Abstract
Poly(A) polymerase (PAP) has a role in two processes, polyadenylation of mRNA precursors in the nucleus and translational control of certain mRNAs by cytoplasmic elongation of their poly(A) tails, particularly during early development. It was found recently that at least three different PAP genes exist in mammals, encoding several PAP isoforms. The in vivo specificity of function of each PAP isoform currently is unknown. Here, we analyse PAP function in Drosophila. We show that a single PAP isoform exists in Drosophila that is encoded by the hiiragi gene. This single Drosophila PAP is active in specific polyadenylation in vitro and is involved in both nuclear and cytoplasmic polyadenylation in vivo. Therefore, the same PAP can be responsible for both processes. In addition, in vivo overexpression of PAP does not affect poly(A) tail length during nuclear polyadenylation, but leads to a dramatic elongation of poly(A) tails and a loss of specificity during cytoplasmic polyadenylation, resulting in embryonic lethality. This demonstrates that regulation of the PAP level is essential for controlled cytoplasmic polyadenylation and early development.
Keywords: cytoplasmic polyadenylation/Drosophila/polyadenylation/poly(A) tail/translational regulation
Introduction
Early steps of development in many species rely on maternally inherited mRNAs because transcription is quiescent at these stages. Therefore, changes in protein synthesis that control early developmental events depend on translational control. One way to regulate translation is by changing the poly(A) tail length of mRNAs in the cytoplasm. Shortening of poly(A) tails correlates with translational repression, whereas lengthening of poly(A) tails induces translation (Richter, 2000; Wickens et al., 2000). In Drosophila embryos, cytoplasmic polyadenylation is crucial for initiation of development; it activates translation of several molecules essential for axis formation, such as the anterior morphogen Bicoid (Salles et al., 1994), Hunchback (Wreden et al., 1997) and Toll (Schisa and Strickland, 1998). Cytoplasmic polyadenylation has also been proposed to regulate translation of the posterior determinant Oskar during Drosophila oogenesis (Chang et al., 1999).
The molecular mechanism of cytoplasmic polyadenylation has been analysed extensively in Xenopus oocytes, and some aspects of the reaction are similar to that of nuclear polyadenylation. Nuclear polyadenylation consists of endonucleolytic cleavage of pre-mRNAs followed by the synthesis of a poly(A) tail onto the upstream cleavage product (Zhao et al., 1999). Poly(A) addition can be reconstituted in vitro from three purified mammalian factors: poly(A) polymerase (PAP), cleavage and polyadenylation specificity factor (CPSF) and poly(A)-binding protein II [PABP2, the nuclear poly(A)-binding protein]. CPSF is a complex of four proteins that binds the polyadenylation signal AAUAAA located upstream of the cleavage site. Recognition of the poly(A) site also requires cleavage stimulation factor (CstF) that binds to a GU/U-rich element downstream of the cleavage site and interacts with CPSF. PAP catalyses the polyadenylation reaction, but is also required for efficient cleavage of pre-mRNAs in vitro (Christofori and Keller, 1989; Takagaki et al., 1989). PAP by itself does not recognize pre-mRNAs specifically. Specificity requires the AAUAAA element and CPSF that binds PAP through its 160 kDa subunit (Murthy and Manley, 1995). Even in the presence of CPSF, PAP activity remains weak; it is again stimulated by binding of PABP2 to the poly(A) tail (Wahle, 1991). Together, CPSF and PABP2 stimulate PAP activity by holding PAP on the RNA such that a full-length poly(A) tail is synthesized in a single processive event (Bienroth et al., 1993). When the poly(A) tail has reached its complete length, elongation is no longer processive and becomes slow and distributive. PABP2 is required for this poly(A) tail length control (Wahle, 1995).
Cytoplasmic polyadenylation in Xenopus relies on two sequences: the nuclear polyadenylation signal, AAUAAA, and an upstream U-rich element called the cytoplasmic polyadenylation element (CPE). CPE-dependent polyadenylation can be recapitulated in vitro in the presence of purified bovine CPSF and PAP (Bilger et al., 1994), indicating a role for CPSF. Indeed, a cytoplasmic form of CPSF has been identified in Xenopus oocytes (Dickson et al., 1999). CPEs are bound by CPEB, a major component of the reaction (Hake and Richter, 1994; Mendez and Richter, 2001). Cytoplasmic polyadenylation during Xenopus oocyte maturation is triggered by phosphorylation of CPEB, which stimulates a direct interaction between CPEB and the 160 kDa subunit of CPSF (Mendez et al., 2000). Thus, the role of CPEB during cytoplasmic polyadenylation would be to recruit CPSF into an active polyadenylation complex containing a PAP.
In Drosophila, the role of CPEs has not been addressed, and the polyadenylation signal is dispensable in some cases, since embryonic cytoplasmic polyadenylation occurs on a bicoid engineered mRNA deleted for this element (Salles et al., 1994). Although genes encoding the four subunits of CPSF are present in the Drosophila genome (Mount and Salz, 2000), their role in cytoplasmic polyadenylation has not been determined. The Drosophila homologue of CPEB is the Orb protein. orb encodes germline-specific proteins different in male and female, and its function has been determined in the female germline (Lantz et al., 1992, 1994; Christerson and McKearing, 1994). Strong orb mutants arrest oogenesis early, before the formation of the 16-cell cyst that would normally differentiate into nurse cells and one oocyte. Using a weaker allele, orbmel, Orb was shown to be required for anchoring of oskar mRNA at the posterior pole of the oocyte (Christerson and McKearing, 1994). However, this could result from a failure in oskar mRNA translation as Oskar protein is required for anchoring its own mRNA at the posterior pole. A more recent study suggests that Orb could have a function analogous to that of CPEB in cytoplasmic polyadenylation. In orb mutant egg chambers, the level of Oskar protein is decreased and poly(A) tails of oskar mRNAs are shortened (Chang et al., 1999).
Another key component required in a functional cytoplasmic polyadenylation complex is a PAP. In vertebrates, multiple PAP isoforms have been identified. Initially, two PAP isoforms were described, PAP I (70 kDa) and PAP II (83 kDa), that differ in their C-terminus (Raabe et al., 1991; Wahle et al., 1991). Analysis of PAP mRNAs in mouse revealed that these two PAP isoforms are generated by alternative splicing (Zhao and Manley, 1996). Truncated forms of PAP RNAs corresponding to the 5′ half of the gene have also been identified in several species (Wahle et al., 1991; Ballantyne et al., 1995; Gebauer and Richter, 1995; Zhao and Manley, 1996). However, these truncated RNAs are thought not to be translated in vivo, and the corresponding proteins produced in baculovirus or in Escherichia coli are inactive in vitro (Wahle et al., 1991; Martin and Keller, 1996; Zhao and Manley, 1996). In addition to the PAP gene, two new PAP-encoding genes have been identified recently in mammals, neo-PAP (or PAPγ) and TPAP. The neo-PAP gene encodes a single protein that shows 60% identity to human PAP II and has identical properties to those of PAP II in in vitro assays (Kyriakopoulou et al., 2001; Perumal et al., 2001; Topalian et al., 2001). TPAP is encoded by an intronless gene (Kashiwabara et al., 2000; Lee et al., 2000). Interestingly, TPAP is expressed specifically in testis, and the protein is specifically cytoplasmic in spermatogenic cells where cytoplasmic polyadenylation is active (Kashiwabara et al., 2000). TPAP was therefore proposed to be responsible for cytoplasmic polyadenylation in mouse testis, although this has not been addressed directly. Although the function of these four PAP isoforms has not been investigated in vivo, it seems plausible that they have specific functions. Results on TPAP suggest that different PAPs are responsible for nuclear and cytoplasmic polyadenylation in vertebrates.
