Abstract
Cdh1p is a substrate-specific subunit of the anaphase-promoting complex (APC/C), which functions as an E3 ubiquitin ligase to degrade the mitotic cyclin Clb2p and other substrates during the G1 phase of the cell cycle. Cdh1p is phosphorylated and thereby inactivated at the G1/S transition predominantly by Cdc28p–Clb5p. Here we show that Cdh1p is nuclear during the G1 phase of the cell cycle, but redistributes to the cytoplasm between S phase and the end of mitosis. Nuclear export of Cdh1p is regulated by phosphorylation and requires active Cdc28p kinase. Cdh1p binds to the importin Pse1p and the exportin Msn5p, which is necessary and sufficient to promote efficient export of Cdh1p in vivo. Although msn5Δ cells are viable, they are sensitive to Cdh1p overexpression. Likewise, a mutant form of Cdh1p, which is consitutively nuclear, prevents accumulation of Clb2p and leads to cell cycle arrest when overexpressed in wild-type cells. Taken together, these results suggest that phosphorylation-dependent nuclear export of Cdh1p by Msn5p contributes to efficient inactivation of APC/CCdh1.
Keywords: APC/C/Cdh1p/cell cycle regulation/Msn5p/nuclear export
Introduction
In eukaryotic cells, cell cycle progression is controlled by cyclin-dependent kinases (CDKs) and regulated protein degradation. In this pathway, E3 ubiquitin ligases select protein substrates and cooperate with ubiquitin-activating (E1) and ubiquitin-conjugating (E2) enzymes to assemble polyubiquitin chains on substrates, thereby targeting them for degradation by the 26S proteasome (Hershko and Ciechanover, 1998). The anaphase-promoting complex (APC/C; also called the cyclosome) and the Skp1p–cullin–F-box protein complex (SCF) are conserved E3 ubiquitin ligases involved in regulation of the cell cycle. The SCF is required for entry into S phase, while the APC/C controls anaphase onset and mitotic exit (Peters, 1998). APC/C is composed of at least 10 subunits, including Cdc16p, Cdc23p and Cdc27p, which remain tightly associated with each other throughout the cell cycle. Mammalian Cdc16p and Cdc27p are found in the nucleus and accumulate on the mitotic spindle and at centrosomes (Tugendreich et al., 1995).
APC/C exists in two functionally distinct forms, APC/CCdc20 and APC/CCdh1 (Zachariae and Nasmyth, 1999). Cdh1p/Hct1p and Cdc20p are proposed to be substrate-specific adaptors, which interact with the substrates and target them to the core APC/C complex for ubiquitylation (Schwab et al., 1997; Visintin et al., 1997). Cdc20p is required for proteolysis of the anaphase inhibitor Pds1p at the onset of anaphase (Cohen-Fix et al., 1996), but dispensable for proteolysis of the mitotic cyclin Clb2p. Conversely, Cdh1p is dispensable for Pds1p proteolysis in mitosis, but necessary and rate limiting for proteolysis of Clb2p (Schwab et al., 1997). Proteolysis of Clb2p starts during anaphase, persists during G1 (Amon et al., 1994) and is terminated by activation of Cdc28p–Clb5p as cells enter S phase (Huang et al., 2001; Yeong et al., 2001). During G1, APC/CCdh1 activity is required to prevent accumulation of proteins that interfere with bud emergence, disturb spindle assembly or lead to premature DNA replication (Peters, 1999). Cdh1p is present throughout the cell cycle, but its binding to APC/C is blocked by CDK phosphorylation (Zachariae et al., 1998; Kramer et al., 2000). Indeed, cells overexpressing a non-phosphoryl atable Cdh1p mutant protein (Cdh1p-m11) arrest with elongated buds at the G2/M transition, because they are unable to accumulate mitotic cyclins. In wild-type cells, Cdh1p remains hyperphosphorylated and inactive throughout S, G2 and M phase. At the end of mitosis, Cdh1p is dephosphorylated by a drop in CDK activity initiated by release of the phosphatase Cdc14p from the nucleolus (Visintin et al., 1998; Jaspersen et al., 1999). In contrast to Cdh1p, Cdc20p is unstable during the G1 phase of the cell cycle, while its protein levels accumulate during G2 and M phase (Prinz et al., 1998; Huang et al., 2001). Moreover, binding of Cdc20p to the APC/C is stimulated by phosphorylation of APC/C core subunits by CDKs (Kramer et al., 2000).
Spatial organization is crucial for the coordination of many cell cycle transitions, and striking connections between subcellular localization and regulated proteolysis have emerged recently (Pines, 1999). For example, the localization of F-box proteins, the substrate-specific adaptors of SCF E3 ligases, restricts their function to specific subcellular locations (Blondel et al., 2000). Likewise, the yeast Ran-binding protein Yrb1p is required for efficient proteolysis of Pds1p and Sic1p (Baumer et al., 2000), suggesting that nuclear localization is a pre requisite for their destruction. Nuclear–cytoplasmic transport is mediated by importin β-related transport receptors (importins and exportins), which bind their cargoes either directly or via adaptor molecules (Gorlich and Kutay, 1999; Kaffman and O’Shea, 1999). Importins and exportins cooperate with the Ran/Gsp1p GTPase system, which regulates the interactions with their transport substrates.
Surprisingly, both cytoplasmic and nuclear proteins have been identified as APC/C targets, raising the question of whether APC/C functions in both cellular compartments. Alternatively, substrates themselves may shuttle in and out of the nucleus, providing a mechanism to protect them from proteolysis by the APC/C. For example, although Clb2p appears predominantly nuclear, it rapidly shuttles through the cytoplasm and accumulates at the mother bud neck region (Hood et al., 2001). To address these questions, we investigated the subcellular localization of Cdc20p and Cdh1p. Strikingly, while Cdc20p is always nuclear, we found that Cdh1p is exported into the cytoplasm at the onset of S phase, and accumulates at the bud neck. Nuclear export of Cdh1p is mediated by the exportin Msn5p, and regulated by phosphorylation of Cdh1p by Cdc28p. Furthermore, our results suggest that cytoplasmic Cdh1p is inactive, implying that nuclear export of Cdh1p contributes to inactivation of APC/CCdh1 during the cell cycle.
Results
The subcellular localization of Cdc23p, Cdc20p and Cdh1p
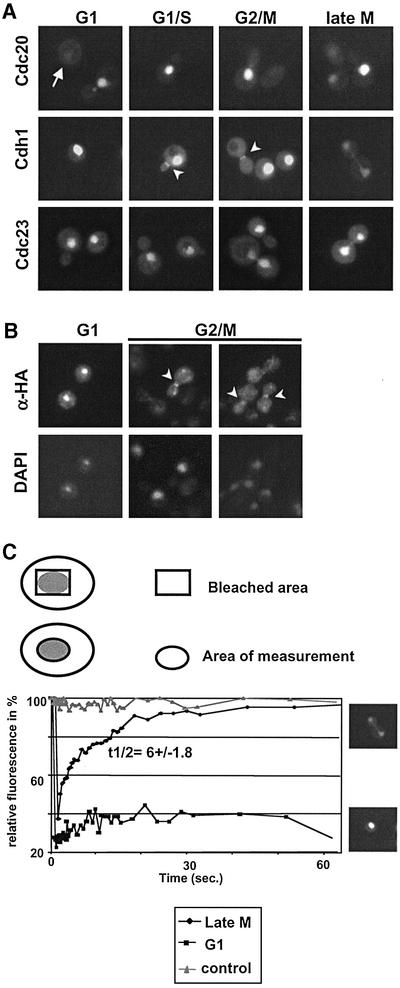
To determine the subcellular localization of Cdc23p, Cdc20p and Cdh1p, we fused GFP to their C-termini, and localized the fusion proteins in wild-type cells after expression from the inducible GAL1,10 promoter. As shown in Figure 1A, Cdc20p–GFP is nuclear at all cell cycle stages (upper row), except in G1 where the protein is degraded and barely visible (arrow) (Prinz et al., 1998; Huang et al., 2001). In contrast, Cdh1p–GFP was found in the nucleus of G1 and G1/S cells and cells at very late stages of mitosis. Surprisingly, however, Cdh1p–GFP was distributed throughout the cytoplasm in small budded cells and in cells at different stages of mitosis (middle row). Quantitation of these results revealed that Cdh1p–GFP was predominantly nuclear in >95% of G1 cells, while <5% of large budded G2/M cells accumulated Cdh1p–GFP in the nucleus. Similar cell cycle-dependent changes were also observed if Cdh1p–GFP was expressed from the endogenous or the ADH1 promoter (data not shown), suggesting that its relocalization was not simply caused by overexpression of Cdh1p–GFP. Interestingly, Cdh1p–GFP was also detected at the mother bud neck shortly after bud emergence (arrowhead), and this staining remained until exit from mitosis. Indirect immunofluorescence experiments using wild-type cells expressing HA3-Cdh1p confirmed the dynamic localization of Cdh1p (Figure 1B), demonstrating that the GFP tag does not affect the subcellular distribution of Cdh1p. Finally, the APC/C core subunit Cdc23p tagged with GFP was nuclear at all cell cycle stages (Figure 1A, lower row), implying that cytoplasmic Cdh1p may not be part of an APC/C complex. Taken together, these results suggest that the subcellular localization of Cdh1p is regulated through the cell cycle, and its nuclear localization correlates with APC/CCdh1 activity.
