Abstract
The lymphoid-specific proteins RAG1 and RAG2 initiate V(D)J recombination by introducing DNA double-strand breaks at the recombination signal sequences (RSSs). In addition to DNA cleavage, the versatile RAG1/2 complex is capable of catalyzing several other reactions, including hybrid joint formation and the transposition of signal ends into a second DNA. Here we show that the RAG1/2 complex also mediates an unusual strand transfer reaction, inverse transposition, in which non-RSS DNA is cleaved and subsequently transferred to an RSS sequence by direct transesterification. Characterization of the reaction products and requirements suggests that inverse transposition is related to both hybrid joint formation and signal-end transposition. This aberrant activity provides another possible mechanism for some chromosomal translocations present in lymphoid tumors.
Keywords: chromosomal translocation/RAG1/2 recombinase/transposition/V(D)J recombination
Introduction
The assembly of immunoglobulin (Ig) and T cell receptor (TCR) genes by V(D)J recombination generates much of the diversity of the antigen receptor repertoire (reviewed in Gellert, 2002). This DNA rearrangement requires the presence of conserved recombination signal sequences (RSSs) marking the boundaries of the coding segments. The assembly is initiated by DNA cleavage by the RAG1 and RAG2 proteins at sites next to the RSS sequences (Oettinger, 1992; McBlane et al., 1995). Efficient recombination requires two different RSSs containing conserved heptamer and nonamer sequences separated by non-conserved spacers of 12 or 23 bp, respectively. This 12/23 rule, enforced at the cleavage step of recombination, ensures appropriate joining of coding segments of V to J, or V to D to J segments (Tonegawa, 1983). The later stages of recombination involve the joining of pairs of coding ends and pairs of signal ends by non-homologous end joining machinery (Bogue and Roth, 1996; Jeggo, 1998; Ma et al., 2002).
The RAG1/2 complex recognizes and binds to RSSs, and in the presence of high mobility group proteins HMG1 or HMG2 is able to form a stable synaptic complex with the 12 and 23 RSSs (Hiom and Gellert, 1998). In normal V(D)J recombination, the RAG1/2–DNA complex catalyzes a series of phosphoryl transfer reactions, as depicted in Figure 1A. First, the DNA at the 5′ boundary of the RSS is nicked by RAG1/2, and then the 3′-OH liberated on the coding flank in the first step attacks the opposite strand to form a hairpin coding end and a blunt signal end (McBlane et al., 1995; van Gent et al., 1995). Efficient coupled cleavage at a pair of 12 and 23 RSSs occurs in the presence of Mg2+ in vitro (Eastman et al., 1996; van Gent et al., 1996), while additional uncoupled DNA cleavage at one RSS is observed in the presence of Mn2+ (McBlane et al., 1995).
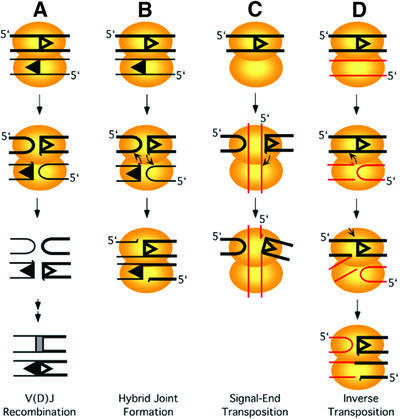
Fig. 1. Reactions mediated by RAG1/2 proteins. RAG1 and RAG2 proteins catalyze various reactions including DNA nicking, hairpin formation and strand transfer. Schematic presentation of (A) V(D)J recombination; (B) hybrid joint formation; (C) signal-end transposition; and (D) inverse transposition are shown. 12 and 23 RSS sequences are indicated by open and filled triangles. Non-RSS and RSS DNA are indicated by red and black lines, respectively. Non-RSS and 23 RSS DNA are depicted by thin lines to distinguish them from 12 RSS. Orientation of dsDNA is indicated by the 5′ end on one of two strands in either case. A half-arrow at the tip of a line represents a terminal 3′-OH. The two combined ovals are a schematic presentation of a synaptic complex.
Besides the reactions mentioned above, RAG1/2 catalyzes other reactions that lead to aberrant junctions or DNA transposition (Figure 1B–D). It has been shown that RAG1/2 mediates rejoining of the cleaved coding and signal ends to generate hybrid or open- and-shut joints in vitro (Figure 1B; Melek et al., 1998). Such joints have also been observed in vivo, both on artificial substrates and at the antigen receptor loci (Lewis et al., 1988; Morzycka-Wroblewska et al., 1988; Lewis and Hesse, 1991). Moreover, RAG1/2 catalyzes the transposition of signal ends into target plasmids or oligonucleotide substrates in vitro, as shown in Figure 1C (Agrawal et al., 1998; Hiom and Gellert, 1998). RAG1/2 can further cleave the branched transposition intermediate by disintegration, an activity similar to that of retroviral integrases (Melek and Gellert, 2000). It has been suggested that this RAG1/2-mediated signal-end transposition (also known as RSS transposition) may be responsible for a subset of the chromosomal translocations found in lymphoid tumors (Hiom et al., 1998; Melek and Gellert, 2000).
Here, we show that RAG1/2 can perform an alternative mode of transposition (Figure 1D). When complexed to an RSS, RAG1/2 can bind and cleave a non-RSS sequence to form a hairpin and a blunt end, and then catalyze the strand transfer of the blunt non-RSS end to the RSS sequence. This new mode of strand transfer uses the transposition donor and target in an inverted manner from the signal-end RSS transposition, and thus we call it ‘inverse transposition’. The mechanism of this reaction has similarities to both RAG1/2-mediated signal-end transposition and hybrid joint formation. As discussed below, this process also provides an alternative mechanism for formation of the inter-chromosomal translocations that are characteristic of B and T cell tumors.
Results
Inverse transposition mediated by RAG1 and RAG2
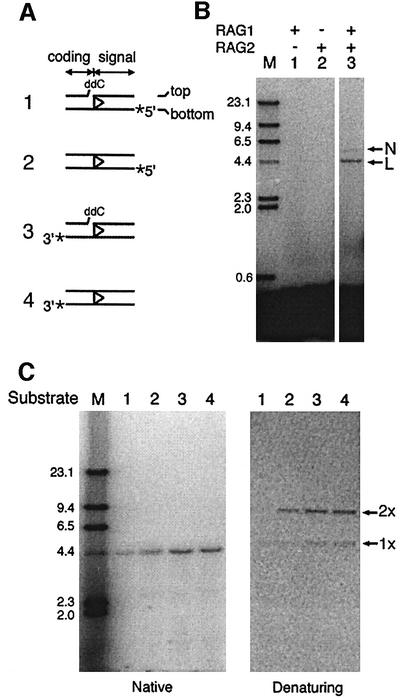
The RAG1/2-mediated RSS transposition previously described covalently connects the 3′-OH on the cleaved signal ends to a target DNA. A plasmid-based assay was developed in earlier studies for the detection of signal-end transposition, using a 32P-labeled RSS-containing oligonucleotide and a higher molecular weight plasmid target (Hiom et al., 1998). After signal-end transposition, label from the 5′ end of the bottom strand (substrate 2, Figure 2A) will become covalently attached to the pBR322 plasmid, leading to retarded migration of radioactivity on a native agarose gel (Figure 2C, lane 2, left panel).