Here, we address the function of Drosophila PAP in vivo. We found that a single PAP isoform is produced in Drosophila and that this single protein is responsible for both types of polyadenylation: nuclear and cytoplasmic. An increase in the level of PAP in vivo does not affect poly(A) tail length during nuclear polyadenylation. In contrast, such an increase during cytoplasmic polyadenylation leads to very long poly(A) tails and embryonic lethality, showing that a regulated level of PAP is essential for the control of cytoplasmic polyadenylation.
Results
Drosophila PAP is encoded by the hiiragi gene
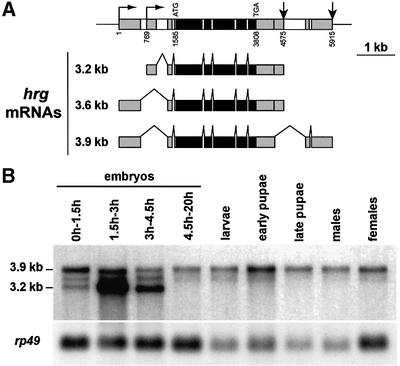
We identified the Drosophila PAP-encoding gene by screening a Drosophila genomic library with a bovine PAP cDNA (Wahle et al., 1991). One positive phage was isolated. Several subclones of this phage were used to screen cDNA libraries from 0–3 h and 12–24 h embryos, and ovaries. Restriction mapping and partial sequencing of 69 positive cDNAs as well as two expressed sequence tags (ESTs) from the Berkeley Drosophila Genome Project (LD11853 and LD05439) allowed us to determine that the PAP-encoding gene produces three different mRNAs that arise from utilization of two alternative transcription start sites and two alternative poly(A) sites (Figure 1A). No alternative splicing was found in the coding sequence and this gene was found to encode a single protein (see below). In addition, no paralogous gene was found in the Drosophila genome either by Southern blot hybridization (not shown) or by examination of the Drosophila genome sequence (Mount and Salz, 2000). During this study, the hiiragi (hrg) gene, which was first identified from its role in formation of the wing margin, was cloned and found to encode the same protein as is described herein (Murata et al., 2001). Therefore, a single PAP, which is encoded by the hrg gene, is produced in Drosophila. Northern analysis shows that the two largest mRNAs (3.9 and 3.6 kb) are present at all development stages (Figure 1B). The shortest mRNA (3.2 kb) arising from utilization of a downstream transcription start site is specific to early embryogenesis and strongly accumulates in 1.5–3 h embryos (Figure 1B). Visualization of hrg mRNAs in embryos by in situ hybridization correlates with a burst of transcription of an embryo-specific transcript before cellularized blastoderm (not shown).
Fig. 1. Structure and expression pattern of hrg. (A) Structure of the hrg gene and mRNAs. Grey boxes are exons in 5′- and 3′-UTRs, black boxes are coding exons and white boxes are introns. Horizontal arrows indicate transcription start sites, and vertical arrows indicate poly(A) sites. The longest cDNA we isolated, NB61, was from a 12–24 h embryo cDNA library. Its sequence was deposited in DDBJ/EMBL/GenBank with accession No. AF231704. Coordinates are from the genomic sequence with the first transcription site starting at 1. (B) mRNA pattern of hrg during Drosophila development. Northern blot of poly(A)+ RNAs hybridized with an RNA probe spanning the 5′ half of the hrg coding sequence and reprobed with the rp49 clone as a loading control.
Structure and in vitro activity of Drosophila PAP
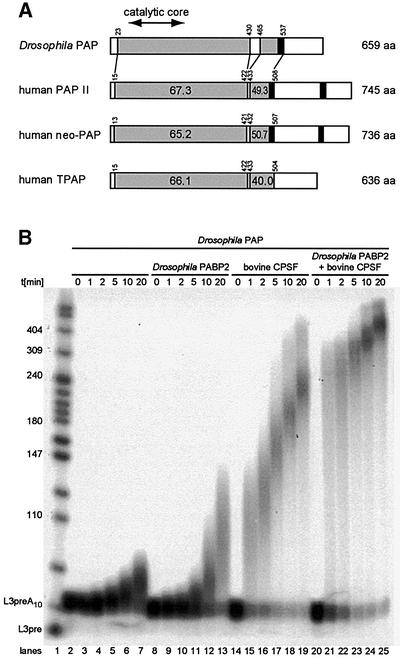
The longest cDNA we isolated (NB61) was sequenced and is predicted to encode a protein of 659 residues that is overall 56% identical and 70% similar to bovine PAP. Comparison between Drosophila PAP and human PAP II, neo-PAP and TPAP is schematized in Figure 2A. The overall identity between Drosophila PAP and each of the three human proteins is similar (45–50%). The N-terminal two-thirds of PAP that contain the catalytic core (67–190) are well conserved between Drosophila and human. The C-terminal region is more divergent, except for a short well-conserved domain (465–537) that contains a primer-binding domain and a nuclear localization signal (NLS) (Martin and Keller, 1996).
Fig. 2. Comparison of Drosophila and human PAPs and in vitro activity of Drosophila PAP. (A) Sequence comparison between Drosophila and human PAPs. Accession Nos are P51003, human PAP II; 15080911, neo-PAP; and 18203318, TPAP. Grey boxes are the conserved regions between Drosophila and human PAPs. Black boxes are the NLS. The percentages of identity with Drosophila PAP are indicated. (B) In vitro activity of Drosophila PAP. In vitro specific polyadenylation assays of a pre-cleaved AAUAAA-containing RNA, in the presence or absence of bovine purified CPSF and Drosophila PABP2. Reactions were started by the addition of ATP and stopped after the indicated times in minutes.
The activity of Drosophila His6-tagged PAP produced in E.coli was assayed in reconstituted polyadenylation reactions. Mammalian PAP requires CPSF and PABP2 in vitro to polyadenylate specifically a pre-cleaved AAUAAA-containing RNA. The ability of Drosophila PAP to carry out polyadenylation was tested in the presence or absence of bovine CPSF and Drosophila PABP2. The RNA substrate, L3pre, was derived from the adenovirus L3 polyadenylation site. It ended at the natural cleavage site and carried a tail of ∼10 A residues so that it could be bound directly by PABP2. We found that Drosophila PAP is almost inactive on its own (Figure 2B, lanes 2–7). PAP activity is slightly enhanced by the presence of Drosophila PABP2 (Figure 2B, lanes 8–13). Bovine CPSF stimulates PAP activity more strongly (Figure 2B, lanes 14–19). This stimulation probably occurs as a result of CPSF tethering Drosophila PAP to the mRNA as described for mammalian PAP. In the presence of both CPSF and PABP2, Drosophila PAP generates poly(A) tails of 200–250 nucleotides within the first minute of the reaction (Figure 2B, lanes 20–25). This increased efficiency is probably due to enhanced processivity. After this burst, poly(A) tail extension slows as described for the bovine PAP (Wahle, 1995). These results show that Drosophila PAP produced in E.coli is active and behaves as its bovine homologue in vitro.
