Abstract
The anti-angiogenic activity of endostatin (ES) depends on interactions with heparan sulfate (HS). In the present study, intact HS chains of ≥15 kDa bound quantitatively to ES whereas N-sulfated HS decasaccharides, with affinity for several fibroblast growth factor (FGF) species, failed to bind. Instead, ES-binding oligosaccharides composed of mixed N-sulfated and N-acetylated disaccharide units were isolated from pig intestinal HS. A 10/12mer ES-binding epitope was identified, with two N-sulfated regions separated by at least one N-acetylated glucosamine unit (SAS-domain). Cleavage at the N-acetylation site disrupted ES binding. These findings point to interaction between discontinuous sulfated domains in HS and arginine clusters at the ES surface. The inhibitory effect of ES on vascular endothelial growth factor-induced endothelial cell migration was blocked by the ES-binding SAS-domains and by heparin oligosaccharides (12mers) similar in length to the ES-binding SAS-domains, but not by 6mers capable of FGF binding. We propose that SAS-domains modulate the biological activities of ES and other protein ligands with extended HS-binding sites. The results provide a rational explanation for the preferential interaction of ES with certain HS proteoglycan species.
Keywords: endostatin/heparan sulfate/heparin/oligosaccharides/sulfate groups
Introduction
Heparan sulfate (HS) proteoglycans, i.e. proteins that carry one or more O-linked HS chains, occur on cell surfaces and in the extracellular matrix. The HS chains bind a variety of proteins, thus affecting their biological activities. For instance, in fibroblast growth factor (FGF) signaling, HS forms part of a ternary complex that involves both FGF and its receptor (reviewed by Casu and Lindahl, 2001). Subtle changes in HS structure contribute to modulation of signaling activity (Pye et al., 1998; Lundin et al., 2000; Nurcombe et al., 2000). The specificity of HS–protein interactions appears to depend primarily on interactions between sulfate and carboxyl groups in HS and positively charged amino acid residues in the protein (Salmivirta et al., 1996). The highly varied fine structure of HS chains involves a multitude of different epitopes that interact with proteins in a selective fashion (Gallagher, 2001; Kreuger et al., 2001; Loo et al., 2001; Jemth et al., 2002).
The HS backbone is assembled from alternating glucuronic acid (GlcA) and N-acetylglucosamine (GlcNAc) units and then is enzymatically modified in serial fashion by enzymes present in the Golgi compartment. The modification reactions include partial N-deacetylation/N-sulfation of GlcNAc units, C-5 epimerization of GlcA to iduronic acid (IdoA) and sulfation at C-2 of IdoA and GlcA, and at C-3 and C-6 of GlcN units (Esko and Lindahl, 2001). The overall control of biosynthetic polymer modification depends in part on the substrate specificities of the enzymes involved, but is otherwise poorly understood. Owing to the difficulty in isolating the HS designed for interaction with a particular protein ligand, the fully sulfated and readily available related polysaccharide, heparin, is often used as a substitute in model interactions.
Endostatin (ES), a 22 kDa polypeptide released by proteolytic cleavage of collagen XVIII, is an anti-angiogenic protein capable of dramatically decreasing the size of experimental tumors (O’Reilly et al., 1997). The mechanism behind this effect remains unclear, but appears to involve interaction with HS (Sasaki et al., 1999; Dixelius et al., 2000; Karumanchi et al., 2001) that somehow blocks the pro-angiogenic activities of FGFs and vascular endothelial growth factor (VEGF) (reviewed by Iozzo and San Antonio, 2001). HS may function as a co-receptor that mediates efficient binding of ES to its assumed high affinity receptor. While many proteins with high affinity for heparin/HS, such as antithrombin or FGFs, recognize short motifs that generally span <6 monosaccharide units (Maccarana et al., 1993; Faham et al., 1996; DiGabriele et al., 1998), we found that ES required heparin fragments as long as 10–12mers for significant binding (Sasaki et al., 1999). In contrast, Karumanchi et al. (2001) recently proposed that a heparin 6mer with a non-sulfated GlcA unit proximal to its reducing end would serve as a specific ligand for ES. The present study was undertaken to define ES-binding epitopes present in the natural HS ligand. Importantly in this regard, the monosaccharide units of the HS chain are organized in distinct domains according to the N-substituents of GlcN units. Such domains, of varying length, are composed of either (i) consecutive N-sulfated (NS-domains); (ii) alternating N-acetylated and N-sulfated (NA/NS domains); or (iii) consecutive N-acetylated (NA-domains) disaccharide units. Previous studies of protein-binding HS domains employed oligosaccharides corresponding to NS-domains, obtained by specific lyase digestion or by chemical N-deacetylation followed by deaminative cleavage (Casu and Lindahl, 2001).
In the present assessment of HS–ES interactions, we chose a novel and less biased approach, aiming for oligosaccharides containing N-sulfated as well as N-acetylated disaccharide units (SAS-domains). The results define an ES-binding HS epitope that contains a single N-acetylated disaccharide unit at a defined site within a 10/12mer N-sulfated domain. The ES-binding SAS oligosaccharides as well as heparin fragments of similar size could counteract the inhibitory effect of ES on endothelial cell migration efficiently, whereas shorter saccharides had no effect. These results are the first to demonstrate the importance of HS domain organization in interactions with a monomeric protein and in modulation of a biological activity.
Results
Binding of poly- and oligosaccharides to ES
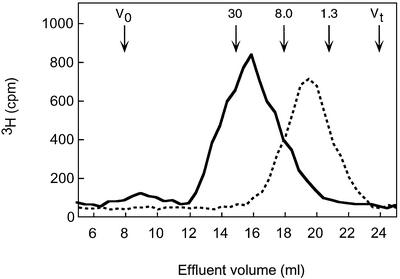
Full-length, N-[3H]acetyl-labeled HS preparations isolated from pig intestinal mucosa and bovine kidney were analyzed by affinity chromatography on immobilized ES, to assess the proportion of chains capable of interacting with the protein. In both cases, ∼50% of the labeled material remained bound to the immobilized ES at physiological ionic strength (data not shown), in accord with previous findings (Sasaki et al., 1999). The ES-bound fractions, and a pool depleted of ES-binding components by three consecutive passages through the ES–Sepharose column, were recovered and analyzed further by gel chromatography (Superose 6; Figure 1). The results were similar for both HS preparations tested, in that the ES-unbound components were small, essentially <15 kDa. Conversely, HS chains >15 kDa were retained almost quantitatively by the ES column.