Fig. 2. Transposition of non-RSS sequence mediated by RAG1 and RAG2 proteins. (A) Schematic drawings of oligonucleotides used in the plasmid assay for inverse transposition. The position of radioactive label on the bottom strand of an RSS-containing oligonucleotide is indicated by an asterisk. The top and bottom strands are defined by the orientation of the RSS sequence. Either intact (substrates 2 and 4) or pre-nicked dideoxy (substrates 1 and 3) oligonucleotides were used. The labeled oligonucleotide was incubated with supercoiled pBR322 DNA in the presence of RAG1/2, HMG1 and Mg2+ at 37°C for 2 h. Reaction samples were deproteinized and analyzed by agarose gel electrophoresis. (B) Strand transfer of the coding sequence to a non-RSS plasmid requires both RAG1 and RAG2 proteins. In addition to 3′-32P-labeled intact substrate (substrate 4), pBR322 plasmid and reaction buffer, lanes 1, 2 and 3 (all from the same experiment) contain purified RAG1, purified RAG2 and co-expressed RAG1/2 proteins, respectively. The reactions were analyzed on native agarose gels. N, nicked circular plasmid; L, linear plasmid; M, molecular marker of λ HindIII ladder. (C) Covalent strand connection of the inverse- transposition products was analyzed by comparing the deproteinized inverse-transposition products on a native gel (1.2%, left panel) and an alkaline denaturing gel (0.8%, right panel). The substrate for each reaction is indicated. 1× and 2× indicate the products with a single and double length of the linear single-stranded plasmid, respectively.
In this assay, signal-end transposition should not transfer radioactive label from the 3′ end of the bottom strand to the plasmid target (Figure 1). Nevertheless, starting with a 3′-labeled 12 RSS (substrate 4, Figure 2A), we detected another type of strand transfer in which the labeled coding flank had been covalently attached to the plasmid. We term this reaction inverse transposition. As shown in Figure 2B, the 3′-labeled coding end was associated with DNA species of high molecular weight. The mobility of the major product corresponded to a pBR322 linear molecule (Figure 2B, lane 3, lower band); a less intense band with the mobility of a nicked circular plasmid was also observed (upper band). Similar high molecular weight products were formed in reactions with a 3′-labeled 23 RSS oligonucleotide (data not shown). This strand transfer depended on both the RAG1 and RAG2 proteins (Figure 2B, lane 3). The individual RAG1 or RAG2 proteins were inactive (Figure 2B, lanes 1 and 2), eliminating the possibility that this activity was due to a contaminant enzyme co-purifying with the RAG proteins.
Like other RAG1/2-mediated activities, inverse transposition required HMG1 protein (van Gent et al., 1997) and divalent metal ions. Among the divalent metal ions tested, only Mg2+ and a mixture of Mn2+and Mg2+ (‘Mn2+ conditions’; Ramsden et al., 1997), but not Ca2+, supported this activity (data not shown). In contrast to the signal-end transposition, there was more inverse transposition in the Mn2+ conditions than in Mg2+ alone. In addition, the reaction was stimulated in the presence of DMSO, which was therefore used in the following experiments (data not shown). These reaction conditions would also support cleavage at the RSS. However, hairpin formation on the coding end adjoining the RSS was not necessary for inverse transposition. A pre-nicked substrate with a dideoxynucleotide at the end of the coding sequence to block hairpin formation (Figure 2A, substrate 3) was still an effective substrate for inverse transposition. This substitution would also prevent the RSS oligonucleotide from acting as the donor in the strand transfer reaction (see below).
Analysis of the inverse-transposition products
Examination of the high molecular weight products from inverse transposition confirmed that the bottom strand of the coding sequence was covalently joined to the plasmid DNA. The covalent strand connection of these products was analyzed by comparing the deproteinized products on native and alkaline denaturing gels (Figure 2C). Strand transfer products were detected on both types of gels in reactions with a 3′-labeled 12 RSS (Figure 2C, substrate 4), suggesting that the coding end was covalently linked to the plasmid. This kind of covalent linkage was also detected when the 3′-labeled pre-nicked substrate carried a dideoxy group at the 3′ nick (Figure 2C, substrate 3), suggesting that the hydroxyl nucleophile for the strand transfer was provided by the non-RSS containing plasmid DNA. In comparison, the product resulting from a 5′-labeled intact RSS oligonucleotide was that expected for RSS transposition: the signal end was covalently linked to the plasmid (Figure 2C, substrate 2). However, when a 5′-labeled pre-nicked substrate carried a dideoxy group at the nick, high molecular weight products were not seen on the denaturing gel (Figure 2C, substrate 1). The dideoxy group blocked hairpin formation at the RSS boundary and prevented liberation of the 3′-OH on the signal end, which is required for covalent linkage of the signal end to the plasmid in the signal-end transposition. The small amount of high molecular weight product observed with this substrate on the native gel represented a branched mole cule generated from inverse transposition. As shown in the third step of Figure 1D, the signal sequence may be held on the plasmid by base pairing to the bottom strand of the coding sequence and would then remain associated in native, but not denaturing, gel electrophoresis.