Expression profile of PAP in Drosophila
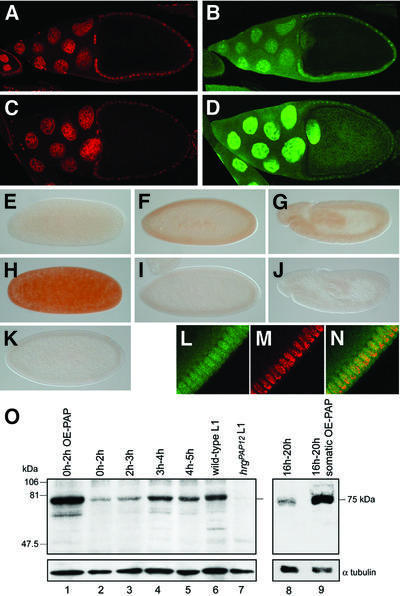
We examined the expression profile of PAP during Drosophila development, using a polyclonal antibody. During oogenesis, PAP is detected in both the nucleus and cytoplasm of nurse cells and follicle cells (Figure 3B). PAP is also present at a low level in oocyte nucleus and cytoplasm. We overexpressed PAP in the female germline using a UASp-hrg transgene under the control of the female germline-specific driver nanos-Gal4:VP16 (nos- Gal4) (Rorth, 1998). In UASp-hrg; nos-Gal4 females, PAP accumulates to a high level in nuclei of nurse cells and oocyte and to a lesser extent in oocyte cytoplasm (Figure 3D). Maternally provided PAP is detected in just laid embryos where the protein is distributed uniformly (Figure 3E). During embryogenesis, the amount of PAP increases until cellularized blastoderm stage (Figure 3F) and remains stable during gastrulation (Figure 3G). The subcellular distribution of PAP was analysed in cellularized blastoderm embryos (Figure 3L–N). PAP accumulates in nuclei and is present at a lower level in the cytoplasm, as was reported for PAP II in human somatic cells (Schul et al., 1998; Kyriakopoulou et al., 2001). A high level of PAP accumulates in early embryos coming from females where PAP is overexpressed in the germline (Figure 3H). In contrast, PAP is not detected in hrgPAP12 mutant embryos (see below) that show no hrg early zygotic transcription (not shown) (Figure 3I and J). This shows that the antibody we generated is specific for PAP. A major protein is detected in Drosophila extracts with this antibody by western blot (Figure 3O). This protein has a mol. wt of 75 kDa, which is the expected molecular weight for Drosophila PAP. Its level increases during the first hour of embryogenesis (Figure 3O, lanes 2–5); it is very abundant in embryos from females overexpressing PAP in the germline (Figure 3O, lane 1) and in late embryos overexpressing PAP ubiquitously (Figure 3O, lanes 8 and 9), and is absent in hrgPAP12 mutant larvae (Figure 3O, lane 7).
Fig. 3. PAP expression during Drosophila development. (A–D) Immunodetection of PAP in ovaries. Confocal images of stage 10 egg chambers from wild-type (A and B) and UASp-hrg; nos-Gal4 (C and D) females, double stained with propidium iodide that stains DNA (A and C in red) and anti-PAP (B and D in green). Posterior is to the right. (E–N) Immunodetection of PAP in wild-type embryos (E–G and L–N), embryos from UASp-hrg; nos-Gal4 mothers (H) or hrgPAP12 mutant embryos (I and J). (K) Embryo stained with the pre-immune serum. (L–N) Confocal images of double staining with anti-PAP (green) and propidium iodide (red) showing that PAP is nuclear and cytoplasmic. (E and H) Just laid embryos. (F, I, K and L–N) Cellularized blastoderm stage. (G and J) Gastrulation. Posterior is to the right. (O) Western blot revealed with anti-PAP. Protein extracts are from 0–2 h embryos coming from UASp-hrg; nos-Gal4 mothers (OE-PAP: PAP overexpression), 0–2 h, 2–3 h, 3–4 h and 4–5 h wild-type embryos, wild-type and hrgPAP12 first instar larvae, and 16–20 h wild-type and UASp-hrg; da-Gal4 embryos (somatic OE-PAP). Extract from 15 embryos or larvae is loaded per lane, the blot was revealed with anti-α-tubulin as a loading control.
These data show that hrg encodes a single form of PAP. This protein is mostly nuclear in somatic cells and is present in the cytoplasm of oocytes and early embryos where cytoplasmic polyadenylation takes place.
Characterization of hrg mutants
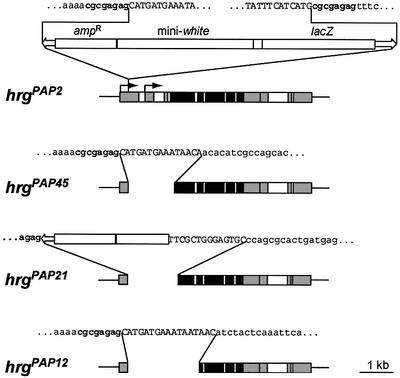
We mapped the hrg gene by in situ hybridization to position 56E5–6 on chromosome II. We screened a collection of 21 P{lacW}-induced lethal mutants on chromosome II (Torok et al., 1993), containing the P insertion in the vicinity of region 56E5–6, by Southern hybridization. In three of these stocks, l(2)k07618, l(2)k07609 and l(2)k07626, the P element was found to be inserted in the 5′-untranslated region (UTR) of hrg. DNA from these three stocks shows the same restriction profile, suggesting that it contains the same P-element insertion. The insertion was mapped in the l(2)k07618 stock and is located 261 bp downstream of the hrg 5′-most transcription start site. Although this insertion was isolated in a P-induced lethal mutant collection, it does not cause lethality. The mutation inducing lethality in the l(2)k07618 stock was removed by recombination. This stock was named hrgPAP2. hrgPAP2 is viable and fertile, although ∼50% of homozygous hrgPAP2 individuals die as first instar larvae. New hrg mutants were generated by imprecise excision of the P-element in hrgPAP2. Three homozygous lethal mutants, hrgPAP45, hrgPAP21 and hrgPAP12, that show a deletion in the coding sequence were used in further studies (Figure 4). The first six residues and the first 44 residues are deleted in hrgPAP45 and in hrgPAP21, respectively. In hrgPAP12, more than the N-terminal third of PAP (243 residues), including the catalytic core, is deleted. In all three mutants, most of the 5′-UTR of the longest mRNAs as well as the embryonic transcription start site are missing. The three mutants are lethal from late embryonic to second instar larval stages, with hrgPAP45 showing the weakest phenotype and hrgPAP12 the strongest. They do not complement each other and are, therefore, alleles of the same gene. Late embryos of all three mutants show no strong phenotype; however, they present a slight defect in head skeleton (distortion of the dorsal bridge, not shown). Lethality of hrgPAP45 and hrgPAP21 is rescued with a hrg genomic transgene. However, only 20% for hrgPAP45 and 5% for hrgPAP21 of the expected rescued progeny survive to adulthood, suggesting that hrg in the transgene is not fully expressed. Lethality of the strongest allele hrgPAP12 is not rescued with the genomic transgene, but is rescued with the transgene UASp-hrg expressed ubiquitously with the driver daughterless-Gal4 (da-Gal4). We verified that hrg mutants described earlier (Murata et al., 2001) are alleles of the gene described in this study. hrgP1 is not lethal but shows a notched wing phenotype. hrgPAP12 does not complement hrgP1, as hrgP1/hgPAP12 adults have a pronounced notched wing phenotype.