Fig. 1. Cell cycle-dependent localization of Cdh1p, Cdc20p and Cdc23p. (A) Wild-type cells (EY957) expressing the indicated GFP fusion proteins from the GAL promoter were grown at 30°C until mid-log phase, and analysed by GFP microscopy. Cells at different stages of the cell cycle are shown. The arrowheads point to Cdh1p–GFP localized to the mother bud neck. Note that Cdh1p is nuclear during G1, but predominantly cytoplasmic during S phase, G2 and mitosis. (B) Indirect immunofluorescence of HA3-Cdh1p expressed from the GAL promoter using 11HA antibodies. The arrowhead points to HA3-Cdh1p localized to the mother bud neck. (C) Nuclear recovery of Cdh1p–GFP was measured by FRAP as schematically represented in the upper panel. Nuclear recovery was measured in G1 cells (squares), or in late mitosis (late M) during relocalization of Cdh1p (diamonds). The nuclear/cytoplasmic exchange rate was quantified as described in Materials and methods, and is shown as t1/2 with SD. An unbleached nucleus was included as control (triangles).
We used fluorescence recovery after photobleaching (FRAP) to measure directly whether Cdh1p–GFP shuttles between the nucleus and the cytoplasm. This technique exploits the observation that the GFP chromophore can be inactivated irreversibly by a short exposure to a laser light of 488 nm wavelength without significantly affecting the structure or function of the protein (Ellenberg and Lippincott-Schwartz, 1998). Thus, the nucleus of cells in late mitosis (late M, inset) or of unbudded G1 cells (G1, inset) was photobleached, and the recovery of nuclear fluorescence was quantified and plotted against the time (in seconds) after photobleaching (Figure 1C). For control, nuclear fluorescence of Cdh1p–GFP was measured in cells that were not photobleached (triangles). Interestingly, nuclear fluorescence of Cdh1p–GFP after photobleaching was rapidly restored in mitotic cells (diamonds), while its nuclear recovery was significantly slower in G1 cells (squares). We conclude that Cdh1p–GFP can shuttle between the nucleus and the cytoplasm, and the exchange rate is regulated in a cell cycle-dependent manner.
Nuclear export of Cdh1p–GFP requires its hyperphosphorylation by Cdc28p
The activity of Cdh1p has been shown to be regulated by phosphorylation (Zachariae et al., 1998), predominantly by the Cdc28p–Clb5p kinase (Huang et al., 2001; Yeong et al., 2001). To test whether redistribution of Cdh1p–GFP at bud emergence correlates with hyperphosphorylation, we analysed the localization and phosphorylation state of Cdh1p in cells arrested in G1 with α-factor, in S phase with hydroxyurea (HU) or in mitosis with nocodazole. As shown in Figure 2A, Cdh1p was predominantly cytoplasmic when it was hyperphosphorylated. Moreover, the levels of Cdh1p were significantly lower in G1 cells (lower panel), as compared with cycling cells or at stages where Cdh1p is phosphorylated. To monitor inactivation of Cdh1p after bud emergence, we arrested cells before bud emergence by depletion of the G1 cyclins (Cln1p, Cln2p and Cln3p). As expected, Cdh1p was unphosphorylated in these G1-arrested cells, and Cdh1p–GFP accumulated in the nucleus (Figure 2B). Cdh1p was rapidly hyperphosphorylated after re-expression of Cln2p, while the unphosphorylatable Cdh1p-m11 mutant (Zachariae et al., 1998) remained unchanged, although the cells released with similar kinetics. Interestingly, hyperphosphorylation of wild-type Cdh1p correlated with its redistribution to the cytoplasm, suggesting that it may trigger nuclear export. To test whether hyperphosphorylation of Cdh1p is required for its nuclear export, we compared the localization of wild-type Cdh1p–GFP and Cdh1p-m11–GFP in cells arrested at the metaphase–anaphase transition by depolymerization of the mitotic spindle (nocodazole) or inactivation of the APC/C subunit Cdc16p (cdc16-1), and cdc14-3 cells, which arrest after completion of anaphase (Figure 2C). Strikingly, Cdh1p-m11–GFP was nuclear in both strains, while wild-type Cdh1p–GFP was predominantly cytoplasmic. Immunoblotting confirmed that Cdh1p-m11 is unphosphorylated under these conditions (lower panel). To determine whether Cdc28p activity is required for nuclear export, we constructed a cdc16-1 cdc28-as1 double mutant strain, where Cdc28p can be inhibited rapidly and specifically by the drug C3-1′-naphthylmethyl PP1 number 9 (Na-PP1; Bishop et al., 2000). Cells were arrested in mitosis by shifting the temperature to 37°C, before Na-PP1 was added for 90 min to inactivate Cdc28p. Indeed, Cdh1p was dephosphorylated under these conditions, while it remained in its hyperphosphorylated form in the absence of the inhibitor (Figure 2D). In addition, the levels of Cdh1p decreased (lower panel), suggesting that Cdc28 activity may be needed for stabilization of Cdh1p. Importantly, while Cdh1p–GFP was cytoplasmic in control cells, Cdh1p–GFP accumulated in the nucleus after inhibition of Cdc28p (Figure 2D). Taken together, these results strongly suggest that phosphorylation of Cdh1p by Cdc28p triggers its nuclear export.
Fig. 2. Phosphorylation of Cdh1p by Cdc28p regulates its subcellular localization. (A) The localization of Cdh1p–GFP was analysed by GFP microscopy in wild-type cells arrested in G1 with α-factor, in S phase with HU or in mitosis with nocodazole (NOCO). The phosphorylation state of HA3-Cdh1p was analysed by immunoblotting (lower panel). The arrow points to the position of unphosphorylated Cdh1p-HA. Immunoblotting with antibodies against actin confirms equal loading of the gel (lower blot). (B) cln1,2,3 pMETCLN2 (EY569) cells expressing Cdh1p–GFP from the ADH1 promoter (upper panel) or HA3-Cdh1p from the GAL1 promoter (lower panel) were arrested in G1 by depletion of Cln2p (time = 0), and released by expression of Cln2p. Aliquots were analysed after the times indicated by GFP microscopy (upper images) or immunoblotting (lower blot). Cells expressing non-phosphorylatable HA3-Cdh1p-m11 were included as a control. (C) cdc16-1 (YMP190), cdc14-3 (YMP809) or wild-type (K699) cells arrested with NOCO expressing either wild-type Cdh1p or non-phosphorylatable Cdh1p-m11 fused to GFP (upper panels) were arrested at the metaphase–anaphase transition or after anaphase, respectively, by shifting the temperature to 37°C for 2 h, and analysed by GFP microscopy. Numbers indicate the percentage of cells with nuclear accumulation of Cdh1p–GFP; at least 200 cells were included in the analysis. For control, the phosphorylation state of HA3-Cdh1p or HA3-Cdh1p-m11 was analysed in wild-type or cdc16-1 cells by immunoblotting (upper blot). Immunoblotting with antibodies against actin controls for the loading of the gel (lower blot). (D) Inhibition of Cdc28 kinase activity is sufficient for nuclear accumulation of Cdh1p–GFP. cdc16-1 cdc28-as1 cells (YMJ1055) expressing Cdh1p–GFP were arrested in mitosis by shifting to 37°C. After 90 min, the culture was divided: one half was incubated with the drug Na-PP1 specifically to inhibit Cdc28p-as1 (+Na-PP1), while only the solvent DMSO was added to the other half (–Na-PP1). The localization of Cdh1p–GFP was analysed by GFP microscopy after 90 min. Numbers indicate the percentage of cells with nuclear accumulation of Cdh1p–GFP; at least 200 cells were included in the analysis. For control, the phosphorylation state of HA3-Cdh1p was examined in cdc16-1 cdc28-as1 cells by immunoblotting (lower panel). An immunoblot against actin (arrow) controls for equal loading (lower blot).
