Abstract
Mitochondrial citrate synthase (mCS) is the initial enzyme of the tricarboxylic acid (TCA) cycle. Despite the key position of this protein in respiratory metabolism, very few studies have addressed the question of the effects of the absence of mCS in development. Here we report on the characterization of 15 point mutations and a complete deletion of the cit1 gene, which encodes mCS in the filamentous fungus Podospora anserina. This gene was identified genetically through a systematic search for suppressors of the metabolic defect of the peroxisomal pex2 mutants. The cit1 mutant strains exhibit no visible vegetative defects. However, they display an unexpected developmental phenotype: in homozygous crosses, cit1 mutations impair meiosis progression beyond the diffuse stage, a key stage of meiotic prophase. Enzyme assays, immunofluorescence and western blotting experiments show that the presence of the mCS protein is more important for completion of meiosis than its well-known enzyme activity. Combined with observations made in budding yeast, our data suggest that there is a general metabolic checkpoint at the diffuse stage in eukaryotes.
Keywords: citrate synthase/development/filamentous fungi/meiosis/organelles
Introduction
Mitochondrial citrate synthase (mCS) is the initial enzyme of the tricarboxylic acid (TCA) cycle, which plays a central role in aerobic energy production and metabolite interconversions. One of the key compounds synthesized in the TCA cycle is α-ketoglutarate, a precursor of glutamate which is itself a precursor for the synthesis of other amino acids. Lack of CS activity leads to glutamate auxotrophy and to respiratory defects. Thus, in strict aerobes, a functional TCA cycle is required for viability. In plants and fungi, a CS activity (pCS) is also found in peroxisomes (glyoxysomes) in which it catalyzes the first step of the glyoxylate cycle, which participates in gluconeogenesis. There is a tight metabolic interaction between the two organelles due to the shuttling of some metabolites between peroxisomes and mitochondria (Tolbert, 1981; Gancedo and Serrano, 1989). The key position of mCS in respiratory metabolism and its high level of conservation through evolution explain why the genes (or cDNAs) encoding this enzyme have been cloned in many eukaryotes from fungi to plants and animals. This protein was also crystallized nearly 20 years ago (Remington et al., 1982). It is thus surprising that inactivation of these genes, or inhibition of their expression, have been reported so far in only two eukaryotes: the yeast Saccharomyces cerevisiae and potato plants.
The S.cerevisiae nuclear genome contains three genes encoding CS isoforms (Suissa et al., 1984; Kim et al., 1986; Jia et al., 1997). The CIT2-encoded protein (Cit2p) is located in peroxisomes and participates in the glyoxylate cycle (Lewin et al., 1990). The CIT1 (Suissa et al., 1984; Rickey and Lewin, 1986) and CIT3 (Jia et al., 1997) products (Cit1p and Cit3p, respectively) are located in mitochondria. Cit1p is the major mitochondrial isoform (Jia et al., 1997) and participates in the TCA cycle. It is worth noting that S.cerevisiae Δcit1 strains do not show any significant respiratory defects when grown on non-fermentable carbon sources other than acetate (e.g. glycerol and lactate). The precise role of Cit3p has not yet been elucidated. Deletion of CIT3 does not cause any respiratory defect, and the Δcit1Δcit3 strains are still able (albeit very slowly) to grow on glycerol medium (Jia et al., 1997). The fact that strains devoid of mCS maintain respiratory abilities may be explained by metabolic cross-feeding of citrate produced by pCS (Cit2p): expression of the CIT2 gene is strongly increased in the absence of CIT1 (Liao et al., 1991). However, the presence of pCS is not able to suppress the inability of cit1 mutants to grow on acetate-containing media.
In potato plants, mCS activity was reduced to 6% of wild-type activity by using an antisense RNA approach (Landschutze et al., 1995). Surprisingly, despite this strong reduction in mCS activity, no significant changes were observed during vegetative growth and development. Furthermore, there were no differences in respiration rates between wild-type and transgenic plants. In contrast, a severe effect on flower formation was observed. In the most strongly inhibited lines, flowers were either not formed or were aborted at an early stage of development. In these latter cases, the ovaries appeared disintegrated (Landschutze et al., 1995). These striking features of plants displaying a dramatic reduction of mCS activity remain unexplained.
We report here a third example of mCS defects in a eukaryotic organism. The cit1 gene encoding mCS in the filamentous fungus Podospora anserina has been characterized. Sixteen mutant strains bearing either a point mutation or a complete deletion of the gene have been submitted to extensive analyses combining molecular, immunological, cytological and genetic approaches. The striking and unexpected property of the cit1 mutants is their developmental defect: in mutant × mutant crosses, meiosis hardly proceeds beyond a particular stage of the meiotic prophase, the diffuse stage. As stressed in the Discussion, this stage is a major landmark of oogenesis for a number of animal species and of meiosis commitment for S.cerevisiae. Altogether, these observations and our data lead us to propose that there is a general metabolic checkpoint at the diffuse stage in eukaryotes and that lack (or decreased production) of mCS activates this checkpoint.
Results
Identification and cloning of the cit1 gene
In P.anserina, the pex2 gene (formerly car1) encodes the PEX2 protein, involved in peroxisome biogenesis (Distel et al., 1996). The pex2 mutants are unable to grow on long-chain fatty acids (e.g. oleic acid) as sole carbon sources (Berteaux-Lecellier et al., 1995). A systematic search for mutations able to restore growth of the pex2 mutants on oleic acid led to the isolation of 63 extragenic suppressors of the pex2 mutations. These suppressors fall in six genes called suo, for suppressors on oleic acid (Ruprich-Robert et al., 2002). Twenty-five percent (16/63) of the suo mutations lie in the suo4 gene, which was re-named cit1 after its sequencing (see below). In a pex2+ background, most cit1 mutations cause a developmental defect: they impair development of fruiting bodies (perithecia) and sporulation when homozygotes in a cross (see below for a detailed description). In addition, with the exception of two alleles, mutant ascospores, obtained from heterozygous crosses, do not ripen and exhibit a low germination level. Otherwise, these mutants have no visible vegetative defects. In particular, their growth rates are similar to those of wild-type strains on current media, including those containing oleic acid as sole carbon source (Ruprich-Robert et al., 2002).
The cit1 mutants are recessive, and their developmental defect was used to clone the cit1 gene by complementation of the cit1-1 mutant (see Materials and methods). Sequence analysis revealed that this gene encodes an mCS, the initial enzyme of the TCA cycle. The P.anserina protein shows 69, 60 and 45% identity with the mCS from S.cerevisiae (Cit1p), pig and Arabidopsis thaliana, respectively. The highest similarities were observed with the enzymes from filamentous fungi. The Neurospora crassa (Ferea et al., 1994) and P.anserina polypeptides exhibit >90% identity across an overlap of 469 amino acids. This strong similarity extends into the putative mitochondrial targeting sequences (67% identity across a 27 amino acid overlap). Furthermore, the P.anserina cit1 gene contains three introns located at the same positions as three of the four introns observed in the N.crassa cit1 gene (see DDBJ/EMBL/GenBank accession Nos M84187 and AJ296102 for the N.crassa and P.anserina sequences, respectively). The P.anserina CIT1 polypeptide shows 79% identity with its Aspergillus nidulans homolog (Park et al., 1997; accession No. U89675). The P.anserina genome probably contains a single copy of the cit1 gene as suggested by hybridization experiments at low stringencies (Southern and northern blots, data not shown).
The abundance of the cit1 mRNA was measured in northern blots after growth of the wild-type strain in current culture conditions and in the presence of oleic acid. We observed that cit1 is highly expressed: the level of its mRNA does not differ significantly from the level of the AS1 messenger which encodes a protein of the cytosolic ribosomes (Dequard-Chablat and Sellem, 1994). Moreover, oleic acid does not increase the transcription level of cit1 (data not shown).
