Abstract
Biological clocks and circadian timing in cells
All living organisms have time-measuring devices that affect their development, generation time, lifespan and lifestyle. These biological timers can be categorized into hourglasses and oscillators; the latter can be further classified as ultradian, circadian and infradian. Most physiological processes in mammals are influenced by a complex circadian timing system in which the master pacemaker in the brain synchronizes numerous subsidiary oscillators in peripheral cells. The phase of the master clock—located in the suprachiasmatic nucleus (SCN) of the ventral hypothalamus—must be readjusted every day by light cues to stay tuned to the actual geophysical time. In both SCN neurons and peripheral cells, the circadian clockwork is constructed from interconnected feedback loops in gene expression that function in a cell-autonomous fashion. A variety of output pathways translate these gene-expression cycles into physiological and behavioural rhythms.
Experiments on a variety of organisms have shown that their maximal lifespan is determined by an hourglass timer (Fig 1, left panel), which measures different timespans in each species and is further modulated by the genetic makeup and lifestyle of the individual. Speciesspecific hourglass clocks also determine the duration of embryonic development, the timespan required to reach sexual maturity and the time at which a woman reaches menopause. Hourglass mechanisms thus determine the timing of biological events that happen only once in a lifetime—unless the hourglass is flipped around by a mystical force, a process that adherents of the Hindu religion believe occurs after death.
Figure 1.
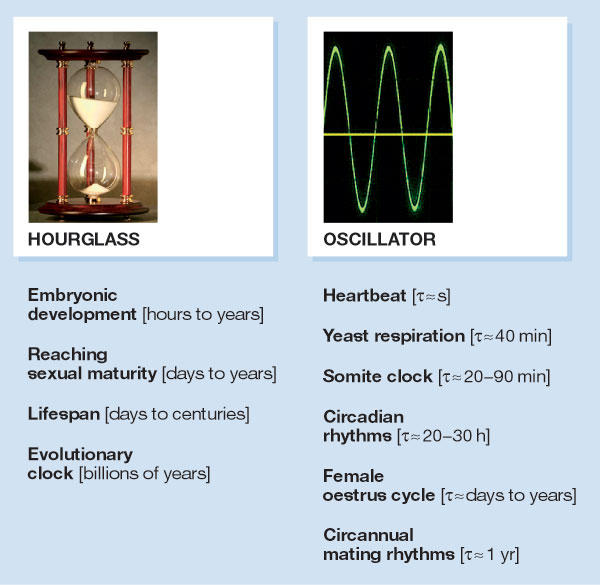
Two types of biological clock. Biological clocks that determine the duration of unique events for an organism or populations of organisms are termed hourglass timers. Examples of such processes are indicated below the hourglass. Oscillators are designed to drive reiterative processes, of which a few examples are listed below the picture. Yeast respiration is controlled by an oscillator that generates 40-min cycles in oxygen consumption. This oscillator also governs global gene transcription and gates DNA replication to a particular time window in the cycle. Somite clocks govern the timing of segmentation during vertebrate embryogenesis. Heartbeat rhythms, yeast respiration cycles and somite deposition rhythms are controlled by ultradian clocks (period length shorter than 20 h), whereas oestrus cycles and circannual mating rhythms are driven by infradian clocks (period length longer than 30 h; Gachon et al, 2004b).
Biological oscillators, conversely (Fig 1, right panel), repeat a given process in the absence of magical hands; they are designed to ensure that a cycle repeats itself once it is completed. If the amplitude of the fluctuation remains more or less constant, the under-lying timer is a 'selfsustained oscillator'; if the amplitude progressively decreases with each consecutive cycle, it is a 'damped oscillator'. Both self-sustained and damped oscillators have important roles in biological timing, and self-sustained oscillators can, in theory, be built by connecting multiple damped oscillators (Roenneberg & Merrow, 2002). These biological oscillators are classified as circadian when their period length is between 20 and 30 hours—circa diem means about a day—ultradian when the period is shorter than 20 hours, and infradian when it is longer than 30 hours. Obviously, this categorization is arbitrary as ultradian oscillators can generate periods between milliseconds and hours, and infradian oscillators can produce periods that range from days to years.