Fig. 1. Size fractionation of HS chains separated with regard to affinity for ES. N-[3H]acetyl-labeled HS from pig intestinal mucosa with (solid line) or without (dotted line) detectable affinity for ES was analyzed on a Superose 6 column. The elution positions of a heparin tetrasaccharide (Mr 1.3 × 103) and of a characterized oligosaccharide pool (Mr 8.0 × 103) are indicated; the position of the arrow corresponding to Mr 30 × 103 is based on extrapolation.
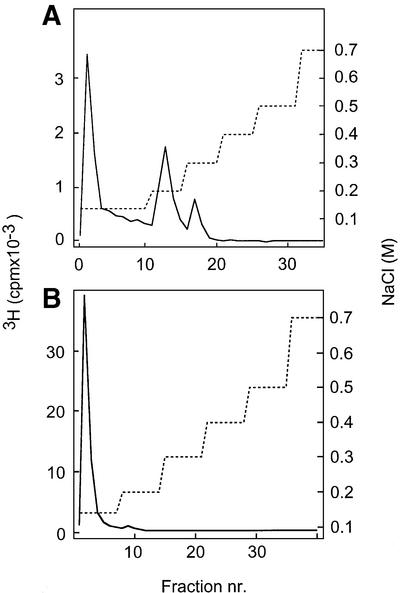
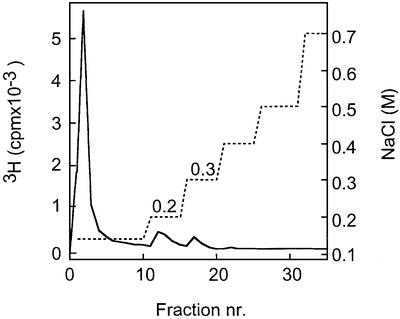
A substantial fraction of heparin 10mers, highly sulfated, was retained by the ES–Sepharose column, and required 0.2–0.3 M NaCl for elution (Figure 2A). In contrast, less sulfated oligosaccharides of similar size, derived from NS-domains of pig intestinal HS and previously shown to bind both FGF-1 and FGF-2 with high affinity (Kreuger et al., 2001), did not bind ES (Figure 2B). We therefore decided to study the overall distribution of variously N-substituted domains in HS to explain the affinity for ES exhibited by the longer HS chains.
Fig. 2. Affinity chromatography of decasaccharides on immobilized ES. 3H-labeled, N-sulfated decasaccharides isolated from heparin (A) or from pig intestinal HS (B) were loaded onto a column (1 ml) of ES–Sepharose and eluted with a stepwise gradient of NaCl (dashed line) as described in Materials and methods. Fractions of 1 ml were collected and analyzed for radioactivity.
Domain organization of HS
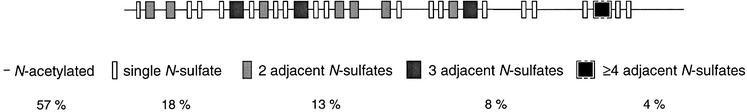
HS from pig intestine was degraded by N-deacetylation followed by deaminative cleavage at pH 3.9 to generate NS-domains of different length. The resultant fragments were 3H-reduced and size separated, and pools of defined size classes were recovered. Labeled di- and tetrasaccharides generated through this procedure are derived from NA-domains and NA/NS-domains, respectively, whereas larger oligosaccharides represent NS-domains. Conversely, the size distribution of NA-domains was assessed following deamination at pH 1.5 (see Materials and methods). The proportions of different domain types in the intact polysaccharide were calculated from the relative abundance of each oligosaccharide class (gel chromatograms not shown; results summarized in Figure 3). Extended NS-domains containing ≥4 consecutive N-sulfated disaccharide units (recovered as ≥10mers following N-deacetylation/deamination of intact HS) accounted for only ∼4% of total saccharide mass, thus an average of one corresponding motif for each ∼100 disaccharide units, or every other HS chain of Mr ∼30 000 Da (∼50 disaccharide units). Moreover, it should be recalled that isolated N-sulfated 10mers derived from the same HS preparation were largely unable to bind ES (Figure 2B). Obviously, motifs other than contiguous N-sulfated domains must be involved in binding to ES, since all HS chains of Mr ∼30 000 Da were retained on the ES column (Figure 1). On the other hand, the hypothetical domain structure in Figure 3 would clearly accommodate extended, sulfated domains containing isolated GlcNAc residues. An ES-binding site of this type would agree with our previous observation that the arginine residues implicated in HS binding occur in two separate clusters at the ES surface (Sasaki et al., 1999). We therefore decided to isolate oligosaccharides with mixed N-substituents from HS, for subsequent testing as ES ligands.
Fig. 3. Domain analysis of HS chains. HS from pig intestine was cleaved by complete N-deacetylation and deamination at pH 3.9. The fragments were 3H-reduced and size separated as described in Materials and methods, and the proportion of each fragment (percentage of total chain mass) was calculated. The approximate number of variously sized NS-domains (gray boxes, darker with increasing number of consecutive N-sulfated disaccharide units) is expressed on the basis of a chain with 100 disaccharide units; the distribution of these domains is arbitrary. N-acetylated oligosaccharide units are represented by a line without boxes. The NS-domain containing ≥4 disaccharide units (within brackets) is not present in all chains (see text). See Figure 10 for actual structures of sugar units.
Isolation and characterization of ES-binding SAS oligosaccharides
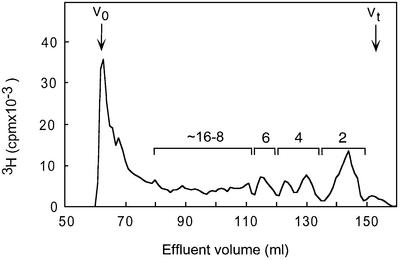
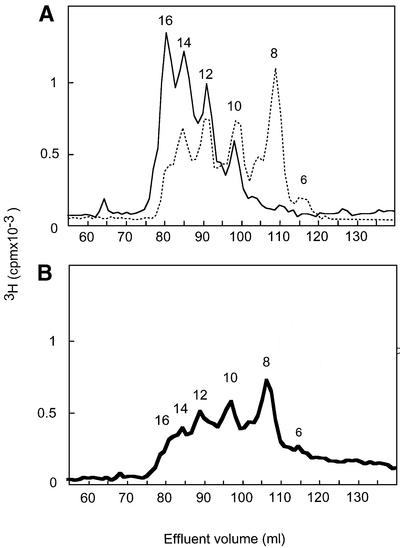
A new type of oligosaccharide library containing combinations of N-sulfated and N-acetylated disaccharide units was generated through partial deaminative cleavage of N-sulfated domains in native HS. The resultant 3H-end-group-labeled fragments (SAS-domains) were separated according to size by gel chromatography (Figure 4), and fractions corresponding to 8–16mers were pooled as indicated, lyophilized and subjected to ES affinity chromatography. Altogether, 15% of these SAS-domains bound to ES, and could be eluted with 0.2 and 0.3 M NaCl (Figure 5). The ES-bound (eluted at 0.3 M NaCl) and unbound SAS-domains were both analyzed with regard to size by gel chromatography on Bio-Gel P-10. The bound fraction contained a larger proportion of long fragments, but showed considerable overlap with the unbound fraction (Figure 6A). Notably, 8mers were abundant in the ES-unbound fraction but were essentially absent from the bound fraction.