Molecular mapping of 16 cit1 mutations and construction of a Δcit1 strain
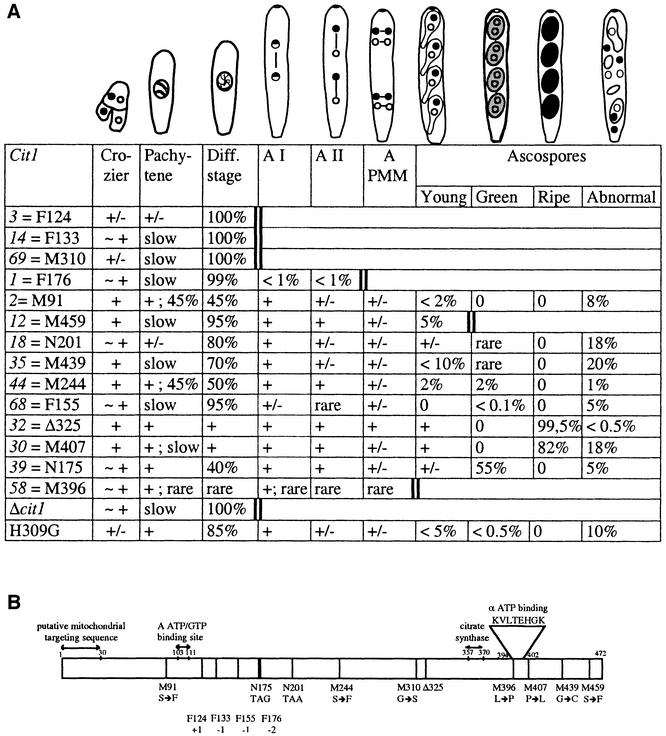
To ascertain that cit1 was indeed the relevant complementing gene, it was sequenced in the 16 mutants available. They all contain a mutation in the cit1 coding region. As shown in Figure 1B, the cit1 mutations are scattered along the coding part of the gene from codon 91 to codon 459. Half of the mutations are either frameshift (5/16) or nonsense (3/16) mutations; they are located between codons 124 and 201. Surprisingly, although all mutants were recovered independently, one frameshift (F176) and one nonsense (N175) mutation were found twice. This may reveal a hot spot for UV mutations: in particular, the frameshift mutations (loss of 2 bp) occurred in a stretch of six pyrimidines. In the following analyses, we used one of the two N175 and F176 mutants, cit1-39 and cit1-1, respectively. With the exception of cit1-2 (codon 91), the missense mutations lie in the second half of the gene (codons 244–459). These positions correspond to amino acid residues highly conserved throughout evolution: at the seven positions, the same residues are found in the wild-type mCS sequences of P.anserina, N.crassa, A.nidulans and pig, while, at six positions, the same residues are also observed in the mCS of S.cerevisiae (CIT1 product) and A.thaliana. The cit1-32 allele was different: it corresponds to the loss of one of two adjacent lysines (positions 324–325). This residue at position 325 is conserved in the wild-type mCS sequences of the three filamentous fungi and the pig. The conclusion of this analysis was that none of these mutants was a bona fide null mutant. In fact, all the mutant polypeptides keep the mitochondrial targeting sequence. Thus, if the mutant proteins are stable enough, they could be imported into mitochondria and disturb the organelle functions independently of their lack of activity. The developmental defects could be explained in at least two, non-exclusive, ways: lack of CS activity and/or poisoning of a cellular function due to the presence of the altered proteins. This conclusion prompted us to obtain a cit1-null mutant (Δcit1).
Fig. 1. Fine analysis of the cit1 mutants. (A) Ascus phenotypes: + indicates that the stage is normal when compared with wild type; +/– means that part of the asci are abnormal as, for example, abnormal nuclear migration during post-meiotic mitosis (PMM); ∼ + indicates that several cells show abnormalities as illustrated in Figure 3; A = anaphase. Percentages were calculated from 100–200 perithecia. A double bold bar indicates the stage when all asci are blocked. (B) Mutation map. M, N and F denote missense, nonsense and frameshift mutations, respectively. Numbers show the codon positions. Δ325 refers to loss of a lysine residue at position 324 or 325. Δcit1 is a complete deletion of the gene (see Figure 2). The cit1 sequence has been submitted to DDBJ/EMBL/GenBank under the accession No. AJ296102.
A strain carrying a complete deletion of the gene was constructed through gene substitution. The cit1 coding sequence was replaced in vitro by the bacterial hph (hygromycin resistance) gene under the control of a eukaryotic promoter (Materials and methods; Figure 2). Our rationale was that, like the original cit1 mutations, the null allele should act as a metabolic suppressor of the pex2 mutants. A linear DNA fragment containing the deleted gene was thus used to transform a pex2-1 strain. Hygromycin-resistant transformants were recovered and tested for their ability to grow on oleic acid as sole carbon source. Twelve transformants (from 240) displayed this property. Southern blot analysis of six of them confirmed that the resident cit1 gene was indeed replaced by the Δcit1 construct (data not shown). It was also ascertained that crosses between these Δcit1 strains and the original cit1-1 mutant yielded barren perithecia.
Fig. 2. cit1 gene replacement. The upper line shows the restriction map of the 5 kb genomic fragment containing the cit1 gene with the length of the restriction fragments indicated in base pairs. The lower line shows the DNA fragment in which cit1 was replaced by the bacterial hygromycin resistance gene (hph) under the control of the A.nidulans trpC promoter (PtrpC). The positions of the oligonucleotides used to amplify the 3′-flanking region of cit1 are shown by arrows. See Materials and methods for more details.
Crosses between the pex2-1 Δcit1 transformants and a wild-type (pex2+ cit1+) strain led to the recovery of pex2+ Δcit1 strains, which were submitted to extensive phenotypic analyses. The Δcit1 mutation does not lead to any visible vegetative defect. As P.anserina is a strict aerobe, it is worth noting that growth, respiration rates and life spans of the Δcit1 strains are similar to those of the wild type (data not shown). Moreover, the mutant mitochondria do not differ from the wild-type organelles with respect to number, size and distribution (Materials and methods). However, as observed for most of the original mutants, Δcit1 ascospores (obtained from heterozygous crosses) do not ripen: they are green (while the wild-type ascospores are black) and their germination efficiency is low (10% instead of near 100% for the wild-type ascospores). Although glutamate auxotrophy is a classic CS-deficient phenotype (Carls and Hanson, 1971; Kim et al., 1986), the P.anserina Δcit1 grows as well as the wild-type strains on minimal medium. The germination defect of the mutant is not due to a conditional (developmental) auxotrophy with respect to this compound: when glutamate was added to the germination medium, the level of Δcit1 germination remained unchanged.
Developmental defects of the cit1 mutants
In heterozygous crosses, cit1 mutant strains can be used as either a male or female partner without any obvious consequences, except that fruiting body (perithecium) development is slightly delayed when the mutants are used as the female partner. In contrast, all perithecia from homozygous mutant crosses are barren, with the exception of three mutants (see below).
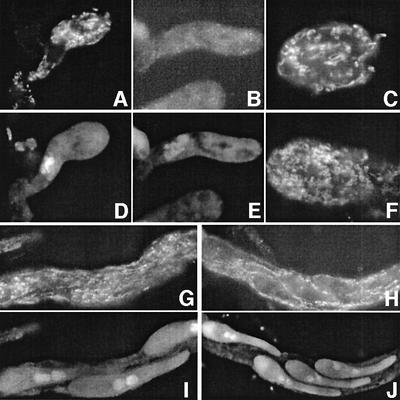
Detailed analyses of the mutant fruiting bodies showed that the first steps of sexual reproduction are normal and that meiosis proceeds normally until late prophase (Figure 1A). Wild-type perithecia contain >150 meiocytes which are not synchronous in their development. Therefore, progression of the successive stages from fertilization to mature asci with four ripe ascospores can be easily observed (Figure 3A). In the mutant perithecia, dicaryotic cells are formed, caryogamy and entry into meiosis occur normally but, as pachytene (Figure 3B) is the most prominent stage seen in mutant perithecia, we infer that pairing and synapsis are slower than in the wild-type strain. Moreover, most meiocytes remain blocked at the diffuse stage (Figure 3C). This stage is characterized by a decondensed appearance of the chromosomes and the loss of their stainability with standard techniques. While the preceding pachytene condition is characterized by a rigid configuration of chromatin, during the following stage, the chromatin exhibits a fuzzy appearance in which individual chromosomes are no longer visible (compare Figure 3B and C). In wild type, this diffuse stage is followed by the re-emergence of condensed, individualized chromosomes in the classical diplotene condition (see Discussion for additional information). The arrest observed in mutant perithecia is not due to a delay in exiting this stage because asci remain blocked even in perithecia followed over 10 days after the end of wild-type perithecium development.
Fig. 3. Meiosis and sporulation of cit1 mutants. (A) Wild type: the upper ascus contains ascospores with two nuclei each (small arrowhead); the nucleus of the lower ascus is at the diffuse stage (large arrowhead). (B) cit1-3: note that the four asci are at the pachytene stage; arrowheads point to two nuclei in which the synapsed homologs are clearly visible. (C) Three asci of cit1-1 at the diffuse stage; the arrowhead points to the prominent nucleolus, characteristic of this stage. (D and E) cit1-39: this mutant forms either normal (wild type-like) (D) or abnormal ascospores (E). (F and G) Δcit1 is completely blocked at the diffuse stage; (F) note the fuzzy appearace of the nucleus in which individual chromosomes are no longer visible (compare with B); (G) lower magnification of a Δcit1 group of eight asci. Bars: 5 µm.