Not all periodic processes in organisms are driven by biological clocks. The circannual mating cycles of the Siberian hamster, for example, are driven by seasonal changes in day length that elicit a corresponding secretion of the 'night hormone' melatonin. Only when the cyclic process persists under constant conditions, such as steady temperature or permanent light or darkness, can one speak of a biological oscillator. Curiously, the same cyclic process can be controlled by environmental conditions in one species and by a biological clock in another species. Thus, whereas the mating behaviour of the Siberian hamster is controlled by day length, the mating cycle of ground squirrels and fruit bats is controlled by a circannual clock that works under constant conditions (Gachon et al, 2004b).
Virtually all light-sensitive organisms from cyanobacteria to man have circadian clocks. This indicates that—at least in unicellular organisms such as cyanobacteria, protozoans and algae—circadian oscillators are built to work in a cell-autonomous fashion. Studies published during the past eight years have also shown this to be true for complex metazoans such as fruit flies (Plautz et al, 1997), (Whitmore et al, 1998) and mammals (Balsalobre et al, 1998; Yamazaki et al, 2000). Thus, many, if not most, body cells of these organisms have their own circadian clocks that continue to oscillate after they have been dissociated from the organism and grown in culture.
As indicated by their name, circadian oscillators can measure a day only approximately. Hence, to stay in resonance with day length, they must be synchronized every day by photic cues. In unicellular and semitransparent animals, each cell with a circadian oscillator has its own photoreceptor that communicates with the clock and sets its phase. The same holds true for larger opaque animals—the circadian phase of Drosophila and zebrafish organs that are kept in tissue culture, for instance, can be entrained by light–dark cycles. The blue-light receptor cryptochrome might be the pigment responsible for this process (Plautz et al, 1997; Whitmore et al, 2000). However, in spite of the ability of peripheral cells to react to light cycles, the Drosophila timing system is organized in a hierarchical manner in which pacemaker cells or tissues determine overall cyclic behaviour and physiology through the cyclic secretion of hormones (Helfrich-Forster, 2005). Obviously, such a hierarchical organization of pacemakers is much more important for opaque organisms in which light cannot reach every cell of the body (Hirota & Fukada, 2004). In mammals, the retina and the master pacemaker located in the SCN are the only structures known to be light-entrained, so the circadian oscillators in the periphery must therefore be synchronized by neuronal and other signals from the SCN.
All living organisms have time-measuring devices that affect their development, generation time, lifespan and lifestyle
Until recently, the rhythm-generating mechanism in all examined organisms was believed to rely on one or more connected feedback loops in gene expression. This model posits that transcriptional activators stimulate the expression of genes encoding repressors that, when reaching a critical threshold level, attenuate transcription of their own genes (Dunlap, 1999). However, this view may have to be revisited. In cyanobacteria, the circadian phosphorylation of KaiC, a key component of the cellular clock, continues for a few days in the absence of transcription and translation (Tomita et al, 2005). It therefore remains possible that the transcriptional and translational feedback loops in clock-gene expression that are observed in higher organisms are a manifestation of the rhythm-generating mechanism, rather than the mechanism itself. Even if this were true, the discovery of clock genes in a variety of organisms now allows the study of circadian clocks at the molecular level with regard to period, phase and amplitude.