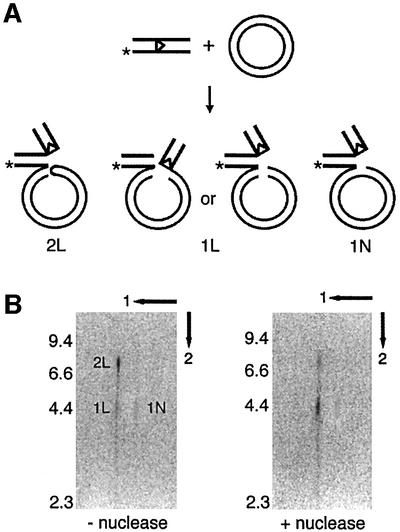
Denaturing gel electrophoresis revealed two species of inverse-transposition products, one with mobility corresponding to the linear single-stranded plasmid (1×) and the other to a species twice as long (2×) (Figure 2C, substrates 3 and 4 on the denaturing gel). Partial nuclease digestion indicated that the 2× species contained a hairpin end in the linear plasmid (Figure 3). When the deproteinized products were fractionated by two-dimensional gel electrophoresis, first on a native and then on an alkaline denaturing gel, the major band corresponding to the linear dsDNA in the first dimension (L species) was resolved into two bands after denaturation (Figure 3B, 2L and 1L). The minor band corresponding to nicked circles in the first dimension (N species) remained as a single species 1N in the second-dimension alkaline gel. 2L was the major product (∼70%) with a chain length (∼8.7 kb) twice that of a linear plasmid (4.3 kb). This double-length species reduced to a plasmid-length band (co-migrating with 1L) when the sample was partially digested with mung bean nuclease after the inverse-transposition reaction (Figure 3B, right panel). Partial digestion by mung bean nuclease has been used previously to identify hairpin structures (Melek and Gellert, 2000). Therefore, 2L is a linear plasmid molecule with a hairpin end, while 1L and 1N represented a topologically linear plasmid-length DNA and a plasmid-length nicked circle, respectively (Figure 3A).
Fig. 3. Hairpin structure on the plasmid DNA resulting from inverse transposition. (A) Inverse-transposition reactions with an intact 3′-32P-12 RSS oligonucleotide (*) and supercoiled pBR322 were carried out in the presence of RAG1/2, HMG1 and 5 mM Mg2+/0.5 mM Mn2+ (Mn2+ conditions) at 37°C for 2 h, as described in Materials and methods. Predicted configurations of the inverse-transposition products are shown. (B) Two-dimensional gel analysis of the inverse-transposition product. The reaction product was treated with mung bean nuclease or with mock digestion before deproteinization, and analyzed by two-dimensional gel electrophoresis. The first dimension was fractionated on a 1.2% native agarose gel, and the second dimension on a 1% alkaline denaturing gel. Three species, 1L, 2L and 1N, were detected. The numbers on the left indicate the positions of molecular markers.
The variety of product configurations suggests that inverse transposition results from several DNA intermediates. The 3′-OH nucleophile on the non-RSS containing plasmid could be generated by a simple nick, or a 3′-OH on the opposite strand would arise if hairpin formation followed nicking. The subsequent transesterification by the hydroxyl groups from these two structures would lead to products 1N and 2L, respectively. Furthermore, 2L could undergo a second strand transfer, with the liberated 3′-OH on the signal end attacking the hairpin structure on the plasmid, and thus generate 1L.
Nicking and cleavage of non-RSS containing DNA by RAG1/2 proteins
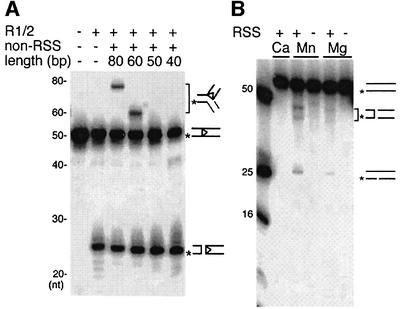
As suggested by the inverse-transposition products, the RAG1/2 proteins should be able to nick and cleave non-RSS DNA to liberate a 3′-OH group and use it as a nucleophile in the subsequent strand transfer reaction. To look for these species, we examined the binding, cleavage and inverse transposition of RAG1/2 proteins on short oligonucleotides that did not contain an RSS sequence. Binding to RAG1/2 proteins was observed by mobility-shift assay with non-RSS DNA oligonucleotides of 50 bp and longer in the absence of RSS DNA. However, binding of non-RSS DNA was not as strong as that of RSS DNA, and was readily competed by RSS oligonucleotides. A small amount of mixed paired complex containing a RSS and a non-RSS DNA was also observed (data not shown). These non-RSS oligonucleotides were further examined to see if they could serve as a strand transfer donor with the 3′-labeled 12 RSS as the target. Higher molecular weight products, corresponding to the 3′-labeled coding flank covalently attached to a fragment of the donor, were observed with DNA oligonucleotides that are long enough to bind to RAG1/2 (Figure 4A, lanes 3–6).
Fig. 4. RAG1/2-mediated activities on non-RSS DNA. (A) Trans position of non-RSS oligonucleotides to a 12 RSS sequence. Inverse-transposition reaction was carried out with 3′-32P-12-RSS (*) oligonucleotides and unlabeled non-RSS DNA oligonucleotides 40–80 bp in length, under the same reaction conditions as described in Figure 3. The drawings indicate the substrate, hairpin cleavage product and inverse-transposition products. The non-RSS oligonucleotides in lanes 3–6 are IhS101/102, MM569/570, MM567/568 and MM565/566, respectively. (B) RAG1/2-mediated cleavage of non-RSS DNA. The 5′-32P-labeled (*) non-RSS DNA oligonucleotide MM569/570 (60 bp) was subjected to RAG1/2 cleavage in the presence or absence of unlabeled 23 RSS. Mg2+ was at 5 mM and Ca2+ at 4 mM. ‘Mn’ indicates the Mn2+ conditions described in Materials and methods. The drawings indicate linear and hairpin cleavage products as well as the substrate.
Furthermore, RAG1/2 was able to cleave non-RSS oligonucleotides, generating shorter-length products in the presence of either Mg2+ or under the Mn2+ conditions. Short products were only observed in reactions that also contained RSS DNA, suggesting that cleavage of the labeled non-RSS oligonucleotide depended on the presence of unlabeled RSS DNA (Figure 4B). Consistent with the results from inverse transposition, cleavage was greater under the Mn2+ conditions than in Mg2+ alone.