Fig. 4. Structure of hrg mutants. Schematic representation of the hrg locus; legend is as in Figure 1. The P-element insertion in hrgPAP2 is represented. The P-element sequence is in uppercase, and the genomic sequence surrounding the P-element is in lowercase. The sequence duplicated upon insertion of P is in bold. For each deletion mutant, the deletion and the remaining sequence are indicated.
Taken together, these data demonstrate that we have induced strong alleles of the hrg gene that encodes Drosophila PAP and that the lack of PAP in Drosophila is lethal.
Role of PAP in pre-mRNA cleavage and polyadenylation in vivo
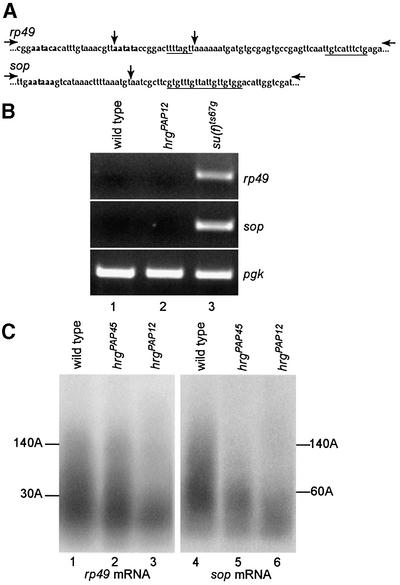
During the mammalian 3′-end processing reaction, PAP has been reported to be required for both the cleavage and polyadenylation steps in vitro. However, in these assays, PAP is not involved in the cleavage of all pre-mRNAs (Takagaki et al., 1989). We determined whether PAP is involved in cleavage and polyadenylation of pre-mRNAs in vivo using hrg mutants. To analyse the cleavage step, we looked, by RT–PCR, for RNA molecules that had not been cleaved at poly(A) sites (Benoit et al., 2002; and references therein). PCR primers were selected on each side of the poly(A) sites of rp49 and sop, two ubiquitously expressed genes that encode ribosomal proteins (Figure 5A), such that if cleavage occurs normally, no or a very low amount of PCR product is expected. Total RNA was prepared from wild-type and hrgPAP12 first instar larvae and controlled by an RT–PCR with primers located in the coding region of pgk, another gene expressed ubiquitously. Figure 5B shows that in hrgPAP12, cleavage occurs normally at the poly(A) sites of rp49 and sop, as in the wild-type (Figure 5B, lanes 1 and 2). As a positive control, we used RNA from suppressor of forked [su(f)] mutant larvae. su(f) encodes the Drosophila homologue of human CstF-77, which we have shown to be required for the cleavage step of the mRNA 3′-end processing reaction in vivo (Benoit et al., 2002). In the su(f) mutant, uncleaved pre-mRNAs accumulate that can be amplified by RT-PCR for both rp49 and sop (Figure 5B, lane 3). These data indicate that, in vivo, PAP is dispensable for the cleavage step of the mRNA 3′-end processing reaction.
Fig. 5. Role of PAP in the cleavage/polyadenylation reaction in vivo. (A) Sequences of rp49 and sop poly(A) site regions. Potential poly(A) signals are in bold. GU/U-rich sequences are underlined. Vertical arrows indicate poly(A) sites determined from cDNA sequences in databases. Horizontal arrows indicate primers used in the PCR. (B) RT–PCR assays. Total RNAs from larvae [first instar at 25°C for wild-type and hrgPAP12, third instar at 29°C for the su(f)ts67g ts mutant] were used for reverse transcription. The control pgk PCR fragment is generated with primers on each side of intron 2 of pgk. The size of the pgk PCR product (434 bp) is the expected size for amplification of pgk RNA after splicing of intron 2. rp49 (369 bp) and sop (316 bp) PCR fragments are obtained with the primers indicated in (A), only if no cleavage occurs at the poly(A) site. (C) PAT assays measuring poly(A) tail length of rp49 and sop mRNAs. Total RNAs were prepared from wild-type or mutant first instar larvae. The lengths of poly(A) tails are indicated.
We next measured poly(A) tail length of rp49 and sop in hrg mutants by poly(A) test (PAT) assays, a PCR-based technique that allows amplification of poly(A) tails (Salles and Strickland, 1999). In wild-type first instar larvae, the longest poly(A) tails of both rp49 and sop mRNAs were found to be up to 140 residues (Figure 5C, lanes 1 and 4). In the weak hrgPAP45 mutant, poly(A) tails of rp49 mRNA are not reduced and those of sop mRNA are reduced to 50% of their length in the wild-type (Figure 5C, lanes 2 and 5). In hrgPAP12 mutant larvae, poly(A) tails of both rp49 and sop are strongly reduced and reach a maximal length of 30 and 60 residues for rp49 and sop mRNAs, respectively (Figure 5C, lanes 3 and 6). This suggests that poly(A) tail synthesis is affected in this mutant.
Therefore, utilization of hrg mutants allowed us to conclude that in vivo PAP is dispensable for the cleavage step, but is required for poly(A) tail elongation during the mRNA 3′-end processing reaction.
Genetic interaction between hrg and orb
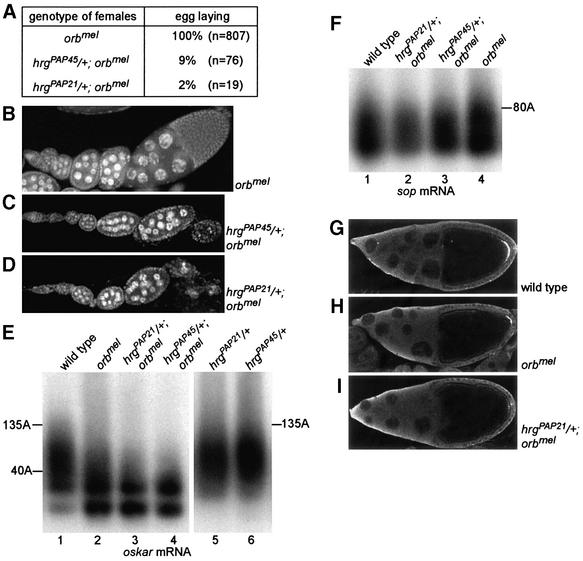
To address a possible role for hrg in cytoplasmic polyadenylation during early development, we induced germline clones homozygous for hrgPAP45, hrgPAP21 or hrgPAP12. No germline clones were obtained for any of these mutants, possibly as a result of a requirement of PAP for cell viability. We therefore studied genetic interactions between hrg and orb, which is known to be involved in cytoplasmic polyadenylation. Females homozygous for the weak orbmel allele produce egg chambers at all stages and lay eggs, 30% of which show a ventralized phenotype (Christerson and McKearing, 1994). We found that hrg lethal mutants act as dominant enhancers of the orbmel phenotype, as hrgPAP45/+; orbmel and hrgPAP21/+; orbmel females lay almost no eggs (Figure 6A). In these females, oogenesis stops most frequently at stage 7/8, after which egg chambers degenerate (Figure 6C and D), even though one or two stage 14 oocytes per ovary can be observed. Poly(A) tails of oskar mRNA were shown previously to be shortened in orb mutant ovaries (Chang et al., 1999). We analysed the defect of these poly(A) tails in hrg–/+; orbmel mutants by PAT assays. We measured oskar mRNA poly(A) tails to be up to 135 residues in wild-type ovaries (Figure 6E, lane 1). These poly(A) tails are weakly reduced in orbmel (Figure 6E, lane 2), but severely reduced in hrg–/+; orbmel ovaries, their maximal length reaching 40–50 residues (Figure 6E, lanes 3 and 4). These short poly(A) tails do not result from the oogenesis defect in hrg–/+; orbmel females, as unrelated mutants that stop oogenesis early show wild-type poly(A) tails of oskar mRNA (Chang et al., 1999). These poly(A) tails were also found to be of wild-type length in hrgPAP21/+ and hrgPAP45/+ ovaries (Figure 6E, lanes 5 and 6). This shows that the strong shortening of oskar poly(A) tails in hrg–/+; orbmel mutants does not result from an additive effect of two phenotypes, but from a synergistic effect of the two mutants due to a simultaneous decrease in PAP and Orb protein levels. This strongly suggests that hrg and orb are involved together in cytoplasmic polyadenylation. This was confirmed by measurements of poly(A) tails of a control mRNA, sop, which is thought not to be regulated by cytoplasmic polyadenylation. Poly(A) tails of sop mRNAs are unaffected in orbmel as well as in hrg–/+; orbmel mutant ovaries (Figure 6F). We verified that shortening of oskar mRNA poly(A) tails in hrg–/+; orbmel mutants leads to a reduction of Oskar protein level, by immunostaining of ovaries with anti-Oskar. Oskar accumulates at the posterior of the oocyte from stage 9 onwards. As described before, the amount of Oskar decreases in orbmel oocytes (Figure 6G and H). This amount decreases again in hrg–/+; orbmel oocytes to a barely detectable level (Figure 6I).