The existence of circadian clocks in virtually all light-sensitive organisms shows that they must offer an evolutionary selective advantage. But, so far, it has been difficult to support this theory with decisive experiments. Although fruit flies and mice with mutations in clock or clock-controlled genes can display diseases or diminished fecundity (Beaver et al, 2002; Fu & Lee, 2003), it remains to be shown whether these conditions are related to perturbations of their circadian rhythm. Indeed, it is likely that clock genes also execute functions that are unrelated to circadian rhythm generation—and the discrimination between clock-related and unrelated phenotypes is not trivial. The most convincing demonstration of the circadian clock's utility has been accomplished in studies with cyanobacteria. In these organisms, mutations of kaiC that lengthen or shorten period length provide a strong selective advantage when the bacteria are grown in long or short light–dark cycles, respectively. As the same mutations have opposite effects on survival under different circadian growth conditions, it must be the resonance of the clock with the photoperiod that improves fitness (Ouyang et al, 1998). Moreover, arrhythmic kaiC mutant bacteria proliferate at least as efficiently as wild-type strains in mixed cultures that are exposed to constant light (Woelfle et al, 2004).
In mammals, the circadian timing system influences most physiological activities, including sleep–wake cycles, cardiovascular activity, body temperature, acuity of the sensory system, renal plasma flow, intestinal peristaltics, hepatic metabolism and detoxification, and many functions of the endocrine system (Schibler et al, 2003). All these rhythmic functions depend on the SCN, which receives photic information from classic rod and cone photoreceptors and from the melanopsin-expressing ganglion cells of the retina that is transmitted as electrical signals through the retino-hypothalamic tract. The electrical signalling involves the neurotransmitters glutamate and pituitary adenylate cyclase-activating peptide, which trigger the influx of calcium ions. This results in the activation of several protein kinases (protein kinase A, PKA; protein kinase C, PKC; mitogen-activated protein kinase, MAPK) and in the stimulation of immediate-early gene expression. The clock genes Per1 and Per2 are among the genes induced, and the photic regulation of period (PER) protein accumulation may have an essential role in tuning the circadian clock to daylight (Albrecht, 2004).
Individual SCN neurons cultured in vitro display daily oscillations in firing frequency and must therefore contain autonomous oscillators (Liu et al, 1997). However, the period length can vary dramatically between individual cultured cells, which causes a rapid desynchronization of SCN neurons when dissociated. In the intact animal, the cell-autonomous oscillators of SCN neurons must therefore be coupled, as laboratory rodents kept in constant darkness show a strongly rhythmic behaviour for months to years. By contrast, mice and other animals exposed to strong constant light become behaviourally arrhythmic with time, and a recent study indicates that this is due to the uncoupling—and thus desynchronization—of SCN neurons (Ohta et al, 2005).
The discovery of several rhythmically expressed mammalian clock and clock-controlled genes (Fig 2) has made it possible to monitor circadian rhythms in gene expression in any cell type. Such studies have shown that most body cells—and even cells cultured in vitro—harbour circadian oscillators with a similar molecular makeup as those that operate in SCN neurons. Recently, circadian gene expression has been recorded in real time in individual mouse and rat fibroblasts that cyclically transcribe a yellow fluorescent protein reporter gene or a luciferase reporter gene (Nagoshi et al, 2004; Welsh et al, 2004). These studies have revealed that fibroblasts contain highly robust selfsustained oscillators, the function of which even persists during cell division. In SCN-lesioned and behaviourally arrhythmic mice, the circadian clocks in peripheral tissue cells keep ticking, but they do so in an uncoordinated manner with phases that differ widely from tissue to tissue (Yoo et al, 2004). Thus, it is obviously the SCN master pacemaker that synchronizes the countless peripheral clocks in an intact animal. As daily feeding cycles are the dominant Zeitgebers for most tissues, it is likely that the SCN controls the phase of peripheral tissues mainly by imposing rest–activity cycles, which in turn determine the timing of food intake. Although the Zeitgeber signals that are elicited by feeding or fasting have not yet been identified, it has been shown that the phase of circadian clocks in cultured fibroblasts is exquisitely sensitive to nearly every signalling pathway that we and others have tested. The unequivocal identification of Zeitgeber signals that set the phase of peripheral clocks in intact animals will therefore be a formidable challenge (Schibler et al, 2003).
Figure 2.