Nuclear import and export of Cdh1p are mediated by Pse1p and Msn5p
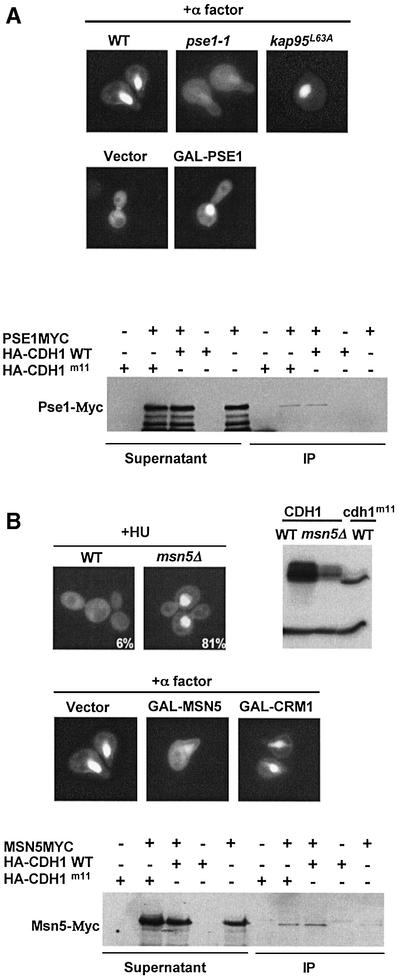
To identify components responsible for regulated nuclear transport of Cdh1p, we first screened mutants defective in known importins or exportins for mislocalization of Cdh1p–GFP (Figure 3). Conversely, we searched for mislocalization of Cdh1p–GFP in wild-type cells overexpressing individual importins or exportins. Interestingly, Cdh1p was predominantly cytoplasmic in α-factor-treated pse1-1 cells (Figure 3A), while overexpression of the importin Pse1p promoted nuclear import of Cdh1p in wild-type cells in G2/M (middle panel). Moreover, Pse1p-myc co-immunoprecipitated specifically with phosphorylated and unphosphorylated Cdh1p (lower panel). Taken together, these results suggest that Pse1p mediates nuclear import of Cdh1p.
Fig. 3. The nuclear localization of Cdh1p is controlled by the importin Pse1p and the exportin Msn5p. (A) Wild-type (K699) cells or temperature-sensitive mutants in the import receptors Pse1p (pse1-1, YBL1) or Kap95p (kap95L63A, YBL14) were arrested in G1 by α-factor, and the localization of Cdh1p–GFP was analysed by GFP microscopy (upper panel). Note that nuclear accumulation of Cdh1p–GFP requires functional Pse1p. Overexpression of Pse1p from the inducible GAL promoter was sufficient for nuclear accumulation of Cdh1p–GFP (middle panels) at cell cycle stages where Cdh1p is predominantly cytoplasmic (vector). Pse1p-myc was able to co-immunoprecipitate (IP) with both wild-type HA3-Cdh1p and unphosphorylatable Cdh1p-m11-HA (lower panel). For co-immunoprecipitation, extracts prepared from cells expressing Pse1p-myc and, as indicated, either no protein (vector), wild-type HA3-Cdh1p or non-phosphorylatable HA3-Cdh1p-m11 were incubated with HA11 antibodies. The immunoprecipitates (IP) were analysed by immunoblot analysis with anti-Myc antibodies (9E10). For control, an aliquot of the extracts before immunoprecipitation (supernatant) was included. (B) The localization of Cdh1p–GFP was examined in wild-type (K699) and msn5Δ (YMJ1171) cells arrested with HU. Numbers indicate the percentage of cells with nuclear accumulation of Cdh1p–GFP; at least 200 cells were included in the analysis. The phosphorylation state of HA-Cdh1p and HA3-Cdh1p-m11 was analysed by immunoblotting in wild-type and msn5Δ cells as indicated (right blot). Overexpression of the exportin Msn5p but not Crm1p from the inducible GAL promoter in wild-type cells arrested with α-factor was sufficient to promote nuclear export of Cdh1p–GFP (middle panel). Co-immunoprecipitation experiments between Msn5p-myc and Cdh1p (lower panel) were performed as described in (A).
Interestingly, Cdh1p–GFP was predominantly cytoplasmic in G1 cells overexpressing the exportin Msn5p (Figure 3B, middle panels), while overexpression of Crm1p or any other exportins (data not shown) had no effect. Conversely, Cdh1p–GFP was detectable in the nucleus of msn5Δ cells at all stages of the cell cycle (data not shown), or when cells were arrested in S phase by the addition of HU (upper panels). Cdh1p was phosphorylated efficiently in msn5Δ cells, indicating that its nuclear accumulation is not the result of a defect in phosphorylation. However, the levels of Cdh1p were significantly reduced in msn5Δ cells, suggesting that nuclear Cdh1p may be less stable. Co-immunoprecipitation (lower panel) and two-hybrid experiments (Figure 4A) confirmed that Msn5p interacted with both wild-type and Cdh1p-m11, implying that phosphorylation of Cdh1p may not directly regulate binding of Cdh1p and Msn5p. Taken together, these results suggest that Msn5p is necessary and sufficient to export Cdh1p during the cell cycle.