Ten of the 14 different mutants exhibit this general picture with a more or less pronounced leakiness (Figure 1A). Three of them, cit1-3, cit1-14 and cit1-69, which correspond to the two earliest frameshift mutations and to a missense mutation, respectively, show no leakiness at all: 100% of asci are blocked at the diffuse stage. Seven other mutants belong to a second subclass in which 70–99% of asci do not proceed further than the diffuse stage. The other asci evolve to later stages and, with the exception of cit1-1 (F176), even form ascospores. However, most of them are abnormal (Figure 1A). Four mutants clearly stand out: cit1-32 (Δ325), cit1-30 (M407), cit1-39 (N175) and cit1-58 (M396). cit1-32 perithecia develop normally and almost all asci form ripe (black) ascospores which are ejected from the fruiting bodies and germinate as observed in the wild type. cit1-30 perithecia develop very slowly but asci do not show any specific arrest and all form ascospores. The ascospores, which are ejected 3 days later than in the wild-type condition, are black and germinate normally. Almost half of the cit1-39 asci also contain four ascospores (Figure 3D) but they are green (unripe) and unable to germinate. In contrast to all other mutants, cit1-58 perithecia contain very few asci, which show normal prophase but are blocked before ascospore formation (Figure 1A). Although the diffuse stage arrest appears as the most prominent landmark of the cit1 mutants, it is worth noting that most mutants also exhibit abnormalities before and/or after this stage, i.e. during crozier development, during nuclear migration events and finally during ascospore formation (Figures 1A and 3E).
Amazingly, there is no clear-cut correlation between the developmental phenotypes of the mutants and the type of the mutations (Figure 1). Cytological analysis of the perithecia from Δcit1 homozygous crosses partially solved this problem. In these crosses, the asci are all arrested at the diffuse stage (Figure 3F and G). Therefore, the leakiness exhibited by some of the cit1 point mutations is likely to be due either to a residual activity of the mutant enzyme or to the presence of an inactive CIT1 protein still able to perform an unknown function required for progression of meiosis (see Discussion). To clarify this point, we constructed a strain able to express a catalytically inactive but structurally unchanged form of CIT1. In S.cerevisiae, a cit1 mutant with the substitution mutation H313G was shown previously to produce such an mCS form (Kispal et al., 1989). The S.cerevisiae Cit1p H313 residue (i.e. H274 in the mature pig enzyme) is one of the conserved residues thought to be essential for mCS activity (Karputas et al., 1990). It corresponds to H309 in P.anserina CIT1. The cit1-H309G allele was obtained by in vitro mutagenesis and introduced into a Δcit1 background (Materials and methods). This new mutant displays the same phenotypic properties as those observed in the original mutants. The cit1-H309G ascospores, from heterozygous crosses, are unripe and exhibit a low germination level. Perithecia from homozygous crosses are barren but contain asci, of which 85% are arrested at the diffuse stage while 15% contain ascospores among which rare wild-type-like four-spored asci are found (Figure 1A). To ensure that cit1-H309G indeed contained an inactive mCS form, we checked the status of the CIT1 protein in this mutant, in comparison with Δcit1 and a subset of the original mutants.
Status of the CIT1 protein in the cit1 mutants
Immunofluorescence experiments using an antibody directed against the S.cerevisiae Cit1p protein (Vélot et al., 1999) were performed on the wild-type strain and a selected set of cit1 mutants. We chose the mutants that exhibit the two extreme developmental phenotypes, i.e. Δcit1 and cit1-32 (Δ325), as well as the cit1-39 nonsense (N175) mutants. In addition, crude extracts of these strains were used for CS activity measurements and immunoblot analysis. As shown in Table I, global CS activity is reduced to 25% of the wild-type activity in both Δcit1 and cit1-39 strains, while it reaches 50% of the control in the cit1-32 mutant. These data lead to three conclusions. First, as deduced from the residual CS activity observed in Δcit1, the cit1 gene does not control all the cellular CS activity. Secondly, the cit1-32 mutant, which exhibits a wild-type phenotype, shows a CS activity intermediate between the wild-type and Δcit1 values. This suggests that the mutant protein is still partially active. Thirdly, the CS activity observed in the nonsense (cit1-39) mutant does not differ significantly from the residual activity present in the null (Δcit1) mutant (ratio 0.9 for cit1-39/Δcit1). As expected, the same ratio was observed between the cit1-H309G and Δcit1 activities. This demonstrates that the stringency of the meiotic phenotypes is not related to the level of the residual CS activity.
Table I. Summary of data obtained on the most relevant strains studied.
| Strain | Mutation | CS activity (%) in crude extractsa |
In situ mCS detection |
Developmental phenotypeb |
||
|---|---|---|---|---|---|---|
| Asci | Spores | Asci containing spores (%) | Ejected spores | |||
| Wild type | 100 | + | + | 100 | Ripe | |
| Δcit | Deletion | 25 | – | 0 | ||
| cit1-1 | F176 | ND | – | 0 | ||
| cit1-39 | N175 | 25 | + | – | 60 | Unripe |
| cit1-30 | M407 | ND | + | + | 100 | Ripe |
| cit1-32 | Δ325 | 50 | + | + | 100 | Ripe |
aSpecific activities were measured on crude extracts and are expressed as a percentage of wild-type activity. Average values were determined from either two (wild-type strain) or three (mutant strains) independent experiments. Measurements were reproducible within 15%.
bSee Figure 1 for more details.
ND, not determined.
Immunofluorescence clearly revealed a mitochondrial protein akin to CS in young wild-type asci (Figure 4A), while no staining was detected in the Δcit1 asci observed at the same developmental stage (Figure 4B). The same result was obtained with the cit1-1 (F176) frameshift mutant (data not shown). In contrast, the staining was similar to wild-type asci all through their development, including ascospores, in cit1-32 (compare Figure 4C and F), cit1-30 (M407) and cit1-H309G (data not shown). The nonsense cit1-39 mutant exhibits two major differences when compared with the wild type. First, in the young mutant asci, staining is weak and limited to a few patches instead of the typical network observed in the young wild-type asci. The second difference concerns the older asci, which contain ascospores: while, in wild type, the mitochondrial network was stained by anti-CIT1 in both the ascus compartment and the ascospores, this network was only seen in the ascus compartment of cit1-39, and not inside the mutant ascospores (compare Figure 4G and I with H and J).
Fig. 4. In situ staining with anti-mCS antibody. (A) A wild-type ascus at the pachytene stage: the clear mCS staining reveals a large mitochondrial network. (B) A Δcit1 ascus: no labeling is observed. (C) A wild-type ascospore: a dense mitochondrial network is observed. (D and E) DAPI staining corresponding to (A) and (B), respectively. (F) A cit1-32 mutant ascospore: the staining pattern is similar to the wild type (compare with C). (G and H) Asci containing three (G, wild type) or four (H, cit1-39) young ascospores: the mitochondrial network is seen in both cases but note that it is not detected inside the cit1-39 ascospores. (I and J) DAPI staining corresponding to (G) and (H), respectively. Bars: 1 µm.
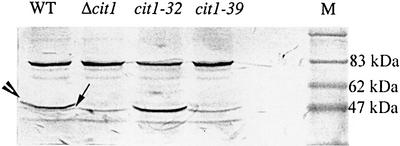
Additional data were obtained with western blotting (Figure 5). Although the polyclonal anti-Cit1p antibody from S.cerevisiae reveals few unspecific bands, the cit1 product can be identified unambiguously by its presence at the expected molecular size (52 kDa) in the wild-type and cit1-32 extracts, and its absence in Δcit1. Neither a truncated form nor a readthrough product can be seen in the cit1-39 extracts. However, in the four extracts, the antibody reveals, just below the CIT1 signal, a faint band, which might correspond to another isoform of CS.
Fig. 5. Immunoblot analysis of crude extracts from cit1 mutants and wild-type strains. Crude protein extracts (80 µg) from the indicated strains were subjected to immunoblot analysis with an anti-Cit1p polyclonal antiserum as described in Materials and methods. The large arrowhead and the thin arrow indicate mCS and a putative isoform, respectively. Numbers on the right indicate the size (in kDa) of the SDS–PAGE standards of lane M (BioLabs).