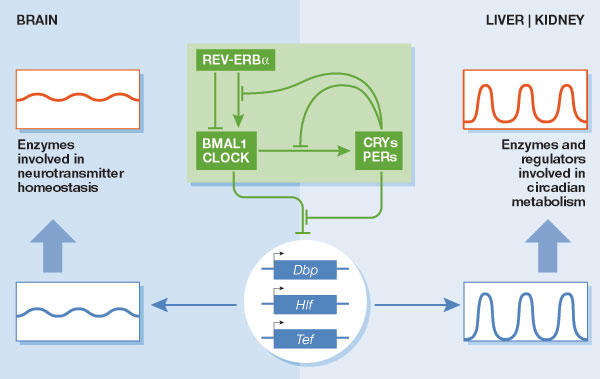
The PAR bZip transcription factors DBP, HLF, and TEF are output regulators of the mammalian circadian oscillator. The molecular circadian oscillator (green box) is composed of two coupled feedback loops in clock-gene expression. The negative limb contains the four repressor proteins PER1, PER2, CRY1 and CRY2, and the positive limb is composed of the two activator proteins CLOCK and BMAL1. The latter are PAS domain helix–loop–helix transcription factors that activate the Per and Cry genes as heterodimers by binding to E-box motifs. Heterotypic CRY–PER protein complexes, once they have reached a critical threshold concentration, attenuate the activity of BMAL1–CLOCK heterodimers and thereby repress their own genes. The same regulatory mechanism also operates in regulating the circadian expression of Rev-erbα, which encodes an orphan nuclear receptor that periodically represses Bmal1 and Clock. As a consequence, REV-ERBα interconnects the circadian transcription of positive and negative limb members. In addition, several protein kinases have important roles in the modulation of the activities of positive and negative limb members (Reppert & Weaver, 2002). The PAR bZip transcription factor family consists of three members, DBP, HLF and TEF. All three of these transcription factors are expressed in brain, liver and kidney. In brain, these proteins accumulate at nearly invariable levels throughout the day, whereas their accumulation strongly oscillates in liver and kidney. The circadian transcription of the Dbp gene is directly regulated by components of the core oscillator, and it remains to be shown whether the same mechanism accounts for cyclic Hlf and Tef expression. In the brain, PAR bZip proteins regulate the expression of pyridoxal kinase (PDXK), and thus the activity of enzymes involved in neurotransmitter metabolism. In liver and kidney, PAR bZip transcription factors regulate circadian amino acid and fatty acid metabolism and circadian detoxification (Gachon et al, 2004a).
Interestingly, with the exception of the SCN and the pineal gland, which produces melatonin, most brain regions have only low amplitudes in their expression of clock genes and clock-controlled genes (Gachon et al, 2004a). In theory, the low amplitude in circadian brain gene expression could be due to an inefficient synchronization of individual cellular oscillators, or to a low amplitude of clock-gene expression in every brain cell. On the basis of the design principles of circadian molecular oscillators—selfsustained feedback loops—I favour the first explanation. Conceivably, the chemical cues that synchronize peripheral oscillators do not efficiently pass the blood–brain barrier and therefore do not reach the threshold levels required for the efficient synchronization of all brain cell clocks.
The dominance of feeding cycles as a Zeitgeber for peripheral clocks suggests that these clocks have important roles in the processing of nutrients and/or in energy homeostasis. Indeed, transcriptome profiling studies have revealed that many cyclically expressed liver genes perform functions related to macromolecular metabolism and detoxification. How are the oscillations that are generated by core clock genes in hepatocytes translated into daily cycles of hepatocyte physiology? Our studies have shown a clock-output pathway that participates in this process.
Most physiological processes in mammals are influenced by a complex circadian timing system...