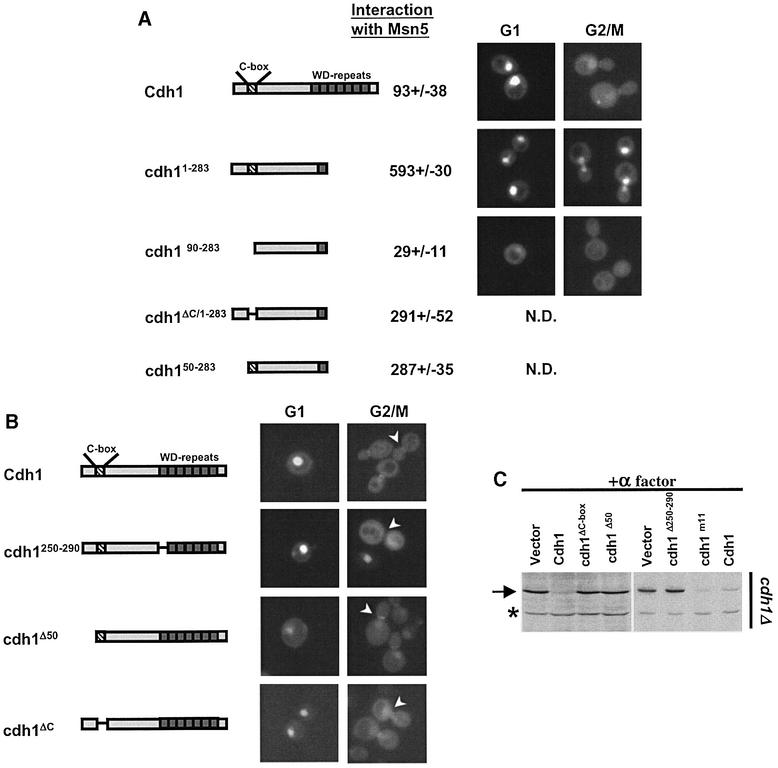
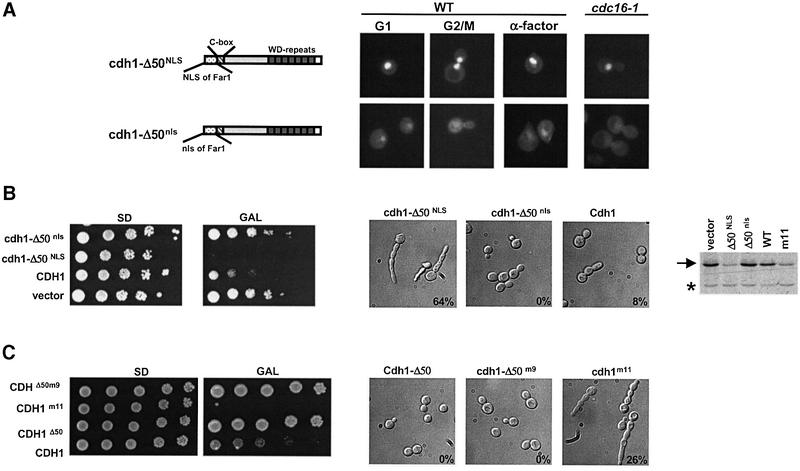
Fig. 4. Characterization of functional domains of Cdh1p for Msn5p binding and subcellular localization. (A) The interaction of Msn5p with full-length Cdh1p or the indicated deletion mutants was analysed by two-hybrid analysis. The numbers indicate Miller units ± SD; the background of the vector controls has already been subtracted. A schematic representation of the various constructs is shown in the left panel. Stippled box: C-box, required for the interaction with the APC/C; hatched bars: WD repeats involved in substrate interaction. The localization of the indicated proteins fused to GFP was analysed by GFP microscopy in wild-type cells at the G1 and G2/M phase of the cell cycle. Note that the 90 N-terminal amino acids are important for binding to Msn5p, but also contain a functional NLS. (B) The localization of various Cdh1p mutant proteins as indicated schematically on the left was analysed by GFP microscopy. Note that an intact substrate interaction site is required for localization of Cdh1p to the mother bud neck (arrowhead). (C) The ability of the indicated proteins to complement the defect of cdh1Δ cells (YMJ1075) to degrade endogenous Clb2p (arrow) in cells arrested in G1 by α-factor was analysed by immunoblotting with Clb2p antibodies.
Deletion analysis of Cdh1p showed that Msn5p interacts with the N-terminal domain of Cdh1p, and required the 90 N-terminal amino acids (Figure 4A). Within this fragment, both an intact C-box, required for APC/C binding (Schwab et al., 2001), and the first 50 amino acids contributed to efficient binding of Msn5p. However, full-length Cdh1p lacking the C-box was still exported in a cell cycle-dependent manner (Figure 4B; data not shown), suggesting that the C-box is not essential for nuclear export in vivo. Besides an Msn5p-binding domain, the first 50 N-terminal amino acids of Cdh1p also contained a functional nuclear localization signal (NLS), as Cdh1p-Δ50 expressed as a GFP fusion was predominantly cytoplasmic (Figure 4B).
We also examined the importance of several functional domains for the localization of Cdh1p to the mother bud neck (Figure 4B, right panel). Full-length Cdh1p deleted for the 50 N-terminal amino acids still accumulated at the mother bud neck. Likewise, the Cdh1p-ΔC-box localized normally during the cell cycle, demonstrating that binding of Cdh1p to the APC/C core complex is required neither for nuclear import or export, nor for its accumulation at the mother bud neck. In contrast, the first WD repeat of Cdh1p, which is likely to be required for binding to Clb2p (Schwab et al., 2001), was necessary for its localization to the mother bud neck, though it did not affect its nuclear localization. Taken together, these results suggest that Cdh1p is targeted to the mother bud neck by binding to its substrates and not to the APC/C core complex. Finally, the Cdh1p-Δ50, Cdh1p-ΔWD and Cdh1p-ΔC-box mutants failed to complement the Clb2p degradation defect of cdh1Δ cells arrested in G1 with α-factor (Figure 4C), indicating that binding to the APC/C as well as nuclear localization of Cdh1p are essential for degradation of Clb2p.
Nuclear export of Cdh1p contributes to inactivation of APC/CCdh1
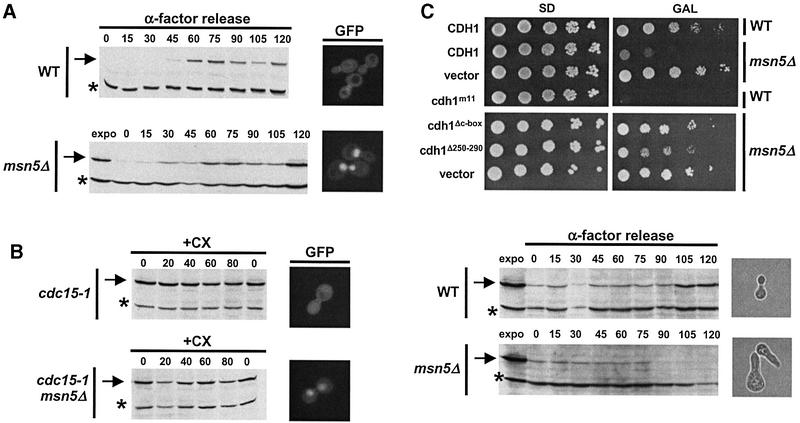
To examine the physiological importance of Cdh1p export, we first characterized msn5Δ cells, which accumulate nuclear Cdh1p at all stages of the cell cycle (Figure 5A). msn5Δ cells are viable (Akada et al., 1996), and the accumulation of Clb2p was comparable in wild-type and msn5Δ cells released from an α-factor arrest. Likewise, the half-life of Clb2p was unaffected in mitotic exit-defective cdc15-1 and cdc15-1 msn5Δ cells (Figure 5B), although Cdh1p accumulated in the nucleus of the latter. In contrast, cdc15-1 cells were able to degrade Clb2p after expression of Cdh1p-m11 (data not shown), excluding the possibility that the Cdc15p function was required for APC/C activity. Taken together, these results indicate that the presence of Cdh1p within the nucleus is not sufficient to induce degradation of Clb2p and thus activation of APC/CCdh1.
Fig. 5. Nuclear Cdh1p has increased activity, but accumulation of endogenous Cdh1p is not sufficient to induce degradation of Clb2p. (A) The accumulation of endogenous Clb2p after bud emergence was examined by immunoblotting in wild-type and msn5Δ cells released from an α-factor block. For control, an extract of exponentially growing cells was included (expo). Note that nuclear Cdh1p is not sufficient to delay accumulation of Clb2p (arrow). The asterisk marks the position of an unknown protein, which is recognized by the Clb2p antibodies. The images on the right show the localization of Cdh1p–GFP in the same strains. (B) The half-life of Clb2p was compared in cdc15-1 (YMP690) or cdc15-1 msn5Δ (YMJ1185) cells after addition of cycloheximide (CX, time = 0). The levels of endogenous Clb2p were examined by immunobloting at the times indicated after addition of CX (in minutes). Note that nuclear Cdh1p is not sufficient to induce degradation of Clb2p (arrow). The images on the right show the localization of Cdh1p–GFP in the same strains. (C) Wild-type (CDH1) or the indicated Cdh1p mutants fused to GFP were overexpressed from the inducible GAL promoter in either wild-type (K699; WT) or msn5Δ (YMJ1171) cells. The plates show 5-fold serial dilutions of the cells spotted in plates containing either glucose (SD; GAL promoter off) or galactose (GAL; GAL promoter on) incubated for 3 days at 30°C. The accumulation of Clb2p (arrow) in wild-type (upper blot) or msn5Δ cells overexpressing Cdh1p from the GAL promoter was examined by immunoblotting after an α-factor release, as described in (A). The phase-contrast images on the right show the morphology of cells at the 120 min time point.
