJ (1991) Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res 40: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R, Moya A, Elena SF (2004) The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci USA 101: 8396–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R, Moya A, Elena SF (2004) The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci USA 101: 15376–15379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana T, Gowda S, Ayllon MA, Dawson WO (2004) Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proc Natl Acad Sci USA 101: 799–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster P, Swetina J (1988) Stationary mutant distributions and evolutionary optimization. Bull Math Biol 50: 635–660 [DOI] [PubMed] [Google Scholar]
- Steinmetz LM et al. (2002) Systematic screen for human disease genes in yeast. Nat Genet 31: 400–404 [DOI] [PubMed] [Google Scholar]
- Vasconcelos DY, Cai XH, Oglesbee MJ (1998) Constitutive overexpression of the major inducible 70 kDa heat shock protein mediates large plaque formation by measles virus. J Gen Virol 79: 2239–2247 [DOI] [PubMed] [Google Scholar]
- Wagner WP, Booth G, Bagheri-Chaichian H (1997) A population genetic theory of canalization. Evolution 51: 329–347 [DOI] [PubMed] [Google Scholar]
- Whitlock MC, Bourguet D (2000) Factors affecting the genetic load in Drosophila: synergistic epistasis and correlations among fitness components. Evolution 54: 1654–1660 [DOI] [PubMed] [Google Scholar]
- Wilke CO (2001) Adaptive evolution on neutral networks. Bull Math Biol 63: 715–730 [DOI] [PubMed] [Google Scholar]
- Wilke CO, Adami C (2003) Evolution of mutational robustness. Mutat Res 522: 3–11 [DOI] [PubMed] [Google Scholar]
- Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C (2001) Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 412: 331–333 [DOI] [PubMed] [Google Scholar]
- Wilke CO, Lenski RE, Adami C (2003) Compensatory mutations cause excess of antagonistic epistasis in RNA secondary structure folding. BMC Evol Biol 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CO, Reissing DD, Novella IS (2004) Replication at periodically changing multiplicity of infection promotes stable coexistence of competing viral populations. Evolution 58: 900–905 [DOI] [PubMed] [Google Scholar]
- Wolf JB, Brodie III ED, Wade MJ (2000) Epistasis and the Evolutionary Process. Oxford, UK: Oxford University Press [Google Scholar]
- Yuste E, Sánchez-Palomino S, Casado C, Domingo E, López-Galíndez C (1999) Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J Virol 73: 2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste E, López-Galíndez C, Domingo E (2000) Unusual distribution of mutations associated with serial bottleneck passages of human immunodeficiency virus type-1. J Virol 74: 9546–9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