Fig. 4. Size separation of SAS fragments on Bio-Gel P-10. SAS fragments were generated through partial deamination (pH 1.5) of pig intestinal mucosa HS, and the products were end labeled by reduction with NaB3H4. The mixture of even-numbered fragments was separated on a Bio-Gel P-10 column and fractions corresponding to 8–16mers were combined into one pool according to the elution positions of heparin standards.
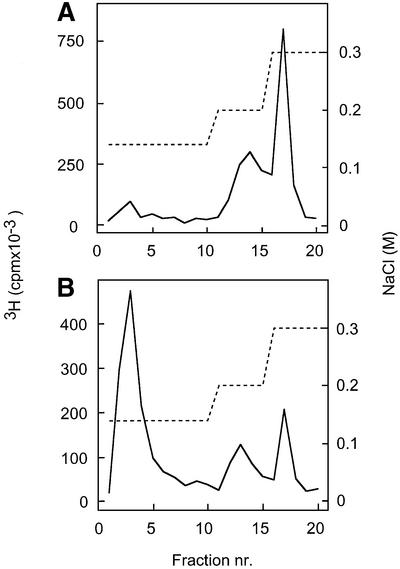
Fig. 5. Affinity chromatography of SAS fragments on immobilized ES. 3H-labeled SAS fragments (8–16mers) were applied to an ES affinity column (1 ml), and eluted with a stepwise gradient of NaCl (dashed line). Fractions of 1 ml were analyzed for radioactivity. Of the total applied SAS oligosaccharides (6000 c.p.m. 3H), ∼15% were retained by the column and eluted with 0.2–0.3 M NaCl.
Fig. 6. Size fractionation of SAS fragments separated by ES affinity chromatography. (A) SAS fragments eluted from ES–Sepharose with 0.3 M NaCl (solid line) or unbound at 0.14 M NaCl (dotted line) were analyzed with regard to size (indicated above the peaks as the number of monosaccharide units/molecule) on a Bio-Gel P-10 column as described in Materials and methods. (B) ES-bound SAS fragments were cleaved at the positions of GlcNAc residues, by N-deacetylation (hydrazinolysis) and deamination (pH 3.9), and the products were separated on Bio-Gel P-10 (V0 ∼65 ml, Vt ∼155 ml).
The ES-bound SAS-domains were examined regarding the occurrence and position of N-acetylated disaccharide units. Analysis by anion-exchange HPLC of a lyase digest revealed ∼25% acetyl and ∼75% sulfate N-substituents, whereas the ES-unbound fraction contained about equal proportions of N-acetylated and N-sulfated disaccharide units (data not shown). Following hydrazinolysis of the ES-bound fraction to deacetylate GlcNAc residues, samples were reacted with nitrous acid at pH 3.9 to induce cleavage at the site of N-unsubstituted GlcN units. Analysis of the products by gel chromatography confirmed that the predominant proportion of ES-bound SAS oligosaccharides indeed had contained one or more N-acetyl groups. The major degradation product was an 8mer, although larger and smaller (less abundant), poorly resolved fragments were also seen (Figure 6B). Finally, the same cleavage products were subjected to repeated affinity chromatography on ES–Sepharose. Contrary to the intact SAS oligosaccharides that were quantitatively bound to the immobilized protein, fewer than one-third of the N-deacetylated and deaminated fraction retained ES binding (Figure 7). Thus, cleavage at the site of N-acetylation abolished ES binding for most of the oligosaccharides.
Fig. 7. Effect of cleavage of ES-bound SAS oligosaccharides on ES affinity. SAS fragments previously recovered by elution with 0.3 M NaCl from immobilized ES (see Figure 5) were reapplied to the ES column either untreated (A) or after cleavage at the position of GlcNAc units (B).
Inhibition of ES anti-migratory activity by heparin and SAS oligosaccharides
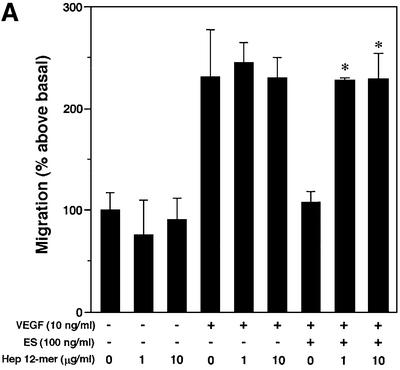
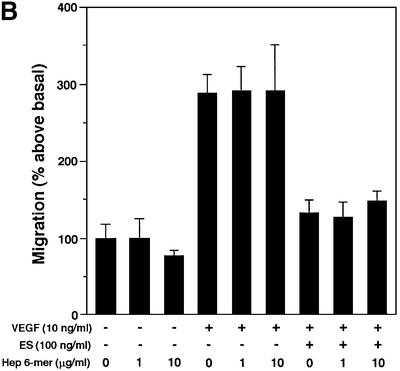
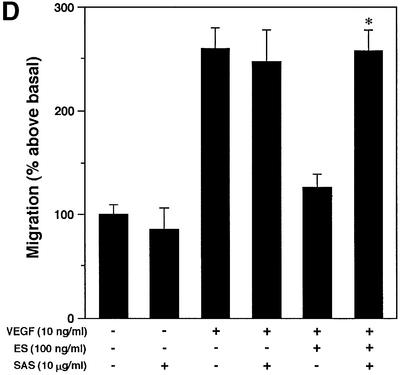
ES is known to inhibit growth factor-induced migration of endothelial cells (Yamaguchi et al., 1999). The modulatory potential of ES-binding SAS and heparin oligosaccharides on VEGF-A165-stimulated migration was assessed in the presence and absence of ES using a mini-Boyden chamber. Inclusion of 100 ng/ml ES completely attenuated the chemotactic response to VEGF-A165 (Figure 8A). Inhibition of migration is dependent on the heparin-binding ability of ES, as the non-heparin-binding R158/184/270A mutant ES failed to inhibit endothelial cell migration (data not shown). As predicted, the inhibitory effect of wild-type ES thus could be neutralized by heparin 12mers (Figure 8A), but not by heparin 6mers (Figure 8B) that previously were found to be unable to bind ES efficiently (Sasaki et al., 1999). The inhibitory effect of the heparin 12mers on ES activity was dose dependent (IC50 ∼0.5 µg/ml heparin, 10 ng VEGF/ml, 100 ng ES/ml), and could be counteracted by increasing the concentration of ES (Figure 8C). Moreover, the ES activity was completely abolished by the inclusion of SAS oligosaccharides (Figure 8D) with affinity for ES similar to that of the heparin 12mers (compare Figures 2A and 7A). Combined, these data strongly support the notion that exogenously added heparin 12mers and SAS oligosaccharides compete with endogenous HS for binding of ES.