However, in contrast to wild-type cells, overexpression of Cdh1p was lethal in msn5Δ cells (Figure 5C). This toxicity required the ability of Cdh1p to interact with both the APC/C core complex and its substrates, as overexpression of neither the Cdh1p-ΔC-box nor Cdh1p-ΔWD affected growth of msn5Δ cells. Indeed, msn5Δ cells overexpressing Cdh1p failed to accumulate Clb2p after release from an α-factor block (Figure 5C). To confirm that lack of Cdh1p export was responsible for this effect, we constructed a functional, but constitutively nuclear form of Cdh1p, by fusing the NLS of Far1p to the N-terminus of Cdh1p-Δ50 (NLS–Cdh1p-Δ50; Figure 6A). The nuclear import signal of Far1p has been characterized previously (Blondel et al., 1999) and, like Cdh1p, is recognized by Pse1p (B.Luke, unpublished results). For control, we fused Cdh1p-Δ50 to a mutated NLS (nls–Cdh1p-Δ50), which is unable to target Far1p to the nucleus (Blondel et al., 1999). As expected, NLS–Cdh1p-Δ50 but not nls–Cdh1p-Δ50 was nuclear throughout the cell cycle, including in cdc16-1 cells arrested in mitosis where wild-type Cdh1p is predominantly cytoplasmic. In contrast to wild-type Cdh1p–GFP (Figure 3B), NLS–Cdh1p-Δ50–GFP was not exported efficiently by overexpression of Msn5p (data not shown), consistent with its reduced ability to interact with Msn5p. These data confirm that binding of Msn5p to the N-terminal motif of Cdh1p is important for its cell cycle-dependent nuclear export. NLS–Cdh1p-Δ50 was able to degrade Clb2p in cdh1Δ cells arrested with α-factor (data not shown), demonstrating that the 50 N-terminal amino acids of Cdh1p are not required to interact functionally with Clb2p and the APC/C Importantly, overexpression of NLS–Cdh1p-Δ50 but not cytoplasmic nls–Cdh1p-Δ50 in wild-type cells was toxic (Figure 6B), suggesting that nuclear Cdh1p exhibits increased activity. The cells arrested with a single highly elongated bud, similar to wild-type cells expressing non-phosphorylatable Cdh1p-m11. This phenotype is characteristic for cells with low Cdc28p–Clbp activity (Rua et al., 2001). Indeed, cells overexpressing NLS–Cdh1p-Δ50 were unable to accumulate Clb2p (Figure 6B, blot). The failure of Cdh1p-Δ50 to arrest the cell cycle was not due to cytoplasmic phosphorylation, as the non-phosphorylatable Cdh1p-Δ50-m9 was also inactive (Figure 6C). These results demonstrate that nuclear Cdh1p is required to degrade Clb2p, implying that nuclear export of Cdh1p by Msn5p may contribute to inactivation of APC/CCdh1 after bud emergence (Figure 7).
Fig. 6. Nuclear Cdh1p is required for degradation of Clb2p. (A) The NLS of Far1p (NLS) or, for control, a non-functional mutant form (nls) was fused to the N-terminus of Cdh1p-Δ50–GFP as schematically indicated on the left. The proteins were expressed as GFP fusions in either wild-type (K699; left panels) or cdc16-1 cells (YMP190; right panels) and analysed by GFP microscopy. Where indicated, wild-type cells were arrested in G1 by addition of α-factor for 2 h. Note that NLS–Cdh1p-Δ50 (upper panels) but not nls–Cdh1p-Δ50 (lower panels) is nuclear at all stages of the cell cycle. (B and C) Wild-type (K699) cells overexpressing either no protein (vector), untagged (B) or HA3-tagged (C) wild-type Cdh1p or the indicated Cdh1p mutants from the inducible GAL promoter were analysed for their ability to form colonies by serial dilution on plates containing glucose (SD; GAL promoter off) or galactose (GAL; GAL promoter on). The plasmids used in (C) were integrated at the URA3 locus of K699 cells. The morphology of the cells was analysed by differential interference contrast microscopy (DIC, right panels). Numbers indicate the percentage of cells with elongated buds. The accumulation of endogenous Clb2p (arrow) was analysed by immunoblotting with specific antibodies (blot). The asterisk marks the position of an unknown protein, which is recognized by the Clb2p antibodies and serves as a loading control.
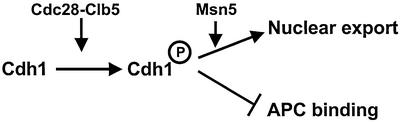
Fig. 7. Nuclear export of Cdh1p may contribute to the inactivation of APC/CCdh1. Model for the inactivation of APC/CCdh1 after bud emergence. Phosphorylation of Cdh1p by Cdk1 (predominantly Cdc28p–Clb5p) prevents its interaction with the APC/C core complex (Zachariae et al., 1998), and promotes nuclear export by Msn5p. Cytoplasmic sequestration of Cdh1p prevents the assembly of functional APC/CCdh1 complexes during cell cycle stages other than G1, thereby contributing to the inactivation of APC/CCdh1.
Discussion
Phosphorylation regulates the subcellular localization of Cdh1p
We found that Cdh1p is regulated at the level of subcellular localization: Cdh1p is nuclear in G1, but predominantly cytoplasmic in S, G2 and M phase of the cell cycle. In addition, Cdh1p accumulates at the mother bud neck shortly after bud emergence. FRAP analysis showed that Cdh1p rapidly shuttles between the cytoplasm and the nucleus at cell cycle stages other than G1, suggesting that nuclear transport of Cdh1p is regulated in a cell cycle-dependent manner. The evidence presented here suggests that cytoplasmic accumulation of Cdh1p is regulated at the level of phosphorylation. First, hyper phosphorylation of Cdh1p correlates with its cytoplasmic localization. Secondly, cytoplasmic Cdh1p redistributes into the nucleus after inactivation of Cdc28p and, finally, a non-phosphorylatable mutant form of Cdh1p is constitutively nuclear. Recent work suggested that Cdh1p is phosphorylated after bud emergence predominantly by Cdc28p–Clb5p (Huang et al., 2001; Yeong et al., 2001). We thus suggest that phosphorylation of Cdh1p triggers its nuclear export and cytoplasmic accumulation.
Cdh1p is exported by the exportin Msn5p
We have identified Pse1p and Msn5p as the probable factors which control the nuclear–cytoplasmic exchange of Cdh1p, although we have not demonstrated that they interact in a Ran/Gsp1p-dependent manner. Pse1p is also required for the import of the transcription factor Pho4p and the scaffold Far1p (Kaffman and O’Shea, 1999). Likewise, several proteins have been shown to be targets of Msn5p, and are thought to be exported in a regulated manner. For example, Msn5p exports Far1p (Blondel et al., 1999) and Ste5p (Mahanty et al., 1999) in response to mating pheromones, and Msn5p keeps Pho4p cytoplasmic in cells grown in rich phosphate conditions (Kaffman et al., 1998). Interestingly, phosphorylation of Pho4p has been shown to promote binding of Pho4p to Msn5p. At present, we do not know how phosphorylation controls nuclear export of Cdh1p. Surprisingly, unphosphorylated Cdh1p interacted with Msn5p, implying that phosphorylation may not regulate this interaction directly. It is possible that phosphorylation of Cdh1p regulates the interaction of Msn5p with the nuclear pore complex or additional adaptor proteins, or that the nuclear export signal (NES) of Cdh1p is only exposed in the phosphorylated conformation. Consistent with this notion, the N-terminal domain of Cdh1p interacted with Msn5p much more efficiently than with the full-length protein. Alternatively, phosphorylation of Cdh1p may not increase nuclear export, but rather inhibit its nuclear import.
Despite the growing number of Msn5p and Pse1p targets, it has been difficult to define a common NES and NLS, respectively. Analysis of Cdh1p mutants revealed that binding to Msn5p required a small N-terminal domain but, at the primary sequence level, we were unable to align this sequence with the putative NES of Far1p, Pho4p and Ste5p. It appears therefore that Msn5p may interact with its targets through a common tertiary structure rather than a linear motif or, alternatively, Msn5p may interact with these proteins through distinct binding sites.