The three proline- and acid-rich (PAR) basic leucine zipper (bZip) transcription factors—albumin-D-site-binding protein (DBP), hepatic leukaemia factor (HLF) and thyrotroph embryonic factor (TEF)—accumulate according to high-amplitude circadian rhythms in the SCN, as well as in liver, kidney and other peripheral tissues. Genetic loss-of-function studies revealed that mice deficient for only one of these circadian transcription factors have a normal life expectancy and display only mild phenotypes. However, mice that lack all three PAR bZip transcription factors have a dramatically shortened lifespan. About half of these animals succumb to lethal seizures during the first three months after birth, and the surviving animals age at an accelerated rate and die prematurely from as yet unknown causes (Gachon et al, 2004a). Preliminary evidence suggests that PAR bZip transcription factors are involved in the regulation of circadian metabolism of endobiotic and xenobiotic compounds in various tissues, including liver and kidney. Biochemical and genetic studies have shown that Dbp is regulated directly by core components of the circadian clock, both in the SCN and in peripheral tissues, and the same may hold true for Tef and Hlf. However, PAR bZip triple knockout mice are behaviourally rhythmic and display nearly wild-type cycles in core clock gene expression. Hence, DBP, HLF and TEF are output regulators rather than central components of the molecular oscillator (Gachon et al, 2004a).
By contrast to the SCN and peripheral tissues, most brain regions express PAR bZip transcription factors at invariable or only slightly oscillating levels. Transcriptome profiling studies have suggested a possible reason for this observation. In both brain and liver, expression of the pyridoxal kinase (PDXK) enzyme is regulated by PAR bZip transcription factors, and as expected for a PAR bZip target gene, Pdxk expression is flat in the brain and cyclic in the liver. PDXK converts vitamin B6 derivatives into pyridoxal phosphate (PLP), an essential coenzyme of the decarboxylases and transaminases that are involved in neurotransmitter and amino-acid metabolism. As moderate decreases in neurotransmitters such as γ-aminobutyric acid (GABA), dopamine and serotonin can provoke generalized lethal seizures, strong oscillations in brain PDXK levels—and hence in brain PLP and neurotransmitter levels—would probably be harmful. By contrast, circadian amino-acid metabolism in the liver might be beneficial to the animal (Gachon et al, 2004a). Figure 2 illustrates how the molecular circadian oscillator can drive different outputs through the regulation of PAR bZip transcription factors in different tissues.
The work on circadian physiology has already revealed potential applications in two related clinical research fields: chronopharmacology and chronotherapy. The main objective of these fields is to establish temporal drug delivery regimens that take into consideration the daytime-dependent activity and toxicity of drugs. To a significant degree, chronopharmacology relies on the observation that drug metabolism can be subject to robust circadian regulation. Indeed, many drug-metabolizing enzymes, such as cytochrome P450 enzymes and carboxylesterases, are expressed according to daily cycles in liver and other tissues. The same is true for aminolevulinic acid synthase (ALAS1; Kaasik & Lee, 2004) and cytochrome P450 oxidoreductase (Oishi et al, 2003), two enzymes that regulate the activity of all cytochrome P450 enzymes. The former controls the synthesis of haem, the coenzyme of cytochrome P450 enzymes, and the latter has an essential part in the catalysis of monooxygenase reactions by transferring electrons from NADPH to the haem-bound iron of cytochrome P450 enzymes. The cyclic regulation of detoxifying enzymes has not yet been studied in detail, but several transcription factors known to regulate members of the cytochrome P450 gene family are expressed in a circadian manner. These include constitutive androstane receptor (CAR), peroxisome-proliferator-activated receptor α (PPARα) and PAR bZip proteins (Lemberger et al, 1996; Barbier et al, 2004; Dickins, 2004).
Curiously, the same cyclic process can be controlled by environmental conditions in one species and by a biological clock in another species
Cancer chronotherapy attempts to explore both the daytime-dependent metabolism of drugs and the difference in proliferation control between tumour cells and normal cells with high turnover rates, such as intestinal epithelium or bone marrow cells. In contrast to normal proliferating cells in which cell cycle progression is influenced by the circadian clock, many tumour cells have lost their circadian rhythm and daytime-dependent cell-cycle progression (Canaple et al, 2003). Thus, it might be advantageous to deliver antiproliferative drugs at times when they are least toxic to normal cells. Chronotherapy is still in its infancy and is not yet widely applied in the clinic. However, with the encouraging results that are now being obtained, this may well change in the near future (Lis et al, 2003; Mormont & Levi, 2003).