Fig. 6. Role of hrg in cytoplasmic polyadenylation. (A) Phenotype of hrg–/+; orbmel mutant females. The number of eggs laid by 50 females for 3 days is indicated. (B–D) DAPI staining of ovaries showing that the ovarian phenotype is strongly enhanced when orbmel is combined with hrg–/+. The genotype of females is indicated. (E and F) PAT assays measuring poly(A) tail lengths of oskar (E) or sop (F) mRNAs in wild-type and mutant ovaries. The genotype of females and the lengths of poly(A) tails are indicated. (G–I) Immunostaining of stage 10 egg chambers with anti-Oskar antibody showing that virtually no Oskar accumulates in hrg–/+; orbmel mutant oocytes. Oskar accumulates as a cap at the posterior of the wild-type oocyte. Posterior is to the right. The genotype of females is indicated.
Taken together, these results show that hrg and orb cooperate in poly(A) tail lengthening during cytoplasmic polyadenylation and that alteration of this process affects protein accumulation and oogenesis.
Overexpression of PAP in the female germline causes embryonic lethality of the progeny
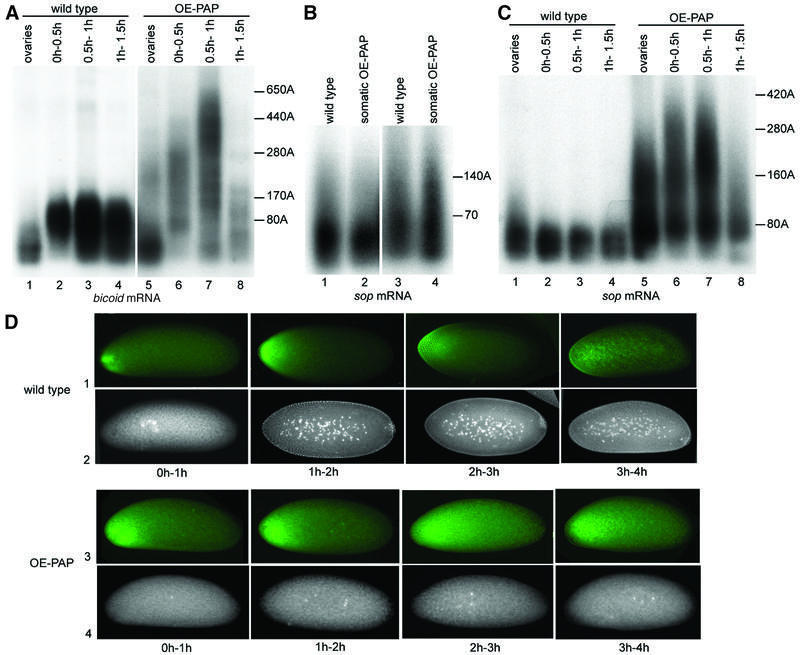
To address whether cytoplasmic polyadenylation could be affected by the level of PAP, we overexpressed PAP in the female germline using the UASp-hrg transgene and nos-Gal4 (Figure 3D). This overexpression does not cause gross alteration of oogenesis, but is extremely detrimental to embryogenesis, leading to 99% lethality of the progeny (n = 5947). These embryos stop development early, before cleavage of nuclei (Figure 7D, row 4). We analysed cytoplasmic polyadenylation in these embryos by measuring poly(A) tails of bicoid mRNA that is regulated by this process during embryogenesis (Salles et al., 1994). In the wild-type, poly(A) tails of bicoid mRNA lengthen from 80 residues in oocytes to 170 residues in 1 h embryos (Figure 7A). This elongation of the poly(A) tails induces Bicoid protein synthesis in early embryos (Figure 7D, row 1) (Salles et al., 1994). Following overexpression of PAP, poly(A) tails of bicoid mRNA strongly increase in length, with a pool of mRNAs bearing a 250 residue poly(A) tail in oocytes and most mRNAs having a poly(A) tail between 300 and 600 residues in 1 h embryos (Figure 7A). The fact that bicoid mRNA poly(A) tails lengthen in 0–1 h embryos, at a stage when there is no transcription, shows that the process affected by PAP overexpression is cytoplasmic polyadenylation. This was confirmed by showing that when PAP is overexpressed ubiquitously in somatic cells with the da-Gal4 driver, poly(A) tails of sop mRNA are not affected (Figure 7B). Therefore, poly(A) tail length control during nuclear polyadenylation is not altered by PAP overexpression, although the level of somatic overexpression is in the same range as that of germline overexpression (Figure 3O, lanes 1 and 2, and 8 and 9). Surprisingly, although sop mRNA does not undergo cytoplasmic polyadenylation in wild-type embryos (Figure 7C, wild-type panel), overexpression of PAP in the female germline leads to a strong lengthening of sop mRNA poly(A) tails by cytoplasmic polyadenylation (Figure 7C). Similar results were found for rp49 mRNA (not shown). This indicates that the increasing PAP level affects both poly(A) tail length control and specificity during cytoplasmic polyadenylation. We correlated Bicoid protein accumulation and bicoid mRNA poly(A) tail length by immunostaining of ovaries and embryos with anti-Bicoid. Poly(A) tail elongation of bicoid mRNA in oocytes, following PAP overexpression, does not induce translation as no Bicoid is detected in UASp-hrg; nos-Gal4 oocytes (not shown). Therefore, in oocytes, long poly(A) tails are not sufficient to induce bicoid mRNA translation. In embryos where PAP is overexpressed, poly(A) tail lengthening correlates with a precocious accumulation of Bicoid and with an increase in Bicoid protein level (Figure 7D, row 3).