Redundant mechanisms ensure efficient inactivation of APC/CCdh1 after bud emergence
It is likely that nuclear export of Cdh1p sequesters Cdh1p away from the APC/C core complex, thereby contributing to its inactivation. Cdc23p is nuclear at all stages of the cell cycle, and Cdh1p does not co-immunoprecipitate with Cdc23p during cell cycle stages where it is predominantly cytoplasmic (Zachariae et al., 1998). Overexpression of a cytoplasmic, but non-phosphorylatable and thus active form of Cdh1p (Cdh1p-Δ50-m9) did not interfere with cell growth, suggesting that nuclear localization of Cdh1p is crucial for its function. However, we cannot exclude that Cdh1p plays an active role in the cytoplasm, which does not require the APC/C core complex. Cdh1p accumulates at the mother bud neck shortly after bud emergence, where both its substrates Clb2p (Hood et al., 2001) and the checkpoint kinase Hsl1p (Barral et al., 1999) are found. Hsl1p disappears from the ring shortly after bud emergence (Burton and Solomon, 2001), implying that it may be degraded at a time when Cdh1p is thought to be inactive. However, we found that Hsl1p was degraded efficiently in cdh1Δ cells expressing Cdh1p-m11 or NLS–Cdh1p-Δ50 (data not shown), implying that Hsl1p is probably ubiquitylated by nuclear Cdh1p. Cdh1p binds to its substrates independently of the APC/C (Burton and Solomon, 2001; Pfleger et al., 2001; Schwab et al., 2001), suggesting that it may recruit substrates to the APC/C complex. A similar mechanism has been proposed previously for F-box proteins (Blondel et al., 2000), which function as substrate-specific adaptors in SCF complexes. Interestingly, a mutant form of Cdh1p unable to interact with its target Clb2p failed to localize to the mother bud neck, suggesting that binding to substrates targets Cdh1p to this structure. We thus speculate that Clb2p is transported into the nucleus in a complex with Cdh1p, which in turn interacts with the APC/C core complex to ubiquitylate Clb2p.
We propose that multiple mechanisms function in a redundant manner to inactivate Cdh1p after bud emergence, and to keep it inactive until exit from mitosis. Phosphorylation of Cdh1p functions as a switch preventing binding of Cdh1p to the APC/C core complex and promoting its nuclear export by Msn5p (Figure 7). The binding site for Msn5p overlaps with the C-box of Cdh1p, raising the possibility that Msn5p and the APC/C may compete for binding to Cdh1p. However, Cdh1p-m11 was not toxic when expressed from the endogenous promoter in wild-type cells (data not shown), suggesting that additional mechanisms exist to inactivate Cdh1p. Interestingly, we observed that nuclear (unphosphorylated) Cdh1p is significantly less abundant compared with its cytoplasmic (phosphorylated) forms. The reason for this difference remains to be investigated, but it appears that nuclear Cdh1p is unstable and rapidly degraded by a ubiquitin-dependent mechanism (M.Jaquenoud and M.Peter, unpublished results). An F-box protein has been identified recently in Xenopus (Reimann et al., 2001) and Drosophila (Grosskortenhaus and Sprenger, 2002), which binds to the Cdh1p homologue Fizzy and functions as a negative regulator of APC/CFizzy activity in vivo.
Orthologues of Cdh1p have been found in most species including plants. In human and mouse, multiple isoforms of Cdh1p have been identified, which vary in their expression pattern and substrate specificity (Wan and Kirschner, 2001). Like Cdh1p, Ste9p in Schizosaccharomyces pombe is regulated by phosphorylation (Blanco et al., 2000), but no nuclear export of Ste9p has been reported. The localization and regulation of mammalian Cdh1p-like proteins remain to be investigated, although it is likely that phosphorylation is also required for their inactivation (Kramer et al., 2000). We expect that the basic mechanistic strategies used by yeast to regulate Cdh1p, including modulation of its binding to the APC/C, re-localization and perhaps destruction, are likely to be conserved, although the precise implementation of those strategies may vary. Multiple inhibitory mechanisms generally may be used to ensure critical cell cycle transitions (Nguyen et al., 2001), which provides the possibility to integrate additional controls including checkpoint pathways.
Materials and methods
Strains constructions and genetic manipulations
Yeast strains are described in Table I. The genotypes of the yeast strains are W303 (ade2-1, trp1-1, can1-100, leu2-3 112, his3-11,15, ura3, ssd1-d2), unless noted otherwise. Standard yeast growth conditions and genetic manipulations were used as described (Guthrie and Fink, 1991). GALHA3-CDH1 constructs were linearized with EcoRV for integration at the URA3 locus. YMJ1055 (cdc16-1 cdc28::cdc28AS1) was obtained by crossing YMP190 and YMJ1054. pse1::PSE1MYC13 was tagged endogenously using a PCR-based method described previously (Longtine et al., 1998).
Table I. Yeast strains.
| Strains | Relevant genotypes | Sources |
|---|---|---|
| MJ989 | ura3::GAL HA3-CDH1URA3 | W.Seufert |
| MJ1180 | ura3::GAL HA3-CDH1ΔC-box URA3 | W.Seufert |
| MJ990 | ura3::GAL HA3-CDHm11 URA3 | W.Seufert |
| MJ1180 | ura3::GAL HA3-CDH1Δ50 URA3 | This study |
| MJ1242 | ura3::GAL HA3-CDHm9Δ50 URA3 | This study |
| YMP190 | cdc16-1 | K.Nasmith |
| MJ991 | cdc16-1, ura3::GAL HA3-CDH1 URA3 | This study |
| MJ992 | cdc16-1, ura3::GAL HA3-CDH1m11 URA3 | This study |
| YMP809 | cdc14-3 | T.Hyman |
| YMP690 | cdc15-1 | R.Deshaies |
| MJ1185 | cdc15-1, msn5::URA3 | This study |
| MJ1055 | cdc16-1, cdc28::cdc28AS1 | This study |
| MJ1175 | cdc16-1, cdc28::cdc28AS1, ura3::GAL HA3-CDH1 URA3 | This study |
| BL1 | pse1-1 | P.Silver |
| BL14 | kap95::HIS3(pSWkap95-L63A RS315) | P.Silver |
| MJ1171 | msn5::URA3 | This study |
| MJ1034 | msn5::HIS3, ura3::GAL HA3-CDH1 URA3 | This study |
| MJ1038 | pse1::PSE1-MYC13 | This study |
| MJ1065 | pse1::PSE1-MYC13 HIS3, ura3::GAL HA3-CDH1URA3 | This study |
| MJ1066 | pse1::PSE1-MYC13 HIS3, ura3::GAL HA3-CDH1m11 URA3 | This study |
| EY569 | cln1::HisG, cln2, cln3::LEU2, METCLN2 TRP1 | E.Schwob |
| MJ1228 | cln1::HisG, cln2, cln3::LEU2, METCLN2 TRP1, ura3::GAL HA3-CDH1 URA3 | This study |
| MJ1229 | cln1::HisG, cln2, cln3::LEU2, METCLN2 TRP1, ura3::GAL HA3-CDH1m11 URA3 | This study |
| MJ1075 | cdh1::LEU2 | A.Amon |
| MJ1182 | cdh1::LEU2, ura3::GAL HA3-CDH1 URA3 | This study |
| MJ1183 | cdh1::LEU2, ura3::GAL HA3-CDH1ΔC-box URA3 | This study |
| MJ1184 | cdh1::LEU2, ura3::GAL HA3-CDH1Δ50 URA3 | This study |
DNA manipulations and two-hybrid assays
Plasmids are described in Table II. Standard procedures were used for recombinant DNA manipulations (Ausubel et al., 1991). PCRs were performed with the Expand polymerase kit as recommended by the manufacturer (Roche). Oligonucleotides were synthesized by Genset (Paris, France), and are listed in Table III. Details of plasmid constructions are available on request. The following sequences encompassing the NLS of Far1p (Blondel et al., 1999) were fused to Cdh1p-Δ50: NLS, MKTPTRVSFEKKIHTPPSGDRDAERSPPKKFL RGLSGKVFRKTPEFKKQQ; and nls, MKTPTRVSFEKKIHTPPSG DRDAERSPPAAFLRGLSGKVFAATPEFKKQQ. Two-hybrid assays were performed in EGY48 containing the LacZ reporter plasmid pSH18.34 (Gyuris et al., 1993). Miller units are the average of at least three independent experiments with standard deviations (Brown et al., 1997). Expression of Cdc23p–GFP or Cdc20p–GFP from the GAL promoter was able to complement temperature-sensitive cdc23-1 and cdc20-1 mutants, respectively, suggesting that the fusion proteins are functional.