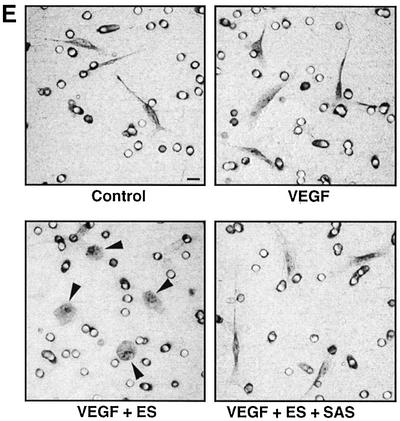
Fig. 8. Inhibitory effect of oligosaccharides on anti-migratory activity of ES. Primary BCE cells were analyzed for their ability to migrate in response to VEGF-A165 in the presence or absence of ES and heparin- or HS-derived oligosaccharides. (A) Cell migration induced by VEGF is inhibited by ES. Heparin 12mers alone have no effect on cell migration, and do not interfere with the activity of VEGF, but counteract the inhibitory effect of ES. (B) Heparin 6mers are unable to inhibit the activity of ES. (C) The inhibition of ES by heparin 12mers is dose dependent. Notably, the effect can be reversed by increasing the concentration of ES. (D) ES-bound SAS oligosaccharides inhibit the activity of ES, but do not alone affect basal or VEGF-induced cell migration. Each bar shows the mean ± SD of triplicate wells. *Significantly different from VEGF-A165 + ES at P < 0.03 (unpaired Student’s t-test). (E) Morphology of migrated cells. Cells treated with ES display a rounded morphology, in contrast to cells treated with ES together with SAS oligosaccharides, which assume the same spindle-shaped morphology as the control and VEGF-treated cells. The circular structures represent pores in the membrane. Bar: 10 µm.
VEGF-A165 previously has been reported to interact with heparin (Tessler et al., 1994). However, in the present study, heparin oligosaccharides at the concentrations used to inhibit the activity of ES did not interfere with VEGF activity, nor did they exert any noticeable effect on cell migration in the absence of VEGF or ES (Figure 8). The majority of the endothelial cells that had migrated towards VEGF in the chemotaxis chamber were spindle shaped or elongated (Figure 8E). Treatment with ES induced striking morphological changes such that cells assumed a flat, rounded shape (Figure 8E, arrowheads). Cells that had been exposed to ES combined with SAS oligosaccharides were morphologically identical to untreated cells, as expected if these oligosaccharides neutralize the effect of ES. The ES-induced change in cell morphology agrees with our previous data (Dixelius et al., 2000), and reflects interference of ES with cell–matrix as well as cell–cell interactions (Dixelius et al., 2002).
Discussion
Initial experiments showed that a heparin-derived N-sulfated decasaccharide could bind ES at physiological ionic conditions, contrary to HS oligosaccharides of similar length (Figure 2). On the other hand, intact HS chains exceeding ∼15 kDa bound quantitatively to ES (Figure 1). These findings raised the question as to the nature of the ES-binding site in the HS molecule. Crystallographic analysis of ES revealed a compact globular folding, with arginine-rich clusters exposed on the surface (Hohenester et al., 1998). Mutation of certain of the arginine residues, individually or in combination, resulted in loss of heparin binding and led to the identification of six arginine residues critical for this interaction (Sasaki et al., 1999). Loss of heparin binding was accompanied by loss of the inhibitory ability of ES on endothelial cell migration and on angiogenesis (Dixelius et al., 2000). The arginine residues implicated in heparin binding are clustered in two regions (R193, R194, and R155, R158, R270, R184, respectively) (Hohenester et al., 1998). Model building indicated that a saccharide fragment spanning both arginine-containing clusters should contain ∼10 monosaccharide units (Sasaki et al., 1999). The ability of the heparin 10mer to bind ES presumably reflects the O-sulfation pattern of the (fully N-sulfated) saccharide, the IdoA and GlcNSO3 units carrying 2-O- and 6-O-sulfate groups, respectively, throughout the length of the sequence. These two types of O-sulfate substituents previously were shown to be important for ES binding (Sasaki et al., 1999). Sufficient sulfation would thus be provided toward the ends of the heparin oligosaccharide to enable interaction with both arginine clusters (Figure 9).
Fig. 9. Assumed interactions between ES and heparin/HS motifs. (A) Heparin decamers are homogeneously O-sulfated, and thus capable of binding to both clusters of arginine residues at the ES surface (R in oval shape). (B) HS decamers derived from NS-domains are O-sulfated preferentially at central positions and are therefore unable to bind efficiently to the arginine clusters. HexA may be either GlcA or IdoA; 2-O-sulfate and 6-O-sulfate groups are designated 2S and 6S, respectively. The structures of the various sugar units are given in Figure 10.
However, the HS side chains of proteoglycans, shown to promote ES action on endothelial cells (Karumanchi et al., 2001), differ in sulfation pattern from that of heparin. The less abundant O-sulfate groups in HS chain NS-domains thus are not uniformly distributed, but located preferentially at the internal portions of the domains (Merry et al., 1999; Kreuger et al., 2001), apparently with some correlation between overall O-sulfate density and domain length (Safaiyan et al., 2000) (Figure 9). Moreover, an estimation of the domain distribution in the HS chain revealed a relative lack of NS-domains of sufficient length to provide an adequately O-sulfated ligand sequence suitable for interaction with both arginine clusters (Figure 3). It therefore seemed reasonable to envisage an ES-binding site composed of two smaller NS-domains, one corresponding to each arginine cluster, separated by a short N-acetylated segment.