Fig. 7. Overexpression of PAP in the female germline enhances cytoplasmic polyadenylation. (A–C) PAT assays measuring poly(A) tail lengths of bicoid (A) and sop (B and C) mRNAs. (A and C) Total RNAs were prepared from ovaries or 0–0.5 h, 0.5–1 h and 1–1.5 h embryos from wild-type or UASp-hrg; nos-Gal4 (OE-PAP: PAP overexpression) females crossed with wild-type males. (B) Total RNAs were prepared from wild-type or UASp-hrg; da-Gal4 (somatic OE-PAP) 16–20 h embryos (lanes 1 and 2) or early pupae (lanes 3 and 4). Poly(A) tail lengths are indicated. (D) Double staining with anti-Bicoid antibody (rows 1 and 3) and DAPI (rows 2 and 4) of 0–1 h, 1–2 h, 2–3 h and 3–4 h embryos from wild-type or UASp-hrg; nos-Gal4 (OE-PAP) females crossed with wild-type males. Posterior is to the right. Embryos where PAP is overexpressed show up to a 2-fold increase in Bicoid level (row 3) (quantification was with MetaMorph). These embryos contain one to a few nuclei only (row 4).
These data demonstrate that a tight regulation of PAP level is essential to control cytoplasmic polyadenylation and to early development.
Discussion
Data in this article allow us to draw two important conclusions. (i) A single isoform of PAP is able to perform both reactions of nuclear and cytoplasmic polyadenylation in vivo. (ii) A controlled level of PAP is essential for specificity of cytoplasmic polyadenylation and for poly(A) tail length control during cytoplasmic polyadenylation.
A single PAP in Drosophila
We and others found that there is a single PAP-encoding gene, hrg, in the Drosophila genome (Mount and Salz, 2000; Murata et al., 2001; this study). Although hrg produces three mRNAs, they all encode the same protein. Western blots on Drosophila extracts confirm the presence of a single PAP isoform in Drosophila. As expected for a gene responsible for such a fundamental process as polyadenylation, hrg is essential for viability. Strong hrg mutants are lethal at late embryonic and larval stages. hrg is also probably essential to cell viability as strong hrg mutant germline clones do not survive. We showed that PAP encoded by this gene is involved in both nuclear polyadenylation of rp49 and sop mRNAs in somatic tissues and cytoplasmic polyadenylation of oskar mRNA in oocytes. This indicates that although the reactions of nuclear and cytoplasmic polyadenylation are not identical, a single PAP is responsible for both in Drosophila.
Recently, a new class of cytoplasmic PAPs was discovered that is not related in sequence to conventional PAPs (Wang et al., 2002). These proteins are widespread in eukaryotes and three homologues exist in Drosophila. A member in nematodes functions in germline and embryonic development; therefore, this class of proteins was proposed to play a role in cytoplasmic polyadenylation during development, in addition to conventional PAPs.
Role of PAP in nuclear polyadenylation
We found that Drosophila PAP indeed has a poly(A) polymerase activity in vitro, in reconstituted specific polyadenylation assays. Stimulation of Drosophila PAP activity by bovine CPSF indicates that Drosophila PAP and bovine CPSF interact. In mammalian PAP, the region thought to be involved in interaction with CPSF overlaps the first NLS (Thuresson et al., 1994) and this domain is conserved in Drosophila.
We showed that Drosophila PAP has a role in vivo in nuclear polyadenylation. In hrg mutant larvae, poly(A) tails of rp49 and sop mRNAs are short. These short poly(A) tails probably result from the decay of rp49 and sop transcript pools and a lack of newly polyadenylated mRNAs. In vitro studies led to the general belief that PAP is required for the cleavage of pre-mRNAs at poly(A) sites (Zhao et al., 1999). However, we found that it is not the case in vivo, at least for two different pre-mRNAs. Overexpression of PAP in somatic tissues does not alter poly(A) tail length of sop mRNAs. This indicates that the amount of PAP is not limiting in vivo for nuclear polyadenylation. This is not unexpected as, during the polyadenylation reaction, PABP2 plays an important role in stimulating PAP. Once PABP2 is bound to the newly synthesized 10 residue poly(A) tail, a single PAP molecule is required to polymerize the complete poly(A) tail (Bienroth et al., 1993). PABP2 also controls the length of the poly(A) tail, and this function of PABP2 may prevent the recruitment of new PAP molecules in the complex, and the poly(A) tail to lengthen even if more PAP is present.
Role of PAP level in cytoplasmic polyadenylation
We showed that hrg, in conjunction with orb, is involved in cytoplasmic polyadenylation of oskar mRNA during oogenesis. The control of oskar mRNA translation is very complex. oskar mRNA is transported to the posterior pole of the oocyte and translation does not start before this posterior localization (Kim-Ha et al., 1995; Markussen et al., 1995; Rongo et al., 1995). An essential determinant in translational repression during oskar mRNA transport is the Bruno protein (Kim-Ha et al., 1995; Webster et al., 1997). Although the mechanism underlying translational repression by Bruno currently is unknown, it has been shown, in a cell-free system, to be independent of poly(A) tail length (Lie and Macdonald, 1999). Therefore, although oskar mRNA was shown to undergo cytoplasmic polyadenylation (Chang et al., 1999), the role of this regulation in the control of oskar mRNA translation is unclear. Our data provide further evidence that cytoplasmic polyadenylation has a role in Oskar expression, since, when this process is impaired, Oskar does not accumulate at the posterior pole of the oocyte. Regulation of oskar mRNA poly(A) tail length probably represents an additional level of control of Oskar expression. We found that in hrgPAP21/+; orbmel mid-oocytes, the amount of oskar mRNA is low (not shown). This suggests that cytoplasmic polyadenylation could be required to unbalance rapid deadenylation and decay of oskar mRNA.
Other mRNAs regulated by cytoplasmic polyadenylation in Drosophila oogenesis have not been identified, but many are to be expected. Strong alleles of orb stop oogenesis early, and a recent study indicates that orb is required for oocyte determination (Huynh and St Johnston, 2000). This suggests that Orb regulates translation of mRNAs that have a function very early during oogenesis. In agreement with this, we found that hrg–/+; orbmel females stop oogenesis at an earlier stage than orbmel females. mRNAs regulated by cytoplasmic polyadenylation during early oogenesis were identified in mouse. They encode two proteins of the synaptonemal complex, a complex required for recombination during meiosis, and these proteins are not produced in CPEB knockout mouse oocytes (Tay and Richter, 2001).