Autonomous ascus development due to intragenic complementation and recombination
mCS is known to act as a dimer. We thus addressed the question of possible cases of intragenic complementation between cit1 mutants. Mutant × mutant crosses were performed in all possible combinations of the 15 mutants, including Δcit1. In addition to crosses involving Δcit1, homoallelic crosses (in which each mutant was crossed with itself) were used as controls. Intragenic complementation, occurring inside the fruiting bodies after fertilization, should lead to the production of ripe, black ascospores, while lack of complementation should give the same results as those observed in the homoallelic crosses. The cit1-32 (Δ325) allele appeared dominant with respect to all other mutants: all crosses involving this mutant produced large amounts of black ascospores. The leakiness of cit1-30 (M407) and cit1-39 (N175) in crosses with other mutants (except cit1-32) was never increased and sometimes decreased. Complementation was observed only in crosses between missense mutants. The amounts of ascospores from these crosses varied, according to the mutant combinations, from a few percent to 100% of the wild-type controls. In all cases, the ascospores exhibited a wild-type phenotype. The highest levels of complementation were observed in combinations involving cit1-12 (M459), i.e. the most C-terminal missense mutant. As expected for a mutant expressing a structurally intact protein, cit1-H309G showed complementation with cit1-12.
In the course of this study, we carefully analyzed all crosses, even those that clearly did not show complementation. Among the latter, we observed the following remarkable data. A number of crosses led to the production of ∼10–4–10–5 asci, which could be explained only by intragenic recombination for the following reasons. First, these asci were absent in the negative controls, i.e. homocaryotic crosses and crosses involving Δcit1. Secondly, these amounts were in the range of the intragenic recombination (conversion) frequencies observed in P.anserina (Touré, 1972). Thirdly, the observed ripe (black) ascospores germinated and their analysis demonstrated that they contained a wild-type (cit1+) nucleus. The explanation is that intragenic recombination, which occurs during the early meiotic prophase (before the meiotic block characteristic of the cit1 mutants), leads to a wild-type cit1 allele which is fully expressed (probably during the diffuse stage) and permits an autonomous development of the relevant ascus up to its normal end, namely ascospore formation. However, in these asci, the ripening of the ascospores is limited to those containing the recombinant wild-type nucleus. Genetic analysis of the recombinant ascospores also showed that the mat locus, which does not map on the same chromosome as cit1, exhibited the same second division segregation frequency as in a wild-type cross (proof of the occurrence of at least one crossing-over between the centromere and the mat locus) and segregated normally in the recombinant cit1+ nuclei. Also, the number of chiasmata visible in the mutants that are not completely blocked is similar to that observed in wild type. These observations led to the conclusion that the general patterns of recombination and chromosome segregation were normal in the meiocytes that underwent intragenic recombination. However, to ensure that recombination occurs before the mutant arrest, even in the Δcit1 meiocytes, we used an antibody raised against the Rad51 protein. In yeast, where all meiotic recombination events are initiated by double-strand breaks, immunostaining foci of Rad51p appear only when those breaks have occurred. The Δcit1 strain shows the same amount of foci and the same temporal order of appearance and disappearance as wild type.
cit1 and peroxisomes
All cit1 mutations were screened as partial suppressors of the metabolic defect of pex2 mutants, i.e. as mutations alleviating their inability to grow on oleic acid as sole carbon source. The Δcit1 mutation also exhibits this property. To shed light on the relationship between cit1 and peroxisomes, two experiments were performed.
First, peroxisomes were examined by immunofluorescence in the Δcit1, cit1-1 (F176), cit1-3 (F124), cit1-35 (M439) and cit1-39 (N175) single mutant strains and in the Δcit1 pex2 double mutant strains (Materials and methods). The data were clear-cut: the cit1 mutant asci showed the same peroxisomal pattern as wild-type asci (Berteaux-Lecellier et al., 1995), at least until they arrest in the mutant context (see above and Figure 1), and Δcit1 did not restore peroxisome biogenesis in a pex2 mutant background (data not shown).
Secondly, catalase activities were measured in wild-type, cit1-1, cit1-35 and Δcit1 strains according to the following rationale. It was suspected previously that the inability of pex2 mutants to grow on media containing oleic acid as sole carbon source was due to the lethal effect of H2O2 production in the cytosolic compartment of the mutants in which β-oxidation is mislocalized. There are thus three ways to alleviate this pex2 metabolic defect: (i) restoration of a peroxisomal compartment; (ii) a decrease in β-oxidation efficiency; and (iii) an increase in the efficiency of H2O2 detoxification through, for instance, an increase in catalase activity (Ruprich-Robert et al., 2002). Clearly, the cit1 mutations do not belong to the first and second classes of suppressors: peroxisome biogenesis is not restored in the pex2 cit1 double mutants and the cit1 mutants grow as well as the wild type on a medium containing oleic acid as sole carbon source. However, as shown in Table II, catalase activity is increased 2-fold in Δcit1 strains as compared with wild type, in both the presence and absence of oleic acid. The three mutant strains tested gave the same results. Moreover, growth in the presence of oleic acid increases catalase activity in wild-type and mutant strains. As previously shown in budding yeast, such an increase is characteristic of the peroxisomal catalase (Skoneczny et al., 1988).
Table II. Catalase activities (per minute per milligram) in cell-free extracts of wild-type and Δcit1 strains.
| Strain | Carbon sourcea | Expt 1b | Expt 2b | Expt 3b |
|---|---|---|---|---|
| cit1+ | Dextrin | 0.014 (1) | 0.019 (1) | 0.021 (1) |
| cit1+ | Dextrin + OA | 0.025 (1.8) | 0.041 (2.2) | 0.039 (1.9) |
| Δcit1 | Dextrin | 0.026 (1.9) | 0.041 (2.2) | 0.048 (2.3) |
| Δcit1 | Dextrin + OA | 0.082 (5.7) | 0.079 (4.2) | 0.084 (4.0) |
aOA, oleic acid; it was added to the growth medium 16 h before catalase activity measurements.
bCatalase activities are expressed in arbitrary units. The values in parentheses indicate the increase factors with respect to the wild-type activity with dextrin as the carbon source. In spite of the variations in the crude values observed between experiments, the increase factors appear highly reproducible.
Discussion
In the filamentous fungus P.anserina, mutations of the cit1 gene, which encodes mCS, result in several noteworthy features with three intertwined facets. First, cit1 mutant strains are viable and do not show any significant vegetative defects except a low efficiency in ascospore germination. Notably, their respiration rates are similar to wild type. Secondly, cit1 mutations act as partial suppressors of the peroxisomal assembly pex2 mutations. Thirdly, they impair meiosis progression downstream of the diffuse stage, and a null mutant (Δcit1) is completely blocked at this stage.
A second CS isoform ensures viability of the cit1 mutants
In S.cerevisiae, the respiratory abilities of a Δcit1 strain are explained by the up-regulation of the CIT2 gene (encoding pCS), which would compensate for the loss of mCS through cross-feeding between peroxisomes and mitochondria. This process, in which changes in mitochondrial functions result in alteration of nuclear gene expression, was called retrograde regulation (Liao et al., 1991; Chelstowska et al., 1995).
The viability of the P.anserina cit1 mutants is probably due to the activity of a second CS isoform, as suggested by our enzyme assays: they reveal that Δcit1 strains maintain a residual CS activity that represents 25% of the wild-type global activity. This second isoform may be either mitochondrial or, more probably, peroxisomal. A second gene for mCS has been reported so far only for S.cerevisiae where it seems to play a minor role in the TCA cycle (Jia et al., 1997). The genome of N.crassa (http://www.mips.biochem.mpg.de/proj/neurospora/), a filamentous fungus closely related to P.anserina, contains only two genes for CSs, one encoding the mitochondrial isoform (Ferea et al., 1994; contig 2.66) and the other probably encoding the peroxisomal isoform (contig 2.220). Furthermore, an antibody raised against the mCS (CIT1 product) from S.cerevisiae gives no signal in the mitochondria of the P.anserina Δcit1 strains. The fact that P.anserina cit1 mutants do not show glutamate auxotrophy is also consistent with the peroxisomal hypothesis: in budding yeast, glutamate auxotrophy has been reported only in strains carrying a simultaneous deletion of CIT1 and CIT2 (Kim et al., 1986). Potato plants also survive and show normal respiration rates when mCS production is strongly reduced (Landschutze et al., 1995). In P.anserina and potato plants, the critical experiment for the pCS-controlled survival hypothesis requires the inactivation of the gene encoding the pCS isoform. Unfortunately, all our attempts to find this gene have failed so far. This may be explained by the fact that a small number of pCS have been described to date and that they show low similarities to mCS, with the exception of Cit1p and Cit2p from S.cerevisiae (see La Cognata, 1996).