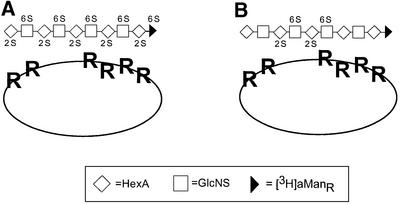
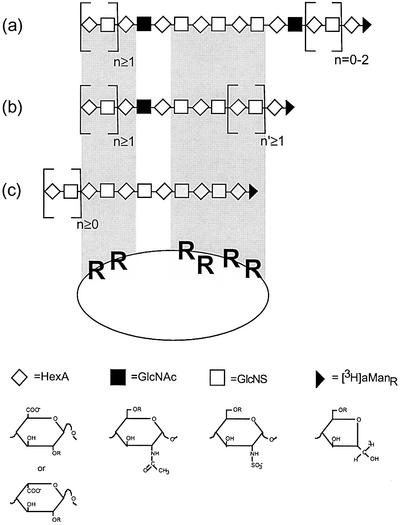
Experiments using the composite SAS-domain library indeed demonstrated the occurrence of such mixed binding sites. A significant proportion (∼15%) of isolated 8–16mers bound to ES (Figure 5) with an apparent affinity similar to that seen for the fully sulfated heparin 10mer (Figure 2A). Size analysis showed that the ES-bound SAS oligosaccharides contained predominantly 12–16mers, with a small proportion of 10mer; no 8mer was found (Figure 6A). Cleavage of the bound fraction at the site of the GlcNAc unit(s) shifted the entire oligosaccharide pattern toward smaller sized molecules, with a predominant 8mer product (Figure 6B). Concomitantly, about two-thirds of the oligosaccharides lost their affinity for ES (Figure 7). These results are interpreted in terms of the model shown in Figure 10. Most of the ES-binding oligosaccharides conform to structure (b), in which a single GlcNAc residue separates two N- and O-sulfated domains. Cleavage at the N-acetylated site released a series of oligosaccharides (Figure 6B), the predominant 8mer fragment being derived from a parent species (b in Figure 10) in which n′ = 1. The less abundant structure (a) is principally similar, but contains a second GlcNAc unit from which minor labeled oligosaccharides (6mer and smaller in Figure 6B) were released upon N-deacetylation/deamination. Finally, minor components of fully N-sulfated structures (c in Figure 10) would correspond to the proportion of oligosaccharides that remained bound to ES also after hydrazinolysis/deamination (Figure 7B). We assume that these fragments had been excised from scarce, more extended NS-domains in the parent HS, and therefore carried O-sulfate groups along their entire length to enable strong interaction with both binding sites in ES. Notably, according to the models presented in Figures 9 and 10, the heparin hexasaccharide previously suggested to interact specifically with ES (Karumanchi et al., 2001) would allow effective interaction with only one of the two clusters of arginine residues at the ES surface. Further refined characterization of ES-binding SAS-domains, e.g. regarding the precise location of a GlcNAc residue within the ES-binding site, will require additional fractionation to obtain binding oligosaccharides of minimal size (and charge) for sequence analysis and comparison with corresponding non-bound species.
Fig. 10. Proposed structures of HS-derived SAS oligosaccharides that interact with ES. O-sulfate groups (not shown) present in NS-domains (within the shaded areas) make contact with arginine residues at the ES surface. Two of these fragments (a and b) contain N-acetylated GlcN residues in different positions (indicated by the filled boxes). Oligosaccharide c is N-sulfated throughout. Structures of the various monosaccharide units are drawn below the corresponding symbols, with ‘R’ indicating the common sites of O-sulfation. For additional information see the text.
The functional importance of the interaction between the saccharide and both arginine clusters at the ES surface is underpinned by the ability of SAS oligosaccharides and heparin 12mers to inhibit the action of ES in a cell migration assay. Saccharides (6mers) too short to span both clusters of arginine residues at the ES surface could not alleviate the inhibitory effect of ES on cell migration (Figure 8). ES mutant experiments showed that both arginine clusters were required for efficient inhibition of FGF-2-induced angiogenesis (Sasaki et al., 1999). Yet the mechanism of inhibition was not clear, as it could involve either competition of ES for HS co-receptor sites required for FGF-2 action or binding of ES to a separate HS domain. The first possibility now appears less likely since HS oligosaccharides with high affinity for FGF-1 and FGF-2 (Kreuger et al., 2001) do not bind to ES (Figure 2B). Additional interactions may be involved, as ES recently was shown to bind integrins (Rehn et al., 2001).
The elucidated SAS-domains found here to interact with ES differ fundamentally from the HS structures previously implicated in binding of monomeric proteins such as FGFs (Turnbull et al., 1992; Maccarana et al., 1993; Faham et al., 1996; DiGabriele et al., 1998; Pellegrini et al., 2000; Schlessinger et al., 2000; Kreuger et al., 2001). The HS- or heparin-derived ligands previously studied generally have been exclusively N-sulfated. So far, antithrombin is the only other example described of a monomeric protein shown to interact with a GlcNAc-containing heparin/HS sequence; however, in this case, the GlcNAc residue is positioned at the very end of the protein-binding pentasaccharide sequence (Bourin and Lindahl, 1993). Notably, N-sulfated domains separated by N-acetylated sequences appear to mediate binding of HS to multimeric proteins such as interleukin-8 (Spillmann et al., 1998), platelet factor 4 (Stringer and Gallagher, 1997) and interferon-γ (Lortat-Jacob et al., 1995). The present study demonstrates that the authentic HS epitopes recognized by ES in vivo are represented by SAS-domains. We predict that such domains will be identified as functionally important ligands to a large variety of monomeric proteins that, like ES, expose extended HS-binding sites. Interestingly, SAS-domains capable of interacting with ES were found to occur preferentially in the longer chains of intestinal HS (Figure 1). The absence of such domains in a fraction of HS chains may relate to the finding by Karumanchi et al. (2001) that ES interaction is selective for certain HS proteoglycans. Structural motifs along HS chains are expressed in a tissue- and temporal-specific manner (Lindahl et al., 1995; van den Born et al., 1995; Maccarana et al., 1996; Feyzi et al., 1998; Nurcombe et al., 2000; Safaiyan et al., 2000). The structure of HS is determined by the concerted action of enzymes present in the Golgi compartment. The first enzymes to act on the nascent HS backbone are N-deacetylase/N-sulfotransferases (NDSTs) that replaces the acetyl group on N-acetylglucosamine residues by a sulfate group. The NDSTs are key enzymes in HS biosynthesis since other modifications (GlcA C-5 epimerization, O-sulfation) depend on the presence of N-sulfate groups. The NDST reaction thus provides the framework for the final domain organization, here implicated in ES recognition. The N-substituent pattern, hence the overall domain organization of HS chains, is subject to intricate biosynthetic control, as reflected by the occurrence of multiple NDST isoforms (Forsberg and Kjellén, 2001). The present study provides a functional context to such regulation, and an incitement to explore further the domain topology of HS and the mechanisms behind domain generation in HS biosynthesis.
Finally, our results have potential implications for clinical therapy. Conceivably, it may be desirable to alleviate the anti-angiogenic affects of ES, for instance in ischemic conditions. SAS oligosaccharides, or appropriately tailored analogs, could induce such inhibition in a selective fashion, thus avoiding the interference with growth factor signaling and other processes that might be compromised by more fully sulfated, heparin-like structures.