A crucial conclusion from our data is that a tightly regulated level of PAP has a major role in cytoplasmic polyadenylation. Overexpressing PAP in the female germline results in a strong elongation of poly(A) tails of bicoid and sop, mRNAs that are and are not regulated by cytoplasmic polyadenylation, respectively. Therefore, increasing the level of PAP alters both poly(A) tail length control and the specificity of cytoplasmic polyadenylation for certain mRNAs. This deregulation leads to early embryonic lethality. That a low level of PAP is important for cytoplasmic polyadenylation regulation correlates with the repression of PAP activity by phosphorylation in Xenopus oocytes during meiotic maturation, when cytoplasmic polyadenylation occurs (Ballantyne et al., 1995; Colgan et al., 1998). In contrast, overexpression of PAP in somatic tissues does not affect poly(A) tail length control during nuclear polyadenylation. This difference has mechanistic implications and suggests that if PABP2 is involved at some step in cytoplasmic polyadenylation, as we found to be the case (B.Benoit and M.Simonelig, in preparation), its role is different from that during nuclear polyadenylation. That an increase of PAP level leads to unregulated very long poly(A) tails suggests that poly(A) tail synthesis during cytoplasmic polyadenylation mainly depends on the ability of PAP to interact with mRNAs and that cytoplasmic polyadenylation never enters a processive state where a single PAP molecule would be sufficient to complete a poly(A) tail in one event. This correlates with the slowness of the reaction during embryogenesis where elongation of bicoid mRNA poly(A) tail extends for 1–1.5 h of development (Salles et al., 1994; this study).
In Xenopus oocytes, CPEB makes the cytoplasmic polyadenylation reaction specific to some mRNAs, and it is probable that Orb has the same role in Drosophila. The loss of specificity of the reaction following PAP overexpression indicates that the PAP level also has an active role in this specificity. In this context, cytoplasmic poly(A) tail elongation of sop and other mRNAs that do not normally undergo this reaction probably does not require Orb, nor another protein that would recognize these mRNAs specifically. This suggests that an active cytoplasmic polyadenylation complex can form in the absence of Orb/CPEB, that would contain CPSF and PAP only. In vitro studies have also led to this conclusion (Dickson et al., 2001). However, whether or not such a complex is actually responsible for cytoplasmic polyadenylation of some mRNAs in vivo under normal conditions, and in that case what makes the reaction specific, represent a challenge for further studies.
Materials and methods
Drosophila stocks and genetics
The w1118 stock was used as a control. Homozygous hrg mutants were selected using the balancer chromosome CyO-pAct-GFP. We used the da-Gal4 (Wodarz et al., 1995) and nos-Gal4 (Rorth, 1998) driver lines, that mediate ubiquitous expression and expression in the female germline, respectively. P-element transformation was performed using a standard method. hrg deletion mutants were screened using the loss of the white+ marker in the P-element. A total of 83 lines were recovered after mobilization of P{lacW} in hrgPAP2, in the presence of transposase. Twenty-three lines are homozygous viable, 22 homozygous lethal and 34 homozygous sublethal. Germline clones were induced by using the FLP/FRT technique (Chou and Perrimon, 1996). No hrgPAP45, hrgPAP21 or hrgPAP12 mutant germline clone was obtained, whereas control wild-type germline clones were obtained.
DNA constructs
The genomic hrg transgene pCaSpeR-hrg was constructed as follows. A PstI–SalI 7.9 kb genomic fragment containing the hrg gene with 0.9 kb upstream of the 5′-most transcription start site and 1.1 kb downstream of the 3′-most poly(A) site was cloned into pBluescript digested with PstI and SalI. The genomic fragment was then digested with XhoI and NotI and cloned into pCaSpeR4 digested by XhoI and NotI. The UASp-hrg transgene was generated as follows. A cDNA fragment containing the complete hrg coding sequence was digested from NB61 with HaeII and NdeI, repaired with Klenow and cloned into pBluescript digested with EcoRV. The cDNA fragment was then digested with NotI and KpnI and cloned into pUASp (Rorth, 1998) digested with NotI and KpnI.
RNA manipulation
Poly(A)+ RNAs for northern blots were purified using the Oligotex mRNA kit (Qiagen). PAT assays were performed as described (Salles and Strickland, 1999) and repeated at least four times with two independent RNA preparations for each experiment. The specific primers were 5′-CTGCCCACCGGATTCAAGAAGT for rp49, 5′-GGATTGCTACACCTCGGGCCGT for sop, 5′-GGCGCAGTGGGCGTGGTACG for oskar, and 5′-CATTTGCGCATTCTTTGACC for bicoid. RT–PCR to assay cleavage of pre-mRNAs were performed as described (Benoit et al., 2002).
Expression and purification of His6-PAP and polyadenylation assays
A His-tagged Drosophila PAP was expressed in E.coli as follows. A 2.6 kb AflIII fragment from NB61 was cloned in the pET30a vector (Novagen) digested by NcoI. The resulting construct contains the whole PAP-coding sequence fused in-frame with a His6 tag at its N-terminus. His6-PAP was expressed in the E.coli BL21 strain and purified as described (Martin and Keller, 1996). Polyadenylation assays were performed as described (Wahle, 1995) except that Drosophila PABP2 was substituted for bovine PABP2 (Benoit et al., 1999).
Antibody preparation, immunostaining and western blots
Anti-PAP polyclonal antibody was raised in rat by Eurogentec against the purified bacterially expressed His6-PAP. Immunostaining and western blots were performed as described (Benoit et al., 1999). Antibody dilutions were: anti-PAP (1/500 or 1/1000), anti-Oskar (1/500) (Kim-Ha et al., 1995), anti-Bicoid (1/200) (Kosman et al., 1998) and anti-α-tubulin (1/1000) (Sigma).
Acknowledgments
Acknowledgements
This work was initiated at Institut Jacques Monod, Paris; we thank D.Anxolabéhère for support during this period. We are grateful to N.Dostatni, P.Rorth, P.M.Macdonald, P.Schedl, J.Reinitz and BDGP for the gift of Drosophila stocks, clones or antibodies, and to A.Ephrussi for sharing unpublished results. This work was supported by the Centre National de la Recherche Scientifique (UPR 1142), the Association pour la Recherche sur le Cancer (No. 9885), the Association Française contre les Myopathies (No. 7411), and by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie to E.W. F.J. and S.Z. held awards from the Ministère de l’Enseignement Supérieur et de la Recherche and from the Fondation pour la Recherche Médicale.