In spite of their ability to grow on several respiratory substrates, yeast Δcit1 strains fail to grow (Kim et al., 1986; Kispal et al., 1988) or display strongly reduced growth (Jia et al., 1997) on acetate medium. McCammon (1996) suggested a toxic effect of this compound in such a mutant background. The same hypothesis could account for the germination defect observed in the cit1 mutant ascospores, especially because, in P.anserina, germination requires acetate, which cannot be replaced by another carbon source.
Increased catalase activity accounts for partial suppression of the pex2 mutations
The inability of the pex2 mutants to grow on long-chain fatty acids (e.g. oleic acid) as sole carbon source has been explained by the fact that peroxisome biogenesis is impaired in these mutants (Berteaux-Lecellier et al., 1995). In this context, β-oxidation occurs in the cytosol and this misplaced process may be toxic for the cell, due to production of H2O2, which cannot be detoxified efficiently in this compartment (Ruprich-Robert et al., 2002). The fact that cit1 mutations partially restore growth of pex2 mutants on oleic acid can be explained neither by restoration of a peroxisomal compartment nor by a decrease in β-oxidation efficiency. In contrast, the ability of the double mutant strains to grow on oleic acid is probably due to the increase in catalase activity observed in the cit1 mutant context: decomposition of the H2O2 produced in the cytosol through β-oxidation of oleic acid would be efficient enough to allow cell survival. As a 4-fold increase in catalase activity is sufficient to counteract the cytotoxic effects of H2O2 in mammalian cells (Santanam et al., 1999), the 2-fold increase found here may also be sufficient. However, growth on oleic acid of the pex2 cit1 double mutants is far from being as efficient as the wild-type growth. In addition, the double mutant strains die when they are stored for a few days on this medium, while the wild type can survive for months (data not shown). We do not know how catalase activity increases in the cit1 mutants. We assume that this could be part of a response to mitochondrial dysfunction, akin to the retrograde regulation described in budding yeast (Chelstowska et al., 1995).
The mCS protein per se is required for progression of meiosis downstream of the diffuse stage
As for flower development in potato plants (Landschutze et al., 1995), the effect of P.anserina cit1 mutations on ascus differentiation remains puzzling (see Table I). Although appealing, an energy problem cannot explain the data reported in the two systems. First, in plants, mitochondrial dysfunctions mostly impair late events in flower development, especially production of functional pollen (Conley and Hanson, 1995). Secondly, in P.anserina, respiration-deficient mutants are unable to differentiate female organs and are thus blocked far upstream of a meiotic defect. This is observed, for example, in a strain carrying an inactivated cox5 gene, which encodes a subunit of cytochrome c oxidase (Dufour et al., 2000). When the respiratory defect of this mutant is partially overcome and, consequently, its energy production enhanced, female fertility is partially restored but most of the asci are arrested at different steps of their development and not at a precise stage (Lorin et al., 2001). Moreover, the meiotic block triggered by the null cit1 mutation occurs when the ascus has nearly reached its maximum size, and both growth and size of the asci are similar in the wild-type and the Δcit1 contexts. A limited energy supply would have been expected to cause a less specific or an earlier block during ascus differentiation and elongation. Such a phenotype is encountered only in the cit1-58 mutant: it differentiates few asci and shows no specific meiotic arrest. This mutant bears a stringent (L→P) mutation in a highly conserved domain (Figure 1B) and the mutant protein might be toxic to mitochondrial metabolism. In contrast, half of the cit1-39 (N175) asci contain ascospores, although the mutant strain harbors a truncated CIT1 protein. The cit1-39 leakiness cannot be explained by a readthrough of the stop codon: this process would produce too few mCS molecules to explain the high frequency of ascospore production. More importantly, the catalytically inactive cit1-H309G mutant appears leaky with respect to the developmental defect. Thus, a limited energy supply cannot account for the developmental phenotypes of the cit1 mutants.
Interestingly, several correlations can be deduced from the mutant phenotypes (Table I). First, the cit1-32 mutant seems to retain enough mCS activity to ensure a near wild-type developmental phenotype. Secondly, the residual CS activity observed in the null mutant (Δcit1), although sufficient for vegetative life, cannot compensate for the lack of mCS during sporulation. Thirdly, lack of mCS activity per se is not responsible for the meiotic block and the subsequent lack of ascospore formation: the same residual CS activity is observed in the null mutant and in cit1-39 (N175), of which more than half of the asci form ascospores. Last, but not least, the phenotypic properties of the cit1-39 mutant lead to the conclusion that the mCS protein is more important than its activity. The truncated protein, which has lost the 300 C-terminal amino acids, is detected in asci containing ascospores but it could not be detected either in the ascospores themselves or during vegetative growth (western blotting). This suggests that the mutant mRNA or the mutant protein are unstable during these phases of the life cycle. However, the possibility that the truncated protein detected in vivo would not be stable anymore in the crude extracts cannot be excluded. In any case, the presence of this truncated protein in the asci correlates with their ability to form ascospores, while lack of the protein in the ascospores correlates with their inability to ripen and to germinate. The cit1-H309G mutant provides definitive evidence for an unknown function for the CIT1 protein: in this mutant, the presence of a catalytically inactive enzyme, which is probably structurally unchanged, leads to a partial bypass of meiotic arrest. All these observations disclose an unexpected role for the mCS protein, in addition to its well-known enzymatic function.
In budding yeast, pCS cannot fully compensate the lack of mCS: Δcit1 strains do not grow on acetate medium. Kispal et al. (1989) and Vélot et al. (1999) have demonstrated that restoration of a full wild-type phenotype (i.e. growth on acetate) requires the presence, inside the mitochondria, of a molecule akin to mCS and able to interact with its partners in the TCA complex: this can be an inactive but structurally unchanged form of mCS (Kispal et al., 1989). A large body of experimental data supports the idea that the sequential TCA cycle enzymes form a supramolecular complex, allowing enzymatic reactions to occur via channeling of metabolites between enzyme active sites (Srere et al., 1997). In addition, using a transdominant genetic approach, Vélot and Srere (2000) have shown that mCS from S.cerevisiae (Cit1p) does interact in vivo with its sequential enzyme malate dehydrogenase. In P.anserina, lack of mCS could alter the TCA complex in a discrete way, impairing a function involved in meiosis progression. Alternatively, mCS per se may be a bifunctional protein as previously shown for other mitochondrial proteins. Interestingly, at least four of these proteins function in the TCA cycle: the NAD+-dependent isocitrate dehydrogenase (Elzinga et al., 1993), two subunits of α-ketoglutarate dehydrogenase, and aconitase (Kaufman et al., 2000). It was shown that these proteins bind either mitochondrial mRNAs or DNA. The possible partners of the P.anserina mCS remain unknown. In any case, with this view, the degree of leakiness of a given cit1 mutant would reflect the capacity of the relevant CIT1 protein to participate still in the TCA complex or to bind another partner. The cit1-39 and cit1-H309G alleles exemplify this idea.
Evidence for a meiotic checkpoint at the diffuse stage
Checkpoints were defined by Hartwell and Weinert (1989) as control mechanisms which ensure the proper order of events in the cell cycle by arresting or delaying progression through the cycle in response to defects in cellular processes. In both mitosis and meiosis, checkpoints avoid the production of defective cells. For instance, meiocytes that fail to complete recombination are arrested at the pachytene stage (Roeder and Bailis, 2000).
Beyond pachytene, the diffuse stage is seen in several animal, plant and fungal species (Barry, 1969; Klasterska, 1976; Zickler, 1977). This stage coincides with dissolution of the synaptonemal complex and is followed by the diplotene stage, with homologs widely separated except at the positions of chiasmata. In many organisms, the diffuse stage is transient, but in other species it can be of very long duration as in amphibian and mammalian oocytes. In the latter cases, it correlates with nuclear growth and/or the development of giant lampbrush chromosomes. In S.cerevisiae, the end of prophase is also the stage when cells are committed to completion of the meiotic division (Esposito and Klapholz, 1981). Lee and Honigberg (1996) have shown that, at least in budding yeast, separate nutritional controls act at initiation of meiosis and for meiosis commitment. When the nutritional conditions required for the transition to meiotic completion are not fulfilled, the cells arrest after synaptonemal complex formation and dissolution (Lee and Honigberg, 1996), thus precisely at the diffuse stage. Thus, completion of meiosis progression requires not only fulfillment of the well-known checkpoints but also appropriate nutritional signals (Honigberg and Lee, 1998). Whether S.cerevisiae cit1 mutants, which are unable to sporulate in an acetate-containing medium, exhibit a precise meiotic arrest in this condition and/or in a sporulation medium containing another respiratory substrate unfortunately remains unknown.