Materials and methods
Materials
Recombinant wild-type mouse ES was expressed using episomal expression vectors in human kidney EBNA-293 cells, and purified on heparin–Sepharose followed by size exclusion chromatography as described previously (Sasaki et al., 1998). Protein concentration was determined by amino acid analysis.
Full-length HS from pig intestinal mucosa was a gift from Dr D.van Dedem (Diosynth, Oss, The Netherlands), and HS from bovine kidney was purchased from Seikagaku. Heparin from pig intestinal mucosa was purified and labeled as described (Kreuger et al., 1999). Bio-Gel P-10 (fine) was from Bio-Rad. Sephadex G-10, Sephadex G-15, PD-10, Superose 6 (pre-packed 1.5 × 30 cm column), a prototype of a Superdex 30 column (2 × 60 cm), NaB3H4 (64 Ci/mmol) and [3H]acetic anhydride (500 mCi/mmol) were all obtained from Amersham Pharmacia Biotech (Uppsala, Sweden). Hydrazine hydrate was purchased from Fluka. All other reagents were of the best grade available.
Bovine capillary endothelial (BCE) cells, a kind gift from Dr Rolf Christofferson, Department of Medical Cell Biology, Uppsala University, were isolated from calf adrenal cortex as described previously (Qi et al., 1999) and used at passage 10–14. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies), 10% newborn calf serum (NCS) and 2 ng/ml FGF-2 (Boehringer Mannheim) on gelatinized dishes.
Chemical depolymerization and radiolabeling of saccharides
N-sulfated oligosaccharides were obtained from HS by chemical N-deacetylation (hydrazinolysis) followed by nitrous acid-induced cleavage (pH 3.9) of the polysaccharide at N-unsubstituted GlcN units, as described previously (Kreuger et al., 1999). The resultant fragments were reduced with NaB3H4 to yield oligosaccharides with a specific activity of 4.4 × 106 d.p.m. 3H/nmol. Conversely, N-acetylated oligosaccharides were obtained by exhaustive nitrous acid treatment at pH 1.5 (Shively and Conrad, 1976).
Oligosaccharides containing mixed acetyl and sulfate N-substituents (SAS fragments) were generated by limited reaction of pig intestinal HS with nitrous acid at pH 1.5 (Shively and Conrad, 1976; Pejler et al., 1988), to induce partial cleavage at N-sulfated GlcN units. To this end, 0.5 mg of HS was dissolved in 0.5 ml of 0.2 mM NaNO2 adjusted with 1 M HCl to pH 1.5, and incubated for 1 h on ice. The reaction was terminated by increasing the pH to 8 with 4 M NaOH. The resultant SAS-domains were reduced (end labeled) with NaB3H4 (Spillmann et al., 1998) and separated by gel chromatography as described below. The fractionated SAS-domains were pooled as indicated in the Results (Figure 4).
Intact HS chains were radiolabeled by 3H-acetylation of free amino groups as described previously (Höök et al., 1982).
Size separation of intact HS chains, NS- and SAS-domains
The molecular size of HS derived from kidney and intestine was estimated by gel chromatography using a Superose 6 column run in 50 mM Tris–HCl pH 7.4, 1 M NaCl (flow rate 0.2 ml/min). SAS-domains, NA-domains and NS-domains were separated according to size on a Bio-Gel P-10 column (1 × 200 cm) in 0.2 M NH4HCO3 (2 ml/h). Individual pools of NS-domains were analyzed further on a Superdex-30 column run in 0.1 M Tris pH 7.4, 1 M NaCl (0.5 ml/min) to confirm their size. Even-numbered oligosaccharides derived from heparin (highly sulfated) and from Escherichia coli K5 capsular polysaccharide (non-sulfated) (Lidholt and Lindahl, 1992) were used as standards to calibrate the sizing columns.
Affinity chromatography
Recombinant mouse ES (1 mg) was coupled to 1 mg of CH-Sepharose (Amersham Pharmacia Biotech) in 0.1 M NH4HCO3 pH 8, 0.1 M NaCl at 20°C for 1 h. Before immobilization, ES was incubated for 10 min with 5 mg of heparin [devoid of free amino groups (Kreuger et al., 2001)] in order to prevent the saccharide-binding epitope of ES from forming bonds with the matrix. The ES–Sepharose (1 ml) was transferred to a column, washed with 2 M NaCl to remove bound heparin and regenerated according to the manufacturer’s instructions. Oligosaccharides were applied in 0.5 ml of 50 mM Tris–HCl pH 7.4, 0.14 M NaCl, at 4°C. Initially, HS from kidney was eluted with a linear gradient (total volume 30 ml) from 0.14 to 0.7 M NaCl in Tris–HCl pH 7.4 (flow rate 1 ml/min). Shorter oligosaccharide fragments and SAS-domains were eluted using a stepwise gradient (total volume 35 ml) from 0.14 to 0.7 M NaCl as indicated in Figure 2. Fractions of 1 ml were collected and analyzed for radioactivity. Larger scale preparative experiments were conducted using an affinity column (5 ml) containing 7 mg of ES.
Analysis of GlcNAc units in SAS-domains
Compositional analysis of SAS oligosaccharides was performed as described (Toyoda et al., 2000), using reversed phase ion-pair chromatography of lyase digests (samples of 5 µg incubated with a mixture of heparin lyase I, II and III). The products were monitored by post-column derivatization with 2-cyanoacetamide, and quantified by reference to standard disaccharides (Sigma).
The location of GlcNAc residues in SAS oligosaccharides was determined by N-deacetylation (hydrazinolysis) (Guo and Conrad, 1989) followed by selective deaminative cleavage (Shively and Conrad, 1976) of the 3H-end-group-labeled sequence at the position of N-unsubstituted GlcN units, and gel chromatography of the resultant labeled fragments. Oligosaccharides (20 000 c.p.m. 3H) were dissolved in 200 µl of hydrazine hydrate with 2 mg of hydrazine sulfate and incubated at 96°C for 4 h. The N-deacetylated samples were then evaporated to dryness and desalted using a PD-10 column before treatment with nitrous acid (20 µl of 125 mM NaNO2, 1.25 M HAc pH 3.9) at room temperature for 20 min. After adjustment of the pH to 8 with 2 M Na2CO3, the cleavage products were reduced with NaBH4. Excess borohydride was eliminated by adding 4 M HAc to pH 4, whereafter the sample was brought to neutral pH with NaOH. The sample was either applied directly to a Bio-Gel P-10 column, or desalted on a PD-10 column before ES affinity chromatography.