Sequence requirement for inverse-transposition activity
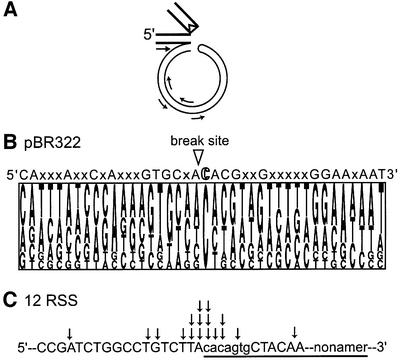
To examine whether the RAG1/2 proteins used pseudo RSS sites on the plasmid for inverse transposition, we analyzed the sequences at the insertion junctions. The insertion junction sequence was PCR amplified with a primer complementary to the bottom strand of the coding sequence and other primers complementary to the pBR322 (Figure 5A); the amplified DNAs were subsequently cloned and sequenced. The sequence analysis revealed several interesting features. First, a 3′ cytosine was strongly preferred at the break point on the plasmid, but there was very weak conservation within the surrounding sequence (Figure 5B). The first seven positions 3′ of the break site have a remote similarity to the RSS heptamer, but no nonamer-like sequence was observed. Nonetheless, when testing the sequence preference for the inverse transposition using the oligonucleotide assay, a sequence derived from Figure 5B was a more effective substrate than a random sequence of the same length (data not shown), suggesting some sequence effect on the inverse transposition. Secondly, for the majority of reaction products sequenced, nucleophilic attack by the 3′-OH of plasmid DNA was not always precisely at the boundary of coding and signal sequences. Among the 17 individual insertion junctions that were sequenced, ∼70% were within 2 bp from the boundary, but a few were more than 5 bp away (Figure 5C).
Fig. 5. Sequence analysis of insertion junctions between coding sequence and non-RSS DNA. (A) Schematic presentation of PCR amplification and cloning of the insertion junction. A primer complementary to the bottom strand of the coding sequence, and one of four primers complementary to the plasmid sequence, were used for PCR amplification and the amplified fragments were cloned into pCR2.1 vector for further sequence analysis. (B) Pictogram analysis of the break site sequences on non-RSS DNA. The size of each letter represents the frequency of appearance for the nucleotide at that position. The sequence analysis of 20 bp on each side of the break point is shown (17 sequences in total). Nucleotides that appear with a frequency >50% at that position are indicated above the pictogram. X represents no significant preference at that position. (C) The insertion sites on the 12 RSS sequence from inverse transposition. Arrows indicate the insertion sites on the 12 RSS, where the bottom strand connects to the 3′ end of the non-RSS DNA. The RSS sequence is underlined while the conserved heptamer and nonamer sequences are indicated by lower case letters. Each arrow represents the insertion site determined from 1 of 17 sequenced clones. The majority of strand transfer (70%) occurs within 2 bp of the boundary. (D) Effect of RSS mutations on inverse transposition. Inverse-transposition activity was measured using the plasmid assay described in Figure 3 with consensus and mutant RSS sequences. The sequence of each mutant RSS tested is shown; upper and lower case letters represent consensus and mutant nucleotides, respectively. The non-RSS oligonucleotide is MM569/570 (see Materials and methods for the exact sequence).
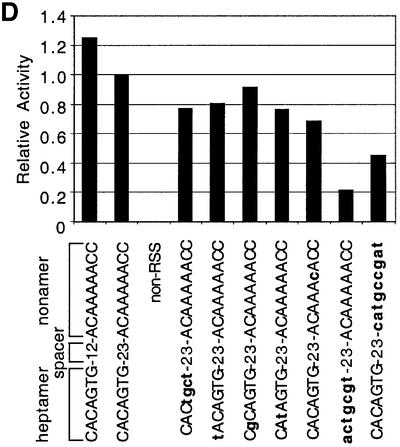
The RSS sequence requirement for the target of inverse transposition is less stringent than that for cleavage or signal-end transposition. We examined the RSS requirements using a set of altered 23 RSS oligonucleotides previously tested for other RAG1/2-mediated activities (Ramsden et al., 1996; Melek and Gellert, 2000). DNA oligonucleotides that did not contain any RSS-like sequence did not act as donors in inverse transposition (Figure 5D). While the effects of mutations on signal-end transposition parallels that on the hairpin formation, inverse-transposition activity was much less affected by these mutations. For example, mutation of the first, second or third position of the heptamer, or complete ablation of the heptamer, greatly diminished hairpin formation (Ramsden et al., 1996), and therefore inhibited signal end transposition (data not shown). In contrast, inverse-transposition activity was only reduced by 10–20% by mutation of one of the first three heptamer positions. Even complete ablation of either the heptamer or nonamer (but not both) allowed ∼20% inverse-transposition activity. These data also confirmed our observation that hairpin formation on the RSS-containing oligonucleotide was not required for inverse transposition.
Formation of a ‘mixed’ synaptic complex for inverse transposition
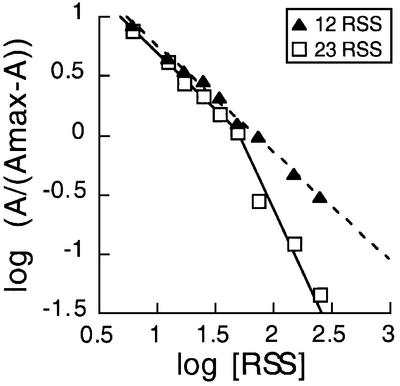
Unlike other RAG1/2-mediated reactions, inverse transposition was inhibited in the presence of a pair of complementary RSSs. The requirement of an RSS and a non-RSS DNA, but not a 12/23 synaptic complex, suggested that the RAG1/2 proteins formed a ‘mixed’ synaptic complex with the non-RSS DNA substituting for one of the RSSs. To verify this hypothesis, we examined the inverse-transposition activity in the presence of competing unlabeled RSS substrates (Figure 6). Inverse-transposition activity was comparable with either the labeled 12 or 23 RSS, but the presence of an excess of unlabeled RSS oligonucleotide inhibited the reaction. To examine this inhibitory effect in more detail, we varied the concentration of unlabeled 12 or 23 RSS competitor in the reactions with a 3′-labeled 12 RSS target and a pBR322 donor. The number of sites involved in competition can be determined graphically by multisite inhibition analysis using a Hill plot (Segel, 1975). In this analysis the logarithm of A/(1 – A) was plotted against the logarithm of RSS concentration in nanomolar units, where A is the relative activity of inverse transposition. With the unlabeled 12 RSS competitor, inhibition showed linear dependence with a slope of approximately –1, suggesting single-site inhibition. On the other hand, the plot with the unlabeled 23 RSS competitor was curved with a limiting slope of approximately –1 at low 23 RSS concentrations, and approximately –2 at high 23 RSS concentrations. This difference between the two competitive inhibitors is due to more favorable formation of a 12/23 or 23/23 complex than a 12/12 complex (Jones and Gellert, 2002). The unlabeled 12 RSS only competed for the labeled 12 RSS binding site in an RAG1/2 complex that was capable of catalyzing inverse transposition, while the unlabeled 23 RSS competed for the binding sites of both the labeled 12 RSS and the plasmid DNA. These results support our model that the strand transfer of inverse transposition occurs in a mixed synaptic complex containing a single RSS and a non-RSS-containing DNA. The results also suggest that the binding site for the non-RSS DNA in this mixed synaptic complex is equivalent to that for an RSS.