Table II. Plasmids.
| Plasmids | Relevant characteristics | Sources |
|---|---|---|
| MJ942 | GAL CDC20 GFP TRP1 CEN | This study |
| MJ943 | GAL CDH1 GFP TRP1 CEN | This study |
| MJ962 | ADH CDH1 GFP URA3 CEN | This study |
| MJ963 | CYC1 CDH1 GFP URA3 CEN | This study |
| MJ1021 | CDH1 GFP TRP1 CEN | This study |
| MJ967 | GAL CDH1m11 GFP TRP1 CEN | This study |
| MJ1022 | GAL CDC23 GFP URA3 CEN | This study |
| BL39 | GAL PSE1 URA3 2µ | This study |
| BL35 | GAL MSN5 URA3 2µ | B.Luke |
| BL32 | GAL CRM1 URA3 2µ | B.Luke |
| BM56 | MSN5 MYC1n TRP1 2µ | M.Blondel |
| MJ1080 | GAL CDH11–250 GFP TRP1 CEN | This study |
| MJ1181 | GAL CDH190–250 GFP TRP1 CEN | This study |
| MJ1082 | GAL CDH1Δ250–290 GFP TRP1 CEN | This study |
| MJ1227 | GAL CDH1Δ–C GFP TRP1 CEN | This study |
| MJ1165 | GAL CDH1Δ50 GFP TRP1 CEN | This study |
| MJ1163 | GAL NLS CDH1Δ50 GFP TRP1 CEN | This study |
| MJ1164 | GAL nls1,2 CDH1Δ50 GFP TRP1 CEN | This study |
| MJ1036 | AD CDH1 pJG4-6 | This study |
| MJ1048 | AD CDH11–283 pJG4-6 | This study |
| MJ1161 | AD CDH1Δ–C/1–283pJG4-6 | This study |
| MJ1138 | AD CDH190–283 pJG4-6 | This study |
| MJ1166 | AD CDH150–283 pJG4-6 | This study |
| BM41 | BD MSN5 pEG203 | M.Blondel |
| MJ1168 | GAL CDH1 TRP1 CEN | This study |
| MJ1206 | GAL CDH1Δ250–290 TRP1 CEN | This study |
| MJ1174 | GAL NLS CDH1Δ50 TRP1 CEN | This study |
| MJ1175 | GAL nls1,2 CDH1Δ50 TRP1 CEN | This study |
| MJ1225 | CDH1m11 TRP1 CEN | This study |
| MJ1226 | CDH1-Δ50NLS TRP1 CEN | This study |
Table III. Oligonucleotides.
| Name | Sequences |
|---|---|
| OTP1032 | 5′ GTATTACAAAGAAGATCTATGCCAGAAAGC 3′ |
| OTP1033 | 5′ CATGAACTTTTATTTTTTTTCTCGAGTCACTGCAGCAAATATTGGCTGG 3′ |
| OTP1034 | 5′ CTTCATCCTAAATTTAGATCTATGTCCACAAACC 3′ |
| OTP1035 | 5′ GATATTACTACTATGAAAACCTCGAGCTAACGGAATTCATTAAATGC 3′ |
| OTP1181 | 5′ CAGATTGTGAAGCATGCGGCCGCATGAATGACG 3′ |
| OTP1182 | 5′ GTGTATGGGCCCTTCACTACATATGCCGTGTATGGGCCCTTCACTACATATGCC 3′ |
| OTP1308 | 5′ CTAAAACTCTATAGAATTCTTTAGC 3′ |
| OTP1309 | 5′ CGATTTTTTTAACCGATGAATTCACTGGCGACG 3′ |
| OTP1503 | 5′ GGGCGTAAGAAGGCAATCTGCAGATGAAGACACC 3′ |
| OTP1504 | 5′ CTTCAATATACCCGAATTCGGGCATCTGTTGC 3′ |
| OTP1505 | 5′ CATCTCCCTCCATGGGATCGAGGGAATTCATGGTATATG 3′ |
| OTP1684 | 5′ GTGGACATGAATTCAAATTTAGGATG 3′ |
| OTP1685 | 5′ GTGGACATCTGCAGAAATTTAGG 3′ |
| OTP1310 | 5′ GCCCTTAATCCATCGCGGCCGCAACTGCAGATGGAATACCAAAAGG 3′ |
| OTP1205 | 5′ GGCAATTTTCAATAGATATCCAGCTGATATTATGGAGAAAGTACATAAATGGTTTGCACGGATCCCCGGGTTAATTAA 3′ |
| OTP1206 | 5′ CCTTATAGAACTACTTAACATTGTTCTTCTTTCTATTAAACGTCTGAATTCGAGCTCGTTTAAAC 3′ |
Immunofluorescence and microscopy
Proteins tagged with GFP were visualized on a Zeiss Axiophot fluorescence microscope using a Chroma GFPII filter. Proteins expressed from the GAL promoter were induced by the addition of 2% galactose for 2 h. For quantitation, at least 200 cells were analysed. FRAP analysis was performed with a Zeiss LSM510 confocal laser scanning microscope, using a LP505 nm filter, essentially as described previously (van Drogen et al., 2001). Briefly, an argon laser with a maximum output of 25 mW was used at 25% capacity. After capture of four initial images (at 0.1% transmission), photobleaching was carried out with 20 iterations at 100% laser transmission. A rectangle was chosen for the bleached area, which included the entire nucleus. The intensity of fluorescence was measured in a 12-bit fashion (4096 greytones), the average of the area of measurement was calculated and the highest value of the four initial images before photobleaching was set to 100% intensity. τ1/2 was calculated as the time needed to reach half of the final intensity after bleaching as described previously in Ellenberg and Lippincott-Schwartz (1998). At least 20 cells were photobleached under each condition.
Indirect immunolocalization experiments were performed as described in Barral et al. (1999). Briefly, cells from log-phase cultures were fixed with formaldehyde for 15 min, washed with 0.1 M phosphate buffer supplemented with 10 mM DTT and EDTA, and spheroplasted for 5 min with 2 mg/ml of Zymolyase 20T. Cells were spotted on a Teflon slide and incubated with 11HA antibodies for 1 h at 37°C. The slides were washed with PBS containing 0.1% BSA and 0.1% Triton before the Alexa488-conjugated secondary antibody (Molecular Probes) was added for 1 h at 37°C.
Cell cycle synchronization
Exponentially growing cln1,2,3Δ METCLN2 (EY569) cells were arrested in G1 by repressing CLN2 for 3 h in selective medium containing 2 mM methionine. After quick washes, cells were released in medium lacking methionine (time = 0), and aliquots were analysed after the times indicated. For α-factor block/release experiments, bar1-1 cells were arrested for 2 h by addition of 30 µg/ml α-factor (LIPAL-Biochemicals, Zurich). After quick washes, cells were released in medium without α-factor (time = 0), and aliquots were analysed by western blot and FACS analysis (data not shown) after the times indicated. Where indicated, cells were arrested for 150 min with 90 µg/ml HU (Sigma), or 50 µg/ml nocodazole (Sigma). Efficient cell cycle arrest was controlled microscopically. For Gal induction in temperature-sensitive strains, 2% galactose was added for 30 min at 25°C before shifting the cells to 37°C for 150 min. For the Na-PP1 experiments, YMJ1055 cells (cdc16-1, cdc28::CDC28AS1) were arrested in mitosis by shifting the temperature to 37°C for 90 min. The culture was divided, and one half was incubated with 1 µM Na-PP1 (PP1 analogue number 9; Bishop et al., 2000), while the solvent DMSO was added to the other half as a control. After 90 min, the cells were analysed by GFP microscopy and immunoblotting.