Chemotaxis assay
Chemotaxis was assayed essentially as described (Yokote et al., 1996) using a 96-well microchemotaxis chamber (Neuro Probe, Cabin John, MD), equipped with a polycarbonate membrane (polyvinylpyrrolidone-free, pore size 8 µm) coated with type I collagen (100 µg/ml; Vitrogen, Cohesion Technologies Inc., Palo Alto, CA). BCE cells isolated from bovine adrenal cortex were starved in DMEM, 0.5% NCS for 24 h, trypsinized and resuspended in DMEM, 0.1% bovine serum albumin (BSA) to 2 × 105 cells/ml, and pre-treated or not for 20 min at 37°C with ES and heparin oligosaccharides at the indicated concentrations. The cell suspension (4 × 104 cells in 200 µl) was transferred to the upper chemotaxis chamber, and the chemoattractant solution (35 µl) containing vehicle (DMEM, 0.1% BSA) with or without VEGF-A165 (Preprotech EC Ltd, London, UK), ES or oligosaccharides at the indicated concentrations was loaded in the lower chemotaxis chamber. The chambers were assembled and incubated for 6 h at 37°C, whereafter the membrane was fixed with methanol and stained with Giemsa solution. Cells that had migrated to the lower surface of the membrane were counted in three separate fields at 100× magnification using an Image analyzer program (Easy Image Measurement 2000, version 2.5; Bergström Instrument AB, Solna, Sweden). Migrated cells were photographed with a Nikon microscope at 400× magnification. All sample conditions were assayed in triplicates.
Acknowledgments
Acknowledgements
We are grateful to Johan Ledin and Lena Kjellén for help with the disaccharide analyses. This work was supported by European Commission Grant QLK-CT-1999.00536 (Program ‘Biologically Active Novel Glycosaminoglycans’), Polysackaridforskning AB and by the Swedish Cancer Foundation (project no 3820-B99-04XBC) to L.C.-W.
References
- Bourin M.-C. and Lindahl,U. (1993) Glycosaminoglycans and the regulation of blood coagulation. Biochem. J., 289, 313–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casu B. and Lindahl,U. (2001) Structure and biological interactions of heparin and heparan sulfate. Adv. Carbohydr. Chem. Biochem., 57, 159–206. [DOI] [PubMed] [Google Scholar]
- DiGabriele A.D., Lax,I., Chen,D.I., Svahn,C.-M., Jaye,M., Schlessinger,J. and Hendrickson,W.A. (1998) Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature, 393, 812–817. [DOI] [PubMed] [Google Scholar]
- Dixelius J., Larsson,H., Sasaki,T., Holmqvist,K., Lu,L., Engström,A., Timpl,R., Welsh,M. and Claesson-Welsh,L. (2000) Endostatin-induced tyrosine kinase signaling through the Shb adaptor protein regulates endothelial cell apoptosis. Blood, 95, 3403–3411. [PubMed] [Google Scholar]
- Dixelius J., Cross,M., Matsumoto,T., Sasaki,T., Timpl,R. and Claesson-Welsh,L. (2002) Endostatin regulates endothelial cell adhesion and cytoskeletal organization. Cancer Res., 62, 1944–1947. [PubMed] [Google Scholar]
- Esko J.D. and Lindahl,U. (2001) Molecular diversity of heparan sulfate. J. Clin. Invest., 108, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faham S., Hileman,R.E., Fromm,J.R., Linhardt,R.J. and Rees,D.C. (1996) Heparin structure and interactions with basic fibroblast growth factor. Science, 271, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Feyzi E., Saldéen,T., Larsson,E., Lindahl,U. and Salmivirta,M. (1998) Age-dependent modulation of heparan sulfate structure and function. J. Biol. Chem., 273, 13395–13398. [DOI] [PubMed] [Google Scholar]
- Forsberg E. and Kjellén,L. (2001) Heparan sulfate: lessons from knockout mice. J. Clin. Invest., 108, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J.T. (2001) Heparan sulfate: growth control with a restricted sequence menu. J. Clin. Invest., 108, 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.C. and Conrad,H.E. (1989) The disaccharide composition of heparins and heparan sulfates. Anal. Biochem., 176, 96–104. [DOI] [PubMed] [Google Scholar]
- Hohenester E., Sasaki,T., Olsen,B.R. and Timpl,R. (1998) Crystal structure of the angiogenesis inhibitor endostatin at 1.5 Å resolution. EMBO J., 17, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höök M., Riesenfeld,J. and Lindahl,U. (1982) N-[3H]acetyl-labeling, a convenient method for radiolabeling of glycosaminoglycans. Anal. Biochem., 119, 236–245. [DOI] [PubMed] [Google Scholar]
- Iozzo R.V. and San Antonio,J.D. (2001) Heparan sulfate proteoglycans: heavy hitters in the angiogenesis arena. J. Clin. Invest., 108, 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemth P., Kreuger,J., Kusche-Gullberg,M., Sturiale,L., Gimenez-Gallego,G. and Lindahl,U. (2002) Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs. Exploring the structural diversity by screening for fibroblast growth factor (FGF) 1 and FGF2 binding. J. Biol. Chem., 277, 30567–30573. [DOI] [PubMed] [Google Scholar]
- Karumanchi S.A. et al. (2001) Cell surface glypicans are low-affinity endostatin receptors. Mol. Cell, 7, 811–822. [DOI] [PubMed] [Google Scholar]
- Kreuger J., Prydz,K., Pettersson,R.F., Lindahl,U. and Salmivirta,M. (1999) Characterization of fibroblast growth factor 1 binding heparan sulfate domain. Glycobiology, 9, 723–729. [DOI] [PubMed] [Google Scholar]
- Kreuger J., Salmivirta,M., Sturiale,L., Giménez-Gallego,G. and Lindahl,U. (2001) Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J. Biol. Chem., 276, 30744–30752. [DOI] [PubMed] [Google Scholar]
- Lidholt K. and Lindahl,U. (1992) Biosynthesis of heparin. The d-glucuronosyl- and N-acetyl-d-glucosaminyltransferase reactions and their relation to polymer modification. Biochem. J., 287, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl B., Eriksson,L. and Lindahl,U. (1995) Structure of heparan sulphate from human brain, with special regard to Alzheimer’s disease. Biochem. J., 306, 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo B.M., Kreuger,J., Jalkanen,M., Lindahl,U. and Salmivirta,M. (2001) Binding of heparin/heparan sulfate to fibroblast growth factor receptor 4. J. Biol. Chem., 276, 16868–16876. [DOI] [PubMed] [Google Scholar]