Fig. 6. Multisite inhibition analysis of the unlabeled RSS competitor in inverse transposition. Inverse-transposition reactions with intact 3′-32P-12 RSS substrate and pBR322 were carried out as described in Figure 2. Inverse-transposition activity was measured as described in Figure 3 with an unlabeled 12 RSS (filled triangles, dashed line) or 23 RSS (open square, solid line) competitor at various concentrations. Log(A/(1 – A)) was plotted against log([RSS]), where A is relative activity and [RSS] is RSS concentration in nanomolar units. The data were fitted to linear dependence of a single slope of –0.98 or two slopes of –0.99 and –1.87, for the 12 or 23 RSS, respectively. The slope in each plot indicates the number of sites involved in competitive inhibition.
Resolution of the branched intermediate by RAG1/2
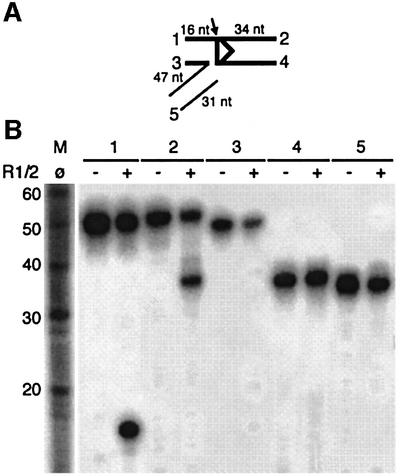
As depicted in Figure 3A, inverse transposition generated a branched molecule consisting of a nicked RSS with its coding flank covalently linked to a non-RSS DNA. It is worth noting that this branched molecule is different from the signal-end transposition intermediate. To test whether RAG1/2 could further process the branched intermediate to yield a linear DNA, a branched molecule pre-assembled with oligonucleotides was used. All strands of the branched intermediate were individually 32P-labeled as depicted in Figure 7A. In the presence of RAG1/2, the top strand of the RSS-containing DNA was nicked at the RSS boundary (Figure 7B, 1 and 2), thus generating a nicked linear DNA consisting of the coding end and non-RSS DNA. We also tested if the 3′-OH group on the RSS heptamer could undergo transesterification and regenerate the RSS-containing DNA oligonucleotides and a broken non-RSS DNA, by a type of disintegration (Chow et al., 1992). The bottom strand of the signal sequence (34 nucleotides) and the bottom strand of the branched structure (47 nucleotides) were 5′- and 3′-end labeled, respectively, such that transesterification would yield a product of ∼50 nucleotides in both cases. However, no products of increased length were observed in either reaction (Figure 7B, 3 and 4), suggesting that nicking at the RSS boundary is the major pathway for resolution of this branched intermediate by RAG1/2.
Fig. 7. Resolution of the branched inverse-transposition intermediate. (A) Schematic presentation of the branched molecule used in this reaction. The position of labeling is indicated by a number, which corresponds to the reaction shown in (B). Thick and thin lines represent the RSS-containing and non-RSS sequences, respectively. An arrow indicates the nick introduced by RAG1/2 proteins. (B) Branched molecules assembled and labeled as shown in (A) were subjected to RAG1/2 protein for cleavage. The reaction conditions were as described in Figure 3. Predicted products of a nicking reaction were 16 and 34 nucleotides for substrate 1 and 2, respectively.
Discussion
RAG1/2 is remarkable for the broad spectrum of reactions that it can catalyze, including DNA nicking and a variety of transesterification reactions (Figure 1). Here we have described a new class of strand-transfer reaction catalyzed by RAG1/2, using a non-RSS DNA as the transposition donor and an RSS sequence as the target, as depicted in Figure 1D. In the presence of an RSS DNA, RAG1/2 can cleave a non-RSS DNA in a two-step mechanism that leads to a double-strand break with a hairpin on one broken end, as is true for normal cleavage at the RSS border (Figure 1A and D). The liberated 3′-OH group on the non-RSS DNA then attacks the RSS DNA and covalently links the cleaved end to the coding sequence. If the RSS-containing sequence is intact during the nucleophilic attack, the strand transfer reaction would yield a three-way branched molecule. We have shown that such a branched intermediate can be further processed by RAG1/2, generating a fusion molecule and two cleaved ends (Figure 7). The latter products are also expected when the RSS sequence has undergone nicking or hairpin formation prior to the nucleophilic attack. These modes of strand transfer contributed ∼70% of the inverse-transposition products in the plasmid assay.
Two other minor species of inverse-transposition products were also observed. If both the RSS and non-RSS substrates are cleaved prior to strand transfer they could potentially undergo an additional reciprocal joint formation, generating two fusion molecules. In the plasmid assay, the reciprocal strand-transfer would generate a linear plasmid-length product (Figure 3A, 1L), which represented 25% of the inverse-transposition products by two-dimensional gel analysis. Finally, a small fraction (∼5%) of the nicked plasmid DNA was able to use its 3′-OH to attack the RSS DNA directly and generate a four-way branched molecule (Figure 3A, 1N). Overall, the majority of inverse transposition yields branched intermediates that could be further resolved into linear DNA by RAG1/2, providing a possible source for RAG1/2- mediated interchromosomal translocation, as discussed in more detail below.
Distinct binding modes of non-RSS DNA to RAG1/2
It is known that RAG1/2 can bind non-RSS DNA, although more weakly than RSS-containing DNA (Ramsden et al., 1996; Hiom and Gellert, 1997). Under the conditions used in this study, RAG1/2 was able to bind to non-RSS DNA oligonucleotides of 50 bp and longer in the presence of HMG and divalent metal ions, with an affinity ∼10-fold lower than an RSS-containing oligonucleotide of the same length. However, RAG1/2-catalyzed cleavage of non-RSS DNA was unexpected, and indeed was only observed in the presence of RSS DNA, suggesting that binding of non-RSS DNA alone does not trigger formation of a catalytically active complex. Instead, RAG1/2 cleavage of non-RSS DNA occurs within a mixed synaptic complex. Binding of an RSS DNA could possibly trans-activate the catalytic residues in the other half of the synaptic complex to cleave a non-RSS DNA. A well-established example for trans-activation is the phage Mu transpososome, in which binding of a MuA subunit to the MuA recognition sequence on one transposon end activates cleavage and strand transfer of the other end (Aldaz et al., 1996; Savilahti and Mizuuchi, 1996). A recent study further demonstrated that with limiting concentrations of Mu DNA, MuA recognition sites can activate a MuA subunit to process and transfer non-Mu DNA into a target (Goldhaber-Gordon et al., 2002). In that reaction, the use of a non-consensus sequence as a transposition donor, and the formation of a mixed synaptic complex containing one consensus and one non-consensus sequence, are similar to the substrate processing step of inverse transposition.