Co-immunoprecipitation and immunoblotting experiments
Standard conditions were used for yeast cell extracts and immunoblotting (Harlow and Lane, 1988). Co-immunoprecipitation experiments were performed essentially as described previously (Blondel et al., 1999). Briefly, cells expressing Pse1p-myc (MJ1038) or Msn5p-myc (plasmid BM56), and either no protein (vector), HA-Cdh1p (MJ1065) or HA-cdh1pm11 (MJ1066), were grown on selective medium and induced for 2 h by adding 2% galactose. Lysis was performed in RIPA buffer [200 mM NaCl, 50 mM Tris pH 7.5, 1% NP-40, 0.5% deoxycholate, 0.1% SDS and the proteases inhibitors PMSF, aprotinin, leupeptin and pepstatin (complete mix from Roche)]. Approximately 10 mg of lysate was incubated for 2 h with 11HA antibodies (Babco, Berkeley), and the immunocomplexes were collected by addition of protein G–Sepharose (Pharmacia). The beads were washed four times with lysis buffer and twice with PBS. The final pellet was transferred into a new tube and bound proteins were eluted in 75 µl of gel sample buffer. Western blotting was performed using α-11HA and α-MYC(9E10) monoclonal antibodies. Polyclonal antibodies against Clb2p were kindly provided by D.Kellogg (UCSC). Antibodies against actin were used as recommended by the manufacturer (Roche).
Acknowledgments
Acknowledgements
We thank B.Luke, S.Seufert, D.Morgan, G.Ammerer, E.Schwob, Y.Barral, M.Tyers and A.Amon for providing plasmids, strains and antibodies, K.Shokat for synthesis of the Na-PP1 compound, Nathalie Garin and Marcel Allegrini for help with microscopy, Nathalie Perrinjaquet for expert technical assistance, C.Schueller for the bioinformatic analysis, and C.Lafourcade for sharing unpublished results. We thank members of the group for stimulating discussion, and Richard Iggo, Viesturs Simanis and Pierre Gönczy for critical reading of the manuscript. M.P. is supported by the Swiss National Science Foundation, the Swiss Cancer League and a grant from the HFSPO.
References
- Akada R., Kallal,L., Johnson,D.I. and Kurjan,J. (1996) Genetic relationships between the G protein by complex, Ste5p, Ste20p and Cdc42p: investigation of effector roles in the yeast pheromone response pathway. Genetics, 143, 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A., Irniger,S. and Nasmyth,K. (1994) Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell, 77, 1037–1050. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1991) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- Barral Y., Parra,M., Bidlingmaier,S. and Snyder,M. (1999) Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev., 13, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer M., Kunzler,M., Steigemann,P., Braus,G.H. and Irniger,S. (2000) Yeast Ran-binding protein Yrb1p is required for efficient proteolysis of cell cycle regulatory proteins Pds1p and Sic1p. J. Biol. Chem., 275, 38929–38937. [DOI] [PubMed] [Google Scholar]
- Bishop A.C. et al. (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature, 407, 395–401. [DOI] [PubMed] [Google Scholar]
- Blanco M.A., Sanchez-Diaz,A., de Prada,J.M. and Moreno,S. (2000) APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J., 19, 3945–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M., Alepuz,P.M., Huang,L.S., Shaham,S., Ammerer,G. and Peter,M. (1999) Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev., 13, 2284–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M., Galan,J.M., Chi,Y., Longaretti,C., Lafourcade,C., Deshaies,R.J. and Peter,M. (2000) Nuclear-specific degradation of Far1p is controlled by the localization of the F-box protein Cdc4p. EMBO J., 19, 6085–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.L., Jaquenoud,M., Gulli,M.P., Chant,J. and Peter,M. (1997) Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev., 11, 2972–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.L. and Solomon,M.J. (2001) D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev., 15, 2381–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters,J.M., Kirschner,M.W. and Koshland,D. (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev., 10, 3081–3093. [DOI] [PubMed] [Google Scholar]
- Ellenberg J. and Lippincott-Schwartz,J. (1998) Fluorescence photo bleaching techniques. In Spector,D., Goldman,R. and Leinwand,L. (eds), Cells: A Laboratory Manual. Cold Spring Habor Laboratory Press, Cold Spring Harbor, NY, pp. 79.1–79.23.
- Gorlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R. and Sprenger,F. (2002) Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev. Cell, 2, 29–40. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to yeast genetics and molecular biology. Methods Enzymol., 194. [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hood J.K., Hwang,W.W. and Silver,P.A. (2001) The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J. Cell Sci., 114, 589–597. [DOI] [PubMed] [Google Scholar]
- Huang J.N., Park,I., Ellingson,E., Littlepage,L.E. and Pellman,D. (2001) Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol., 154, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles,J.F. and Morgan,D.O. (1999) Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol., 9, 227–236. [DOI] [PubMed] [Google Scholar]
- Kaffman A. and O’Shea,E.K. (1999) Regulation of nuclear localization: a key to a door. Annu. Rev. Cell. Dev. Biol., 15, 291–339. [DOI] [PubMed] [Google Scholar]
- Kaffman A., Rank,N.M., O’Neill,E.M., Huang,L.S. and O’Shea,E.K. (1998) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature, 396, 482–486. [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer,N., Podtelejnikov,A.V., Mann,M. and Peters,J.M. (2000) Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell, 11, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Mahanty S.K., Wang,Y., Farley,F.W. and Elion,E.A. (1999) Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell, 98, 501–512. [DOI] [PubMed] [Google Scholar]
- Nguyen V.Q., Co,C. and Li,J.J. (2001) Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature, 411, 1068–1073. [DOI] [PubMed] [Google Scholar]
- Peters J.M. (1998) SCF and APC: the yin and yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol., 10, 759–768. [DOI] [PubMed] [Google Scholar]
- Peters J.M. (1999) Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res., 248, 339–349. [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., Lee,E. and Kirschner,M.W. (2001) Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev., 15, 2396–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. (1999) Four-dimensional control of the cell cycle. Nat. Cell Biol., 1, E73–79. [DOI] [PubMed] [Google Scholar]
- Prinz S., Hwang,E.S., Visintin,R. and Amon,A. (1998) The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol., 8, 750–760. [DOI] [PubMed] [Google Scholar]
- Reimann J.D., Freed,E., Hsu,J.Y., Kramer,E.R., Peters,J.M. and Jackson,P.K. (2001) Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell, 105, 645–655. [DOI] [PubMed] [Google Scholar]
- Rua D., Tobe,B.T. and Kron,S.J. (2001) Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol., 4, 720–727. [DOI] [PubMed] [Google Scholar]
- Schwab M., Lutum,A.S. and Seufert,W. (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell, 90, 683–693. [DOI] [PubMed] [Google Scholar]
- Schwab M., Neutzner,M., Mocker,D. and Seufert,W. (2001) Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J., 20, 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugendreich S., Tomkiel,J., Earnshaw,W. and Hieter,P. (1995) CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell, 81, 261–268. [DOI] [PubMed] [Google Scholar]
- van Drogen F., Stucke,V., Jorritsma,G. and Peter,M. (2001) MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol., 3, 1051–1059. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz,S. and Amon,A. (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science, 278, 460–463. [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig,K., Hwang,E.S., Prinz,S., Tyers,M. and Amon,A. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell, 2, 709–718. [DOI] [PubMed] [Google Scholar]
- Wan Y. and Kirschner,M.W. (2001) Identification of multiple CDH1 homologues in vertebrates conferring different substrate specificities. Proc. Natl Acad. Sci. USA, 98, 13066–13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong F.M., Lim,H.H., Wang,Y. and Surana,U. (2001) Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol. Cell. Biol., 21, 5071–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab,M., Nasmyth,K. and Seufert,W. (1998) Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science, 282, 1721–1724. [DOI] [PubMed] [Google Scholar]