Acknowledgments
The research conducted in my laboratory is supported by the Swiss National Science Foundation through an individual grant and the NCCR programme Frontiers in Genetics, the State of Geneva, the Bonizzi Theler Stiftung and the Louis Jeantet Foundation of Medicine.
References
- Albrecht U (2004) The mammalian circadian clock: a network of gene expression. Front Biosci 9: 48–55 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937 [DOI] [PubMed] [Google Scholar]
- Barbier O, Fontaine C, Fruchart JC, Staels B (2004) Genomic and non-genomic interactions of PPARα with xenobiotic-metabolizing enzymes. Trends Endocrinol Metab 15: 324–330 [DOI] [PubMed] [Google Scholar]
- Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM (2002) Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc Natl Acad Sci USA 99: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaple L, Kakizawa T, Laudet V (2003) The days and nights of cancer cells. Cancer Res 63: 7545–7552 [PubMed] [Google Scholar]
- Dickins M (2004) Induction of cytochromes P450. Curr Top Med Chem 4: 1745–1766 [DOI] [PubMed] [Google Scholar]
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Fu L, Lee CC (2003) The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 3: 350–361 [DOI] [PubMed] [Google Scholar]
- Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, Duboule D, Petit B, Tafti M, Schibler U (2004a) The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev 18: 1397–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U (2004b) The mammalian circadian timing system: from gene expression to physiology. Chromosoma 113: 103–112 [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C (2005) Neurobiology of the fruit fly's circadian clock. Genes Brain Behav 4: 65–76 [DOI] [PubMed] [Google Scholar]
- Hirota T, Fukada Y (2004) Resetting mechanism of central and peripheral circadian clocks in mammals. Zool Sci 21: 359–368 [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC (2004) Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430: 467–471 [DOI] [PubMed] [Google Scholar]
- Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J (1996) Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem 271: 1764–1769 [DOI] [PubMed] [Google Scholar]
- Lis CG, Grutsch JF, Wood P, You M, Rich I, Hrushesky WJ (2003) Circadian timing in cancer treatment: the biological foundation for an integrative approach. Integr Cancer Ther 2: 105–111 [DOI] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM (1997) Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell 91: 855–860 [DOI] [PubMed] [Google Scholar]
- Mormont MC, Levi F (2003) Cancer chronotherapy: principles, applications, and perspectives. Cancer 97: 155–169 [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U (2004) Circadian gene expression in individual fibroblasts: cell-autonomous and selfsustained oscillators pass time to daughter cells. Cell 119: 693–705 [DOI] [PubMed] [Google Scholar]
- Ohta H, Yamazaki S, McMahon DG (2005) Constant light desynchronizes mammalian clock neurons. Nat Neurosci 8: 267–269 [DOI] [PubMed] [Google Scholar]
- Oishi K et al. (2003) Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem 278: 41519–41527 [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA 95: 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA (1997) Independent photoreceptive circadian clocks throughout Drosophila. Science 278: 1632–1635 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935–941 [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M (2002) Life before the clock: modeling circadian evolution. J Biol Rhythms 17: 495–505 [DOI] [PubMed] [Google Scholar]
- Schibler U, Ripperger J, Brown SA (2003) Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms 18: 250–260 [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H (2005) No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251–254 [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14: 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Sassone-Corsi P (2000) Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404: 87–91 [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P (1998) Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci 1: 701–707 [DOI] [PubMed] [Google Scholar]
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH (2004) The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol 14: 1481–1486 [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H (2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science 288: 682–685 [DOI] [PubMed] [Google Scholar]
- Yoo SH et al. (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 101: 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]