Inverse transposition and signal-end transposition both involve strand transfer between an RSS and non-RSS sequence, but with inverted roles for target and donor DNA (Figure 1B and D). Moreover, signal-end transposition is most efficient in the presence of both 12 and 23 RSSs, suggesting that the reaction occurs preferentially in a complex with two RSS ends and a target DNA (Hiom et al., 1998). In contrast, inverse-transposition activity is inhibited in the presence of a complementary RSS (Figure 6). This observation implies that the binding modes for the non-RSS DNA in the two reactions are different (Figure 1C and D). Furthermore, for signal-end transposition, target capture experiments argue that a RAG1/2 synaptic 12/23 complex can bind to non-RSS DNA both before and after cleavage, indicating that the binding site for the non-RSS DNA target in these complexes does not overlap with the binding site for either RSS or its flanking coding sequences (Neiditch et al., 2001). Conversely, for inverse transposition, competitive inhibition analysis suggests that unlabeled 12 RSS could only compete for one of two binding sites (32P-labeled 12 RSS site) in this complex. The same experiment with unlabeled 23 RSS suggests that the 23 RSS can compete for both the labeled 12 RSS and non-RSS binding sites, in keeping with the observation that the 23/23 complex is more stable than the 12/12 complex (Jones and Gellert, 2002). Taken together, these results suggest that the productive binding of non-RSS DNA in the inverse-transposition reaction utilizes one of the two RSS binding sites in a synaptic complex.
Similarity of inverse transposition and hybrid joint formation
RAG1/2-mediated hybrid joints, involving a linkage of a signal end to the coding flank of the partner RSS, have been identified in vivo, and such joints can also be formed in vitro with purified RAG1/2 (Lewis et al., 1988; Morzycka-Wroblewska et al., 1988; Lewis and Hesse, 1991; Melek et al., 1998). One of the mechanisms proposed for RAG1/2-mediated hybrid joining involves cleavage of a pair of RSSs in a synaptic complex, followed by nucleophilic attack of the 3′-OH group from one signal end onto the hairpin at the complementary coding end (Figure 1B) (Melek et al., 1998). Inverse transposition resembles this model in its chemical mechanism and formation of a synaptic complex, but replacing one of the substrates with a non-RSS DNA. During the first stage of the reaction, both non-RSS and RSS DNA may undergo nicking and hairpin formation prior to the strand transfer reaction and subsequently either a single or reciprocal strand transfer takes place. As discussed above, the binding site of non-RSS DNA overlaps with that of an RSS, and both cleavage and strand transfer occur in a synaptic complex; thus, inverse transposition can also be considered as a variant of hybrid joint formation.
As depicted in Figure 1D, inverse transposition uses intact RSS DNA as strand transfer target. In reactions with the pre-nicked dideoxy-RSS substrate that can not undergo hairpin formation, the inverse-transposition product is still formed, but to a slightly lower extent. Additionally, a three-way branched intermediate is observed with reactions starting from either intact or pre-nicked dideoxy-RSS substrates (Figure 2C). This observation prompts speculation whether hybrid joints can also be made by a similar pathway. Using an oligonucleotide assay, we could detect a hybrid joining product starting from an intact RSS and a complementary pre-nicked dideoxy-RSS substrate (I-h.Shih and M.Gellert, unpublished data). This suggests an alternative mechanism for hybrid joint formation where the 3′-OH from one signal end attacks a pre-nicked complementary RSS, instead of a hairpin structure on the complementary coding end.
Inverse transposition and chromosomal translocation
In lymphoid tumors, the high frequency of translocations at RSS sites in the Ig and TCR loci links them to V(D)J-induced breaks (Nowell, 1997). Some of these translocations have been connected to RAG1/2-mediated cleavage. Although some translocations may result from inter-chromosomal V(D)J joining between an Ig or TCR locus and a cryptic RSS sequence at the breakpoint on the partner chromosome (Tsujimoto et al., 1985; Aplan et al., 1990), in many cases no RSS is identified near the breakpoint. Thus, it has been suggested that these events could arise from RAG1/2-mediated signal-end transposition (Hiom et al., 1998; Melek and Gellert, 2000). In this model, after RAG cleavage at an Ig or TCR locus, the 3′ end of a RSS sequence is covalently joined to the target DNA to generate a three-way branched molecule. This intermediate is further processed by hairpin formation on the target DNA, leaving a fusion molecule of target DNA to a RSS sequence, and a cleaved coding end and a target sequence both with a hairpin structure. Joining of the two cleaved ends would then generate a translocation. These translocations could result from either single-ended or two-ended RSS attack, and would be expected to introduce P nucleotide tracts or end processing (deletion or N nucleotide addition) at the junction of the coding end and target DNA.
The RAG1/2-mediated inverse transposition described in the present work provides an alternative mechanism for chromosomal translocation. A translocation resulting from inverse transposition would be expected to have features distinct from signal-end transposition. The junction of the signal end with non-RSS DNA would be likely to add or delete nucleotides, because of the hairpin structure on the non-RSS DNA end. This would not be true of direct signal-end transposition. Furthermore, the coding end fusion would be close to the coding-signal end boundary since the strand transfer of non-RSS DNA usually attacks within 2 bp from the RSS border, but would not be expected to add or delete many nucleotides. Additionally, the break point on the partner chromosome would have a strong preference for sites 5′ of a cytosine. Several sequences at the translocation junctions from T and B cell tumors fit this pattern (Tsujimoto et al., 1987; Bernard et al., 1988; Neri et al., 1988; Adachi et al., 1989). One such example is the translocation t(14;8)(q24; q11) found in an acute lymphoblastic leukemia (Bernard et al., 1988). Sequence analysis of the reciprocal translocation junctions revealed that the break point on the 3′ side of the c-myc locus (chromosome 8) occurs immediately 5′ of a cytosine, and on chromosome 14, the insertion junction is 3 bp 5′ to the RSS boundary. The resulting RSS junction to chromosome 8 has one nucleotide deleted and six N nucleotides added, whereas the reciprocal junction is a perfect fusion of the two sequences without addition or deletion of nucleotides.