To our knowledge, the P.anserina cit1-3 (F124), cit1-14 (F133), cit1-69 (M310) and Δcit1 are the first mutants specifically blocked at the diffuse stage. As it appears unlikely that a mitochondrial enzyme would be a positive regulator of meiosis progression, we propose that this specific arrest discloses a checkpoint, which is activated in the absence of the mCS protein. Lack of CIT1 could cause a specific defect in meiosis, which, in turn, could be sensed by an unknown protein triggering the arrest. The critical experiment would be to identify this protein through mutations allowing Δcit1 cells to complete meiosis but with defective ascospore formation. However, the checkpoint hypothesis is already supported by a fact that fulfills one of the major criteria in the definition of checkpoints (Hartwell and Weinert, 1989): when the arrest is overcome, in the leaky mutants, the late events in ascospore formation are impaired (Figure 1A). cit1-H309G is a paradigm for this problem: all spores are abnormal. Obviously, the catalytically inactive but structurally intact protein encoded by this allele is able to elude the checkpoint and thus permits progression of meiosis. The diffuse stage checkpoint would thus avoid the formation of progeny unable to survive, by arresting the cycle in response to a metabolic deficiency, here lack of mCS, required for ascospore germination. The presence of a control mechanism activated by the absence of CIT1 is demonstrated by our recombination data. In P.anserina, each fertilized female organ (perithecium) is able to differentiate >100 asci. Strikingly, when a wild-type cit1 allele is restored in a cell, the production of mCS is sufficient for the full development of this cell among all others blocked at the diffuse stage. This fact shows that the development of asci containing a wild-type cit1 gene through recombination is completely autonomous. This means that each meiocyte is able to sense the signal required for meiosis progression.
The existence of mechanisms that prevent meiosis progression when metabolic requirements are not met is suggested in other organisms. The diffuse stage arrest in amphibian oocytes is probably also an important control point. Indeed, this step is needed for synthesis and storage of mRNAs and proteins, which are required in the early stages of embryogenesis before the late expression of the zygotic genome. Arrest at this precise stage, when the decondensed state of the chromatin favors transcription, makes sense. Our intragenic recombination data show that transcription can also occur at this stage in other organisms, followed by translation and proper targeting of the proteins. One can suggest that a transient arrest at the diffuse stage could allow the meiocytes to express some of the genes required for viability of the progeny: if this requirement is not achieved, the checkpoint is activated. However, as stressed by Roeder and Bailis (2000) for the pachytene checkpoint, the critical question that remains to be addressed in all systems is the nature of the signal(s) that trigger arrest.
Most studies have focused on checkpoint activation by impairment of the meiotic process per se: defects in DNA replication, homolog synapsis, recombination and chromosome segregation. Our data strongly suggest that the meiocytes also sense their metabolic state and avoid progression of meiosis if this could lead to unproductive progeny.
Materials and methods
Strains and media
Podospora anserina is a filamentous ascomycete whose life cycle and general methods for genetic analysis have been described (Rizet and Engelmann, 1949). The car1 mutants were characterized previously (Simonet and Zickler, 1972, 1978; Berteaux-Lecellier et al., 1995). The car1 gene was re-named pex2 in accordance with the unified nomenclature for peroxisome biogenesis factors (Distel et al., 1996). The cit1 mutants were isolated as revertants of the pex2-3 mutant, as growing sectors which appeared, after UV mutagenesis (100 J/m2), on medium containing oleic acid as sole carbon source. The culture and spore germination media have been reviewed recently (Berteaux-Lecellier et al., 1995). When required, sodium glutamate was added to the spore germination medium in the range of 0.02–0.2% (w/v). Life spans were measured as previously described (Marcou, 1961; Contamine and Picard, 1998) for five subcultures from two strains exhibiting a given genotype.
Cosmids, plasmids and bacterial strains
The genomic library used for the transformation experiments has been described (Berteaux-Lecellier et al., 1995). The integrative cosmid vector (pMOcosX) carries as selectable marker the bacterial (hph) hygromycin-resistant gene under the control of a eukaryotic promoter (Orbach, 1994). Subcloning of the cit1 gene was performed using pUC18, pBluescript (Stratagene). Cloning and plasmid preparations were carried out in either Escherichia coli DH5α (Hanahan, 1983) or XL1-Blue (Stratagene).
Cloning procedures
The cit1 gene was cloned by complementation of the cit1-1 sexual defect using the SIB selection method (Akins and Lambowitz, 1985). The library contained ∼6000 cosmids from the entire genome, divided into 60 pools. In the 20th pool tested, among 360 hygromycin-resistant transformants, one presented a wild-type phenotype. Two successive rounds of SIB selection allowed the isolation of the cosmid carrying the cit1 gene. Localization of the gene was obtained according to the procedure developed by Turcq et al. (1990). An EcoRI–HindIII fragment of 2.7 kb competent to complement the cit1-1 mutant was recovered (Figure 2). Transformation experiments were performed as previously described (Picard et al., 1991) except that protoplasts were made with Glucanex (Novo Nordisk Ferment AG) instead of Novozym.
Sequencing
The 2.7 kb fragment was sequenced on both strands with the universal primer and synthetic oligonucleotides. For gene substitution, a larger XhoI–XhoI fragment of 5 kb was also sequenced (Figure 2). The sequences of the mutant alleles were determined after PCR amplification of the gene in each strain. The PCR fragments were prepared for direct sequencing as described (Rosenthal et al., 1993). Sequencing was performed using an automatic sequencing machine (373A DNA sequencer, Applied Biosystems) using a Dye Terminator, Cycle Sequencing Kit (Abi-Prism, Perkin Elmer).
Cytology
Mitochondria were stained in the vegetative cells (mycelium) with the mitochondrion-specific dye 2-(4-dimethylaminostyryl)-1-methylpyri dinium iodide (DASPMI, Sigma) using the procedure described for yeast (McConnell et al., 1990) and adapted for P.anserina (Jamet-Vierny et al., 1997). Staining of meiocytes and immunofluorescence detection of peroxisomes were performed as described (Berteaux-Lecellier et al., 1995). Here also, we used an antibody against the trifunctional peroxi somal enzyme FOX2 of N.crassa (a kind gift of Dr W.-H.Kunau). Immunodetection of mCS was performed using a polyclonal antibody directed against the yeast Cit1p (Vélot et al., 1999) and diluted 3000-fold. The RAD51 antibody (Ab1, Oncogene), raised against a recombinant human protein, was diluted 400-fold. The secondary antibody was fluorescein isothiocyanate-conjugated goat anti-rabbit IgG diluted 80-fold (Sigma).
DNA procedures
Total DNA was extracted by a miniprep method (Lecellier and Silar, 1994). Standard procedures for Southern blotting on neutral membrane (Appligene) were used. The probes were prepared with a T7 Quick Prime labeling kit (Pharmacia). Low-stringency hybridizations were performed at 37°C, according to Church and Gilbert (1984), and the membranes were washed at 50°C, in 2× SSC.
RNA procedures
Total RNA was isolated from mycelia as described (Lockington et al., 1987). Standard procedures for northern blotting on positive membrane (Appligene) were used. For quantitative northern analyses, autoradiograms were quantified against the levels of AS1 mRNA with a phosphorImager (STORM) using ImageQuant software. The RT–PCR experiment was performed with the Titan one tube RT–PCR kit (Roche Diagnostic). The two oligonucleotides used were chosen in order to hybridize, respectively, on the sequence following the start codon and, on the other strand, before the stop codon. This 1.4 kb template for direct sequencing was prepared as described (Rosenthal et al., 1993).
Construct for gene replacement (Figure 2)
The HindIII–EcoRI fragment of 2.7 kb containing the cit1 gene, in the B-XE plasmid (pBluescript vector containing the 3.9 kb XhoI–EcoRI fragment with the cit1 gene and its 5′-flanking region), was substituted by the 1.4 kb fragment containing the trpC::hph construct from pUChygro. This last vector was constructed by cloning, at the SmaI site of pUC18, the 1.4 kb HpaI fragment carrying the trpC::hph construct from pCB1004 (Carroll et al., 1994) The 3′-flanking region of the cit1 gene was amplified by PCR from the B-XH plasmid (pBluescript vector containing the 3.8 kb fragment HindIII–XhoI) (Figure 2). A NotI restriction site was added at the end of the 3′ primer to permit cloning in the B-XE plasmid between the EcoRI and NotI sites, after digestion of the PCR product, which was then sequenced (Figure 2).