- Lortat-Jacob H., Turnbull,J.E. and Grimaud,J.A. (1995) Molecular organization of the interferon γ-binding domain in heparan sulphate. Biochem. J., 310, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin L., Larsson,H., Kreuger,J., Kanda,S., Lindahl,U., Salmivirta,M. and Claesson-Welsh,L. (2000) Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J. Biol. Chem., 275, 24653–24660. [DOI] [PubMed] [Google Scholar]
- Maccarana M., Casu,B. and Lindahl,U. (1993) Minimal sequence in heparin/heparan sulfate required for binding of basic fibroblast growth factor. J. Biol. Chem., 268, 23898–23905. [PubMed] [Google Scholar]
- Maccarana M., Sakura,Y., Tawada,A., Yoshida,K. and Lindahl,U. (1996) Domain structure of heparan sulfates from bovine organs. J. Biol. Chem., 271, 17804–17810. [DOI] [PubMed] [Google Scholar]
- Merry C.L., Lyon,M., Deakin,J.A., Hopwood,J.J. and Gallagher,J.T. (1999) Highly sensitive sequencing of the sulfated domains of heparan sulfate. J. Biol. Chem., 274, 18455–18462. [DOI] [PubMed] [Google Scholar]
- Nurcombe V., Smart,C.E., Chipperfield,H., Cool,S.M., Boilly,B. and Hondermarck,H. (2000) The proliferative and migratory activities of breast cancer cells can be differentially regulated by heparan sulfates. J. Biol. Chem., 275, 30009–30018. [DOI] [PubMed] [Google Scholar]
- O’Reilly M.S. et al. (1997) Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell, 88, 277–285. [DOI] [PubMed] [Google Scholar]
- Pejler G., Lindahl,U., Larm,O., Scholander,E., Sandgren,E. and Lundblad,A. (1988) Monoclonal antibodies specific for oligosaccharides prepared by partial nitrous acid deamination of heparin. J. Biol. Chem., 263, 5197–5201. [PubMed] [Google Scholar]
- Pellegrini L., Burke,D.F., von Delft,F., Mulloy,B. and Blundell,T.L. (2000) Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature, 407, 1029–1034. [DOI] [PubMed] [Google Scholar]
- Pye D.A., Vivès,R.R., Turnbull,J.E., Hyde,P. and Gallagher,J.T. (1998) Heparan sulfate oligosaccharides require 6-O-sulfation for promotion of basic fibroblast growth factor mitogenic activity. J. Biol. Chem., 273, 22936–22942. [DOI] [PubMed] [Google Scholar]
- Qi J.H., Matsumoto,T., Huang,K., Olausson,K., Christofferson,R. and Claesson-Welsh,L. (1999) Phosphoinositide 3 kinase is critical for survival, mitogenesis and migration but not for differentiation of endothelial cells. Angiogenesis, 3, 371–380. [DOI] [PubMed] [Google Scholar]
- Rehn M., Veikkola,T., Kukk-Valdre,E., Nakamura,H., Ilmonen,M., Lombardo,C., Pihlajaniemi,T., Alitalo,K. and Vuori,K. (2001) Interaction of endostatin with integrins implicated in angiogenesis. Proc. Natl Acad. Sci. USA, 98, 1024–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaiyan F., Lindahl,U. and Salmivirta,M. (2000) Structural diversity of N-sulfated heparan sulfate domains: distinct modes of glucuronyl C5 epimerization, iduronic acid 2-O-sulfation and glucosamine 6-O-sulfation. Biochemistry, 39, 10823–10830. [DOI] [PubMed] [Google Scholar]
- Salmivirta M., Lidholt,K. and Lindahl,U. (1996) Heparan sulfate: a piece of information. FASEB J., 10, 1270–1279. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Fukai,N., Mann,K., Gohring,W., Olsen,B.R. and Timpl,R. (1998) Structure, function and tissue forms of the C-terminal globular domain of collagen XVIII containing the angiogenesis inhibitor endostatin. EMBO J., 17, 4249–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Larsson,H., Kreuger,J., Salmivirta,M., Claesson-Welsh,L., Lindahl,U., Hohenester,E. and Timpl,R. (1999) Structural basis and potential role of heparin/heparan sulfate binding to the angiogenesis inhibitor endostatin. EMBO J., 18, 6240–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J., Plotnikov,A.N., Ibrahimi,O.A., Eliseenkova,A.V., Yeh,B.K., Yayon,A., Linhardt,R.J. and Mohammadi,M. (2000) Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell, 6, 743–750. [DOI] [PubMed] [Google Scholar]
- Shively J.E. and Conrad,H.E. (1976) Formation of anhydrosugars in the chemical depolymerization of heparin. Biochemistry, 15, 3932–3942. [DOI] [PubMed] [Google Scholar]
- Spillmann D., Witt,D. and Lindahl,U. (1998) Defining the interleukin-8-binding domain of heparan sulfate. J. Biol. Chem., 273, 15487–15493. [DOI] [PubMed] [Google Scholar]
- Stringer S.E. and Gallagher,J.T. (1997) Specific binding of the chemokine platelet factor 4 to heparan sulfate. J. Biol. Chem., 272, 20508–20514. [DOI] [PubMed] [Google Scholar]
- Tessler S., Rockwell,P., Hicklin,D., Cohen,T., Levi,B.Z., Witte,L., Lemischka,I.R. and Neufeld,G. (1994) Heparin modulates the interaction of VEGF165 with soluble and cell associated flk-1 receptors. J. Biol. Chem., 269, 12456–12461. [PubMed] [Google Scholar]
- Toyoda H., Kinoshita-Toyoda,A. and Selleck S.B. (2000) Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J. Biol. Chem., 275, 2269–2275. [DOI] [PubMed] [Google Scholar]
- Turnbull J.E., Fernig,D.G., Ke,Y., Wilkinson,M.C. and Gallagher,J.T. (1992) Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. J. Biol. Chem., 267, 10337–10341. [PubMed] [Google Scholar]
- van den Born J., Gunnarsson,K., Bakker,M.A., Kjellén,L., Kusche-Gullberg, M., Maccarana,M., Berden,J.H. and Lindahl,U. (1995) Presence of N-unsubstituted glucosamine units in native heparan sulfate revealed by a monoclonal antibody. J. Biol. Chem., 270, 31303–31309. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N. et al. (1999) Endostatin inhibits VEGF-induced endothelial cell migration and tumor growth independently of zinc binding. EMBO J., 18, 4414–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokote K., Mori,S., Siegbahn,A., Rönnstrand,L., Wernstedt,C., Heldin,C.-H. and Claesson-Welsh,L. (1996) Structural determinants in the platelet-derived growth factor α-receptor implicated in modulation of chemotaxis. J. Biol. Chem., 271, 5101–5111. [DOI] [PubMed] [Google Scholar]