References
- Ballantyne S., Bilger,A., Astrom,J., Virtanen,A. and Wickens,M. (1995) Poly(A) polymerases in the nucleus and cytoplasm of frog oocytes: dynamic changes during oocyte maturation and early development. RNA, 1, 64–78. [PMC free article] [PubMed] [Google Scholar]
- Benoit B., Nemeth,A., Aulner,N., Kühn,U., Simonelig,M., Wahle,E. and Bourbon,H.M. (1999) The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res., 27, 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B., Juge,F., Iral,F., Audibert,A. and Simonelig,M. (2002) Chimeric human CstF-77/Drosophila Suppressor of forked proteins rescue suppressor of forked mutant lethality and mRNA 3′-end processing in Drosophila. Proc. Natl Acad. Sci. USA, 99, 10593–10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienroth S., Keller,W. and Wahle,E. (1993) Assembly of a processive mRNA polyadenylation complex. EMBO J., 12, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilger A., Fox,C.A., Wahle,E. and Wickens,M. (1994) Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev., 8, 1106–1116. [DOI] [PubMed] [Google Scholar]
- Chang J.S., Tan,L. and Schedl,P. (1999) The Drosophila CPEB homolog, Orb, is required for Oskar protein expression in oocytes. Dev. Biol., 215, 91–106. [DOI] [PubMed] [Google Scholar]
- Chou T.B. and Perrimon,N. (1996) The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics, 144, 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christerson L.B. and McKearing,D.M. (1994) orb is required for antero-posterior and dorso-ventral patterning during Drosophila oogenesis. Genes Dev., 8, 614–628. [DOI] [PubMed] [Google Scholar]
- Christofori G. and Keller,W. (1989) Poly(A) polymerase purified from HeLa cell nuclear extract is required for both cleavage and polyadenylation of pre-mRNA in vitro. Mol. Cell. Biol., 9, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan D.F., Murthy,K.G., Zhao,W., Prives,C. and Manley,J.L. (1998) Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J., 17, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson K.S., Bilger,A., Ballantyne,S. and Wickens,M.P. (1999) The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol. Cell. Biol., 19, 5707–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson K.S., Thompson,S.R., Gray,N.K. and Wickens,M. (2001) Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem., 276, 41810–41816. [DOI] [PubMed] [Google Scholar]
- Gebauer F. and Richter,J.D. (1995) Cloning and characterization of a Xenopus poly(A) polymerase. Mol. Cell. Biol., 15, 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake L.E. and Richter,J.D. (1994) CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell, 79, 617–627. [DOI] [PubMed] [Google Scholar]
- Huynh J.R. and St Johnston,D. (2000) The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development, 127, 2785–2794. [DOI] [PubMed] [Google Scholar]
- Kashiwabara S., Zhuang,T., Yamagata,K., Noguchi,J., Fukamizu,A. and Baba,T. (2000) Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev. Biol., 228, 106–115. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kerr,K. and Macdonald,P.M. (1995) Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell, 81, 403–412. [DOI] [PubMed] [Google Scholar]
- Kosman D., Small,S. and Reinitz,J. (1998) Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol., 208, 290–294. [DOI] [PubMed] [Google Scholar]
- Kyriakopoulou C.B., Nordvarg,H. and Virtanen,A. (2001) A novel nuclear human poly(A) polymerase, PAPγ. J. Biol. Chem., 276, 33504–33511. [DOI] [PubMed] [Google Scholar]
- Lantz V., Ambrosio,L. and Schedl,P. (1992) The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding proteins and has localized transcripts in ovaries and early embryos. Development, 115, 75–88. [DOI] [PubMed] [Google Scholar]
- Lantz V., Chang,J.S., Horabin,J.I., Bopp,D. and Schedl,P. (1994) The Drosophila orb RNA-binding protein is required for the formation of egg chamber and establishment of polarity. Genes Dev., 8, 598–613. [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Lee,Y. and Chung,J.H. (2000) An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Lett., 487, 287–292. [DOI] [PubMed] [Google Scholar]
- Lie Y.S. and Macdonald,P.M. (1999) Translational regulation of oskar mRNA occurs independent of the cap and poly(A) tail in Drosophila ovarian extracts. Development, 126, 4989–4996. [DOI] [PubMed] [Google Scholar]
- Markussen F.H., Michon,A.M., Breitwieser,W. and Ephrussi,A. (1995) Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development, 121, 3723–3732. [DOI] [PubMed] [Google Scholar]
- Martin G. and Keller,W. (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases and to other nucleotidyltransferases. EMBO J., 15, 2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Mendez R. and Richter,J.D. (2001) Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol., 2, 521–529. [DOI] [PubMed] [Google Scholar]
- Mendez R., Murthy,K.G., Ryan,K., Manley,J.L. and Richter,J.D. (2000) Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell, 6, 1253–1259. [DOI] [PubMed] [Google Scholar]
- Mount S.M. and Salz,H.K. (2000) Pre-messenger RNA processing factors in the Drosophila genome. J. Cell Biol., 150, F37–F44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Nagaso,H., Kashiwabara,S., Baba,T., Okano,H. and Yokoyama,K.K. (2001) The hiiragi gene encodes a poly(A) polymerase, which controls the formation of the wing margin in Drosophila melanogaster. Dev. Biol., 233, 137–147. [DOI] [PubMed] [Google Scholar]
- Murthy K.G.K. and Manley,J.L. (1995) The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- Perumal K., Sinha,K., D,D.H. and Reddy,R. (2001) Purification, characterization and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J. Biol. Chem., 276, 21791–21796. [DOI] [PubMed] [Google Scholar]
- Raabe T., Bollum,F.J. and Manley,J.L. (1991) Primary structure and expression of bovine poly(A) polymerase. Nature, 353, 229–234. [DOI] [PubMed] [Google Scholar]
- Richter J.D. (2000) The influence of polyadenylation-induced translation on metazoon development and neuronal synaptic function. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Trans lational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 785–806.
- Rongo C., Gavis,E.R. and Lehmann,R. (1995) Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development, 121, 2737–2746. [DOI] [PubMed] [Google Scholar]
- Rorth P. (1998) Gal4 in the Drosophila female germline. Mech. Dev., 78, 113–118. [DOI] [PubMed] [Google Scholar]
- Salles F.J. and Strickland,S. (1999) Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol. Biol., 118, 441–448. [DOI] [PubMed] [Google Scholar]
- Salles F.J., Lieberfarb,M.E., Wreden,C., Gergen,J.P. and Strickland,S. (1994) Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science, 266, 1996–1999. [DOI] [PubMed] [Google Scholar]
- Schisa J.A. and Strickland,S. (1998) Cytoplasmic polyadenylation of Toll mRNA is required for dorsal–ventral patterning in Drosophila embryogenesis. Development, 125, 2995–3003. [DOI] [PubMed] [Google Scholar]
- Schul W., Driel,R.V. and Jong,L.D. (1998) A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp. Cell Res., 238, 1–12. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Ryner,L.C. and Manley,J.L. (1989) Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev., 3, 1711–1724. [DOI] [PubMed] [Google Scholar]
- Tay J. and Richter,J.D. (2001) Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell, 1, 201–213. [DOI] [PubMed] [Google Scholar]
- Thuresson A.C., Astrom,J., Astrom,A., Gronvik,K.O. and Virtanen,A. (1994) Multiple forms of poly(A) polymerases in human cells. Proc. Natl Acad. Sci. USA, 91, 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian S.L., Kaneko,S., Gonzales,M.I., Bond,G.L., Ward,Y. and Manley,J.L. (2001) Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol. Cell. Biol., 21, 5614–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok T., Tick,G., Alvarado,M. and Kiss,I. (1993) P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: isolation of lethals with different overgrowth phenotypes. Genetics, 135, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. (1991) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell, 66, 759–768. [DOI] [PubMed] [Google Scholar]
- Wahle E. (1995) Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem., 270, 2800–2808. [DOI] [PubMed] [Google Scholar]
- Wahle E., Martin,G., Schiltz,E. and Keller,W. (1991) Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J., 10, 4251–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann,C.R., Kadyk,L.C., Wickens,M. and Kimble,J. (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature, 419, 312–316. [DOI] [PubMed] [Google Scholar]
- Webster P.J., Liang,L., Berg,C.A., Lasko,P. and Macdonald,P.M. (1997) Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev., 11, 2510–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Goodwin,E.B., Kimble,J., Strickland,S. and Hentze,M. (2000) Translational control of developmental decisions. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 295–370.
- Wodarz A., Hinz,U., Engelbert,M. and Knust,E. (1995) Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell, 82, 67–76. [DOI] [PubMed] [Google Scholar]
- Wreden C., Verrotti,A.C., Schisa,J.A., Lieberfarb,M.E. and Strickland,S. (1997) Nanos and Pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development, 124, 3015–3023. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′-ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. and Manley,J.L. (1996) Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol. Cell. Biol., 16, 2378–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]