Obtaining a strain bearing the cit1H309G allele
To create the B-XE H309G plasmid, site-directed mutagenesis was performed on the B-XE plasmid, according to the procedure described by Briand et al. (2001). The replacement of the CAC (histidine) codon by the GGC (glycine) codon creates an AciI restriction site. We used a PCR approach on putative mutant clones to amplify the region encompassing the mutated codon. The amplified fragment was then digested by AciI, which allowed the selection of mutant clones. The whole cit1 gene was then sequenced to ensure that no additional mutations were introduced during the mutagenesis. This plasmid was introduced into the Δcit1 strain by co-transformation with the Ppable plasmid (Coppin and Debuchy 2000) which carries, as selectable marker, the phleomycin resistance gene (ble). Transformants were thus selected on medium containing 10 µg/ml phleomycin and then crossed with a wild-type strain to allow their purification. These crosses provided Δcit1‘B-XE H309G’ ascospores whose genotype was determined according to the following rules. First they had to grow on media containing both hygromycin and phleomycin and, secondly, the mutant protein was expected to lead to intragenic complementation in a cross with cit1-12. Several transformants were examined to ensure that the integration sites of the construct did not influence the phenotype of the transformants.
Respiration
Measurements were made polarographically on protoplasts in a Gilson oxygraph equipped with a Clark type O2 electrode in 0.6 M sorbitol, 7.5 mM MgCl2, 10 mM KH2PO4, 10 mM imidazole pH 7.4, 0.2% bovine serum albumin (BSA). The percentage of cyanide (KCN)-insensitive and of salicyl hydroxamic acid (SHAM)-insensitive respiration was measured. KCN and SHAM were added to final concentrations of 1 and 2.5 mM, respectively, to inhibit the cytochrome c oxidase or the alternative pathway (Dufour et al., 2000).
Enzyme assays
Crude extracts were obtained as follows: mycelia were harvested from liquid cultures after 36 h growth and crushed in liquid nitrogen. The resulting powder was suspended in homogenization buffer (10 mM Tris pH 7.5, 1 mM EDTA, 76 mM glycine). After centrifugation at 11 000 r.p.m. at 4°C for 10 min, the supernatant was kept on ice. Enzyme activities for catalase and CS were determined spectrophotometrically (Zeiss spectrophotometer) as described by Baudhuin et al. (1964) and Srere et al. (1963), respectively. Protein concentrations were determined at 562 nm using the BCA protein assay (kit from Pierce) as described by Smith et al. (1985), with BSA as the standard.
Immunoblot analysis
Proteins were separated on a 12% SDS–polyacrylamide gel and transferred to nitrocellulose (Potran BA, Schleicher and Schuell). The nitrocellulose blot was treated strictly as described previously (Vélot et al., 1999) with the same rabbit anti-Cit1p polyclonal antibody (diluted 3000-fold). The secondary antibody (anti-rabbit IgG coupled to alkaline phosphatase) was from Sigma and was diluted 1000-fold.
Acknowledgments
Acknowledgements
We are much indebted to Annie Sainsard-Chanet and Mustapha Cherkaoui-Malki for their help in respiration and catalase activity measurements, respectively, all members of our laboratory for helpful discussions, Dr W.-H.Kunau for his generous gift of anti-FOX2 antibody, Mark McCammon for fruitful discussion, and Françoise James for her technical assistance. We also would like to acknowledge the release of the N.crassa genome sequence to the international community. This work was supported by grants from the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche (ACC-SV4 no. 9504114) and from the Association de Recherche contre le Cancer (no. 9538). G.R.-R. was a fellow of MENESR and ARC.
References
- Akins R.A. and Lambowitz,A.M. (1985) General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol. Cell. Biol., 5, 2272–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry E.G. (1969) The diffuse diplotene stage of meiotic prophase in Neurospora. Chromosoma, 26, 119–129. [DOI] [PubMed] [Google Scholar]
- Baudhuin P., Beaufay,H., Rahman-Li,O., Sellinger,Z., Wattiaux,R., Jacques,P. and De Duve,C. (1964) Tissue fractionation studies. 17. Intracellular distribution of mono amino oxidase aspartate aminotransferase, d-amino acid oxidase and catalase in rat liver tissue. Biochem. J., 92, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berteaux-Lecellier V., Picard,M., Thompson-Coffe,C., Zickler,D., Panvier-Adoutte,A. and Simonet,J.M. (1995) A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell, 81, 1043–1051. [DOI] [PubMed] [Google Scholar]
- Briand J.F., Navarro,F., Rematier,P., Boschiero,C., Labarre,S., Werner,M., Shpakovski,G.V. and Thuriaux,P. (2001) Partners of Rpb8p, a small subunit shared by yeast RNA polymerases I, II and III. Mol. Cell. Biol., 21, 6056–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carls R.A. and Hanson,R.S. (1971) Isolation and characterization of tricarboxylic acid cycle mutants of Bacillus subtilis. J. Bacteriol., 106, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A.M., Schweigard,J.A. and Valent-Central,B. (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl., 41, 22. [Google Scholar]
- Chelstowska A., Jia,Y., Rothermel,B. and Butow,R.A. (1995) Retrograde regulation: a novel path of communication between mitochondria, the nucleus and peroxisomes in yeast. Can. J. Bot., 73, S205–S207. [Google Scholar]
- Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley C.A. and Hanson,M.R. (1995) How do alterations in plant mitochondrial genomes disrupt pollen development. J. Bioenerg. Biomembr., 27, 447–457. [DOI] [PubMed] [Google Scholar]
- Contamine V. and Picard,M. (1998) Escape from premature death due to nuclear mutations in Podospora anserina: repeal versus respite. Fungal Genet. Biol., 23, 223–236. [DOI] [PubMed] [Google Scholar]
- Coppin E. and Debuchy,R. (2000) Co-expression of the mating-type genes involved in internuclear recognition is lethal in Podospora anserina. Genetics, 155, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequard-Chablat M. and Sellem,C.H. (1994) The S12 ribosomal protein of Podospora anserina belongs to the S19 bacterial family and controls the mitochondrial genome integrity through cytoplasmic translation. J. Biol. Chem., 269, 14951–14956. [PubMed] [Google Scholar]
- Distel B. et al. (1996) A unified nomenclature for peroxisome biogenesis factors. J. Cell Biol., 135, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour E., Boulay,J., Rincheval,V. and Sainsard-Chanet,A. (2000) A causal link between respiration and senescence in Podospora anserina. Proc. Natl Acad. Sci. USA, 97, 4138–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga S.D., Bednarz,A.L., van Oosterum,K., Dekker,P.J. and Grivell,L.A. (1993) Yeast mitochondrial NAD(+)-dependent isocitrate dehydrogenase is an RNA-binding protein. Nucleic Acids Res., 21, 5328–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito R.E. and Klapholz,S. (1981) Meiosis and ascospore development. In Strathern,J.N., Jones,E.W. and Broach,J.R. (eds), The Molecular Biology of the Yeast Saccharomyces. Life Cycle and Inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 211–287.
- Ferea T., Contreras,E.T., Oung,T., Bowman,E.J. and Bowman,B.J. (1994) Characterization of the cit-1 gene from Neurospora crassa encoding the mitochondrial form of citrate synthase. Mol. Gen. Genet., 242, 105–110. [DOI] [PubMed] [Google Scholar]
- Gancedo C. and Serrano,R. (1989) Energy-yielding metabolism. In Rose,A.H. and Harrison,J.S. (eds), The Yeasts, Vol. 3. Academic Press, New York, NY, pp. 206–259.
- Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol., 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. and Weinert,T.A. (1989) Checkpoints: controls that ensure the order of cell cycle events. Science, 246, 629–634. [DOI] [PubMed] [Google Scholar]
- Honigberg S.M. and Lee,R.H. (1998) Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet-Vierny C., Contamine,V., Boulay,J., Zickler,D. and Picard,M. (1997) Mutations in genes encoding the mitochondrial outer membrane proteins Tom70 and Mdm10 of Podospora anserina modify the spectrum of mitochondrial DNA rearrangements associated with cellular death. Mol. Cell. Biol., 17, 6359–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y.K., Becam,A.M. and Herbert,C.J. (1997) The CIT3 gene of Saccharomyces cerevisiae encodes a second mitochondrial isoform of citrate synthase. Mol. Microbiol., 24, 53–59. [DOI] [PubMed] [Google Scholar]
- Karputas M., Branchaud,B. and Remington,S.J. (1990) Proposed mechanism for the condensation reaction of citrate synthase: 1.9-Å structure of the ternary complex with oxaloacetate and carboxymethyl coenzyme A. Biochemistry, 29, 2213–2219. [PubMed] [Google Scholar]
- Kaufman B.A., Newman,S.M., Hallberg,R.L., Slaughter,C.A., Perlman,P.S. and Butow,R.A. (2000) In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl Acad. Sci. USA, 97, 7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.S., Rosenkrantz,M.S. and Guarente,L. (1986) Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol. Cell. Biol., 6, 1936–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G., Rosenkrantz,M., Guarente,L. and Srere,P.A. (1988) Metabolic changes in Saccharomyces cerevisiae strains lacking citrate synthases. J. Biol. Chem., 263, 11145–11149. [PubMed] [Google Scholar]
- Kispal G., Evans,C.T., Malloy,C. and Srere,P.A. (1989) Metabolic studies on citrate synthase mutants of yeast. A change in phenotype following transformation with an inactive enzyme. J. Biol. Chem., 264, 11204–11210. [PubMed] [Google Scholar]
- Klasterska I. (1976) A new look on the role of the diffuse stage in problems of plant and animal meiosis. Hereditas, 82, 193–204. [Google Scholar]
- La Cognata U. (1996) Structure and expression of mitochondrial citrate synthases from higher plants. Plant Cell Physiol., 37, 1022–1029. [DOI] [PubMed] [Google Scholar]
- Landschutze V., Willmitzer,L. and Muller-Rober,B. (1995) Inhibition of flower formation by antisense repression of mitochondrial citrate synthase in transgenic potato plants leads to a specific disintegration of the ovary tissues of flowers. EMBO J., 14, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier G. and Silar,P. (1994) Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr. Genet., 25, 122–123. [DOI] [PubMed] [Google Scholar]
- Lee R.H. and Honigberg,S.M. (1996) Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol. Cell. Biol., 16, 3222–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin A.S., Hines,V. and Small,G.M. (1990) Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol. Cell. Biol., 10, 1399–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.S., Small,W.C., Srere,P.A. and Butow,R.A. (1991) Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockington R.A., Scazzocchio,C., Sequeval,D., Mathieu,M. and Felenbock,B. (1987) Regulation of alcR, the positive regulatory gene of the ethanol utilization regulon of Aspergillus nidulans. Mol. Microbiol., 1, 275–281. [DOI] [PubMed] [Google Scholar]
- Lorin S., Dufour,E., Boulay,J., Begel,O., Marsy,S. and Sainsard-Chanet,A. (2001) Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol. Microbiol., 42, 1259–1267. [DOI] [PubMed] [Google Scholar]
- Marcou D. (1961) Notion de longévité et nature cytoplasmique du déterminant de la sénescence chez quelques champignons. Ann. Sci. Nat. Bot., 11, 653–764. [Google Scholar]
- McCammon M.T. (1996) Mutants of Saccharomyces cerevisiae with defects in acetate metabolism: isolation and characterization of Acn– mutants. Genetics, 144, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell S.J., Stewart,L.C., Talin,A. and Yaffe,M.P. (1990) Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J. Cell Biol., 111, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M.J. (1994) A cosmid with a HyR marker for fungal library construction and screening. Gene, 150, 159–162. [DOI] [PubMed] [Google Scholar]
- Park B.W., Han,K.H., Lee,C.Y., Lee,C.H. and Maeng,P.J. (1997) Cloning and characterization of the citA gene encoding the mitochondrial citrate synthase of Aspergillus nidulans. Mol. Cell, 7, 290–295. [PubMed] [Google Scholar]
- Picard M., Debuchy,R. and Coppin,E. (1991) Cloning the mating types of the heterothallic fungus Podospora anserina: developmental features of haploid transformants carrying both mating types. Genetics, 128, 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington S., Wiegand,G. and Huber,R. (1982) Crystallographic refinement and atomic models of two different forms of citrate synthase at 2.7 and 1.7 Å resolution. J. Mol. Biol., 158, 111–152. [DOI] [PubMed] [Google Scholar]
- Rickey T.M. and Lewin,A.S. (1986) Extramitochondrial citrate synthase activity in bakers’ yeast. Mol. Cell. Biol., 6, 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizet G. and Engelmann,C. (1949) Contribution à l’étude génétique d’un ascomycète tétrasporé: Podospora anserina. Rev. Cytol. Biol. Veg., 11, 201–304. [Google Scholar]
- Roeder G.S. and Bailis,J.M. (2000) The pachytene checkpoint. Trends Genet., 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Rosenthal A., Coutelle,O. and Craxton,M. (1993) Large-scale production of DNA sequencing templates by microtitre format PCR. Nucleic Acids Res., 21, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprich-Robert G., Berteaux-Lecellier,V., Zickler,D., Panvier-Adoutte,A. and Picard,M. (2002) Identification of six loci in which mutations partially restore peroxisome biogenesis and/or alleviate the metabolic defect of pex2 mutants in Podospora. Genetics, 161, 1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santanam N., Augé,N., Zhou,M., Keshava,C. and Parthasarathy,S. (1999) Overexpression of human catalase gene decreases oxidized lipid-induced cytotoxicity in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol., 19, 1912–1917. [DOI] [PubMed] [Google Scholar]
- Simonet J.M. and Zickler,D. (1972) Mutations affecting meiosis in Podospora anserina. I. Cytological studies. Chromosoma, 37, 327–351. [DOI] [PubMed] [Google Scholar]
- Simonet J.M. and Zickler,D. (1978) Genes involved in caryogamy and meiosis in Podospora anserina. Mol. Gen. Genet., 162, 237–242. [Google Scholar]
- Skoneczny M., Chelstowska,A. and Rytka,J. (1988) Study of the coinduction by fatty acids of catalase A and acyl-CoA oxidase in standard and mutant Saccharomyces cerevisiae strains. Eur. J. Biochem., 174, 297–302. [DOI] [PubMed] [Google Scholar]
- Smith P.K. et al. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem., 150, 76–85. [DOI] [PubMed] [Google Scholar]
- Srere P.A., Brazil,H. and Gonen,L. (1963) The citrate condensing enzyme of pigeon breast muscle and moth flight muscle. Acta Chem. Scand., 17, S129–S134. [Google Scholar]
- Srere P.A., Sherry,A.D., Malloy,C.R. and Sumegi,B. (1997) Channeling in the Krebs tricarboxylic acid cycle. In Agius,L. and Sherratt,H.S.A. (eds), Channeling in Intermediary Metabolism. Portland Press, London, UK, pp. 201–217.
- Suissa M., Suda,K. and Schatz,G. (1984) Isolation of the nuclear yeast genes for citrate synthase and fifteen other mitochondrial proteins by a new screening method. EMBO J., 3, 1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert N.E. (1981) Metabolic pathways in peroxisomes and glyoxysomes. Annu. Rev. Biochem., 50, 133–157. [DOI] [PubMed] [Google Scholar]
- Touré B. (1972) Double reversal of gene conversion polarity and multiple conversion events in the locus ‘14’ in Podospora anserina. Mol. Gen. Genet., 117, 267–280. [Google Scholar]
- Turcq B., Denayrolles,M. and Bégueret,J. (1990) Isolation of the two allelic incompatibility genes s and S of the fungus Podospora anserina. Curr. Genet., 17, 297–303. [Google Scholar]
- Vélot C. and Srere,P.A. (2000) Reversible transdominant inhibition of a metabolic pathway. In vivo evidence of interaction between two sequential tricarboxylic acid cycle enzymes in yeast. J. Biol. Chem., 275, 12926–12933. [DOI] [PubMed] [Google Scholar]
- Vélot C., Lebreton,S., Morgunov,I., Usher,K.C. and Srere,P.A. (1999) Metabolic effects of mislocalized mitochondrial and peroxisomal citrate synthases in yeast Saccharomyces cerevisiae. Biochemistry, 38, 16195–16204. [DOI] [PubMed] [Google Scholar]
- Zickler D. (1977) Development of the synaptonemal complex and the ‘recombination nodules’ during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora. Chromosoma, 61, 289–316. [DOI] [PubMed] [Google Scholar]