Like signal-end transposition, inverse transposition must be rare in vivo. Normal RAG1/2 action during V(D)J recombination is limited to cleavage at pairs of RSSs, guided by the 12/23 rule (Tonegawa, 1983). Despite the higher affinity and activity of RAG1/2 at RSS sites, non-RSS sequences are in immense excess of the available RSSs, so that other factors must protect against this deleterious strand transfer activity. It is possible that the portions of the RAG1 and RAG2 proteins missing from the core proteins used in our biochemical system act in this way, because the full-length proteins have been shown to be much less effective in forming hybrid joints in vivo (Sekiguchi et al., 2001). Separately, the tight control and regulation of accessibility within rearranging loci could limit the misuse of non-RSS DNA in recombination. In this regard, chromatin structure and cis-acting elements within the rearranging loci could help ensure proper V(D)J rearrangement during lymphoid cell development (Sleckman et al., 1998; Roth and Roth, 2000).
Materials and methods
Materials
Murine RAG1 (384–1008) and RAG2 (1–387) proteins tagged with N-terminal maltose binding protein and C-terminal polyhistidine and myc epitopes were expressed individually or co-expressed, and prepared as described previously (McBlane et al., 1995). HMG1 (1–163) was expressed and purified as described previously (Mo et al., 2000). Deoxynucleotidyl transferase was purchased from GIBCO-Life Technologies (Rockville, MD), and T4 polynucleotide kinase, BSA and all restriction enzymes were purchased from New England Biolabs.
DNA oligonucleotides were purchased from Oligos Etc. (Wilsonville, OR) or synthesized in-house. All the oligonucleotides were gel purified prior to use. Double-stranded substrates were assembled in 100 mM potassium glutamate/10 mM Tris pH 7.0 annealing buffer, heated to 95°C for 10 min and then slowly cooled to room temperature. RSS substrates used in this work were described previously (Hiom and Gellert, 1997; Melek and Gellert, 2000), including intact 12 RSS (DAR39 and DAR40), intact 23 RSS (DG61 and DG62) and pre-nicked dideoxy 12 RSS (MM579, DG10, DAR40). The intact 12 RSS used in sequence analysis of insertion junctions has a longer coding flank (IhS071, 5′-GACGA TGTCAATTATACCATAAGTGTAACCTGTAGATATTGGAATAA TCAACATCATGTTACTGAGAGTGCACCATAAAA-3′, and its com plementary sequence IhS072). Non-RSS DNA substrates used in binding and cleavage assays included complementary oligonucleotide pairs: MM561/MM562, 5′-CAGTTACGTTAGGCCAGATC-3′; MM563/MM564, 5′-ATCGAGGACGCAGTTACGTTAGGCCAGATC-3′; MM565/MM566, 5′-AATCCATAGGCAGTACTGTCGACAGGCCAATGCGAGCGCA-3′; MM567/MM568, 5′-AATAAATAGGCAGTACTGCGACAGGCCAATGCGAGCGCACAATCAATAA-3′; MM569/MM570, 5′-TTACAGCCAGACAGTGGAGTACTACACGCAGTCCT ACTCGAGAATCCATAGGCAGTACT-3′; and IhS101/IhS102, 5′-GA CGATGTCAATTATACCATAAGTGTAACCTGTAGATATTGGA ATAATCAACATCATGTTACTGAGAGTGCACCATAAAA-3′. Oli gonucleotides were 3′- or 5′-32P-end labeled as described previously (Melek and Gellert, 2000). Supercoiled plasmid pBR322 was a gift from Mary O’Dea of this laboratory.
Reaction conditions and data analysis
The reaction buffer for inverse transposition, signal end transposition and cleavage reactions contained 25 mM MOPS (pH 7.0), 4 mM DTT, 75 mM potassium glutamate, 10% DMSO, 0.02 mg/ml HMG, 0.1 mg/ml BSA. RSS substrates (20 nM), plasmid pBR322 (10 µg/ml) and RAG1/2 (50 µg/ml) were assembled in the reaction buffer prior to addition of divalent metal ions. Divalent metal conditions used in these reactions contained 5 mM MgCl2 or a combination of 5 mM MgCl2 and 0.5 mM MnCl2 (‘Mn2+ conditions’). The Mn2+ conditions allowed those reactions normally found in MnCl2 alone (cleavage at a single RSS, etc.) to proceed, but with greater efficiency. When a non-RSS DNA oligonucleotide replaced pBR322 plasmid as inverse-transposition donor, it was present at 20 nM. All reactions were incubated at 37°C for 2 h. The reactions were deproteinized by incubation with proteinase K (1 mg/ml) and SDS (0.2%) at 55°C for 30 min, or phenol/chloroform extracted, and ethanol precipitated prior to gel electrophoresis analysis. Gels were dried down and exposed to phosphorimaging plates. Autoradiography image analysis and quantification was done on a Molecular Dynamics Typhoon 8600 PhosphorImager, with Molecular Dynamics ImageQuant 5.1 software. Competitive inhibition data were plotted and fit to a linear dependence (y = mx + c) by KaleidaGraph 3.0 (Synergy Software).
Gel electrophoresis
The inverse-transposition products from plasmid assay were analyzed with either native agarose electrophoresis in 1× TBE buffer, or denaturing alkaline electrophoresis in 0.1 M NaOH and 5 mM EDTA. Sample preparation and gel electrophoresis conditions for two-dimensional electrophoresis analysis have been described previously (Melek and Gellert, 2000).
Sequence analysis of insertion junctions
Inverse-transposition products were phenol/chloroform extracted and ethanol precipitated prior to PCR amplification. Primers used for amplification included MM578, which is complementary to the bottom strand of coding sequence, and one of four primers complementary to different parts of pBR322, 1238, 1239, 1240 and 1241 (New England Biolabs). PCR and cloning (TA Topo cloning kit, Invitrogen) were carried out as described previously (Hiom et al., 1998). Sequencing reactions of cloned fragments were carried out using a DNA sequencing kit (PE Applied Biosciences) and analyzed by an ABI310 automated sequencer.
Gel mobility shift assay
Reactions were assembled as described for the inverse-transposition reactions in 4 mM CaCl2 and incubated at 37°C for 30 min. An equal volume of 50% glycerol was added to each sample before it was loaded on a 8% native acrylamide gel (crosslinking ratio of 80:1) in 0.5× TBE buffer. Gels were electrophoresed at 20 mA.
Acknowledgments
Acknowledgements
We thank Kiyoshi Mizuuchi for stimulating discussion leading to this work, Eric Greene for critical reading of the manuscript, and Jessica Jones for much advice and technical help.
References
- Adachi M. and Tsujimoto,Y. (1989) Juxtaposition of human bcl-2 and immunoglobulin λ light chain gene in chronic lymphocytic leukemia is the result of a reciprocal chromosome translocation between chromosome 18 and 22. Oncogene, 4, 1073–1075. [PubMed] [Google Scholar]
- Agrawal A., Eastman,Q.M. and Schatz,D.G. (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature, 394, 744–751. [DOI] [PubMed] [Google Scholar]
- Aldaz H., Schuster,E. and Baker,T.A. (1996) The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell, 85, 257–269. [DOI] [PubMed] [Google Scholar]
- Aplan P.D., Lombardi,D.P., Ginsberg,A.M., Cossman,J., Bertness,V.L. and Kirsch,I.R. (1990) Disruption of the human SCL locus by ‘illegitimate’ V-(D)-J recombinase activity. Science, 250, 1426–1429. [DOI] [PubMed] [Google Scholar]
- Bernard O., Larsen,C.J., Hampe,A., Mauchauffe,M., Berger,R. and Mathieu-Mahul,D. (1988) Molecular mechanisms of a t(8;14) (q24;q11) translocation juxtaposing c-myc and TCR-α genes in a T-cell leukaemia: involvement of a V α internal heptamer. Oncogene, 2, 195–200. [PubMed] [Google Scholar]
- Bogue M. and Roth,D.B. (1996) Mechanism of V(D)J recombination. Curr. Opin. Immunol., 8, 175–180. [DOI] [PubMed] [Google Scholar]
- Chow S.A., Vincent,K.A., Ellison,V. and Brown,P.O. (1992) Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science, 255, 723–726. [DOI] [PubMed] [Google Scholar]
- Eastman Q.M., Leu,T.M. and Schatz,D.G. (1996) Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature, 380, 85–88. [DOI] [PubMed] [Google Scholar]
- Gellert M. (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- Goldhaber-Gordon I., Williams,T.L. and Baker,T.A. (2002) DNA recognition sites activate MuA transposase to perform transposition of non-Mu DNA. J. Biol. Chem., 277, 7694–7702. [DOI] [PubMed] [Google Scholar]
- Hiom K. and Gellert,M. (1997) A stable RAG1–RAG2–DNA complex that is active in V(D)J cleavage. Cell, 88, 65–72. [DOI] [PubMed] [Google Scholar]
- Hiom K. and Gellert,M. (1998) Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell, 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- Hiom K., Melek,M. and Gellert,M. (1998) DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell, 94, 463–470. [DOI] [PubMed] [Google Scholar]
- Jeggo P.A. (1998) Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat. Res., 150, S80–S91. [PubMed] [Google Scholar]
- Jones J.M. and Gellert,M. (2002) Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J., 21, 4162–4171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M. and Hesse,J.E. (1991) Cutting and closing without recombination in V(D)J joining. EMBO J., 10, 3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1988) Novel strand exchanges in V(D)J recombination. Cell, 55, 1099–1107. [DOI] [PubMed] [Google Scholar]
- Ma Y., Pannicke,U., Schwarz,K. and Lieber,M.R. (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell, 108, 781–794. [DOI] [PubMed] [Google Scholar]
- McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- Melek M. and Gellert,M. (2000) RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell, 101, 625–633. [DOI] [PubMed] [Google Scholar]
- Melek M., Gellert,M. and van Gent,D.C. (1998) Rejoining of DNA by the RAG1 and RAG2 proteins. Science, 280, 301–303. [DOI] [PubMed] [Google Scholar]
- Mo X., Bailin,T., Noggle,S. and Sadofsky,M.J. (2000) A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res., 28, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morzycka-Wroblewska E., Lee,F.E. and Desiderio,S.V. (1988) Unusual immunoglobulin gene rearrangement leads to replacement of recombinational signal sequences. Science, 242, 261–263. [DOI] [PubMed] [Google Scholar]
- Neiditch M.B., Lee,G.S., Landree,M.A. and Roth,D.B. (2001) RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol. Cell. Biol., 21, 4302–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri A., Barriga,F., Knowles,D.M., Magrath,I.T. and Dalla-Favera,R. (1988) Different regions of the immunoglobulin heavy-chain locus are involved in chromosomal translocations in distinct pathogenetic forms of Burkitt lymphoma. Proc. Natl Acad. Sci. USA, 85, 2748–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell P.C. (1997) Genetic alterations in leukemias and lymphomas: impressive progress and continuing complexity. Cancer Genet. Cytogenet., 94, 13–19. [DOI] [PubMed] [Google Scholar]
- Oettinger M.A. (1992) Activation of V(D)J recombination by RAG1 and RAG2. Trends Genet., 8, 413–416. [DOI] [PubMed] [Google Scholar]
- Ramsden D.A., McBlane,J.F., van Gent,D.C. and Gellert,M. (1996) Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J., 15, 3197–3206. [PMC free article] [PubMed] [Google Scholar]
- Ramsden D.A., Paull,T.T. and Gellert,M. (1997) Cell-free V(D)J recombination. Nature, 388, 488–491. [DOI] [PubMed] [Google Scholar]
- Roth D.B. and Roth,S.Y. (2000) Unequal access. Regulating V(D)J recombination through chromatin remodeling. Cell, 103, 699–702. [DOI] [PubMed] [Google Scholar]
- Savilahti H. and Mizuuchi,K. (1996) Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell, 85, 271–280. [DOI] [PubMed] [Google Scholar]
- Segel I.H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. John Wiley and Sons, New York, NY, pp. 465–469.
- Sekiguchi J.A., Whitlow,S. and Alt,F.W. (2001) Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell, 8, 1383–1390. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Bassing,C.H., Bardon,C.G., Okada,A., Khor,B., Bories,J.C., Monroe,R. and Alt,F.W. (1998) Accessibility control of variable region gene assembly during T-cell development. Immunol. Rev., 165, 121–130. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. (1983) Somatic generation of antibody diversity. Nature, 302, 575–581. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Gorham,J., Cossman,J., Jaffe,E. and Croce,C.M. (1985) The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science, 229, 1390–1393. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Bashir,M.M., Givol,I., Cossman,J., Jaffe,E. and Croce,C.M. (1987) DNA rearrangements in human follicular lymphoma can involve the 5′ or the 3′ region of the bcl-2 gene. Proc. Natl Acad. Sci. USA, 84, 1329–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D.C., McBlane,J.F., Ramsden,D.A., Sadofsky,M.J., Hesse,J.E. and Gellert,M. (1995) Initiation of V(D)J recombination in a cell-free system. Cell, 81, 925–934. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Ramsden,D.A. and Gellert,M. (1996) The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell, 85, 107–113. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Hiom,K., Paull,T.T. and Gellert,M. (1997) Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J., 16, 2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]