Abstract
Lipopolysaccharide (LPS) stimulates production of inflammatory mediators, partly by stabilizing [interleukin-6 (IL-6), cyclooxygenase 2 (COX-2)] and/or stimulating translation [tumour necrosis factor-α (TNF-α)] of their mRNAs. Such regulation depends on AU-rich elements (AREs) within the 3′-untranslated regions and is partially suppressed by SB 203580 (which inhibits SAPK2a/p38). The LPS-induced production of TNF-α and IL-6 is suppressed in MAPKAP-K2-deficient mice (a kinase activated by SAPK2a/p38). Here, we identify 18 macrophage proteins that bind to AREs and show that hnRNP A0 is a major substrate for MAPKAP-K2 in this fraction. MAPKAP-K2 phosphorylated hnRNP A0 at Ser84 in vitro and this residue became phosphorylated in LPS-stimulated cells. Phosphorylation was prevented by SB 203580 and suppressed in macrophages derived from MAPKAP-K2-deficient mice. The mRNAs encoding TNF-α, COX-2 and macrophage inflammatory protein-2 (MIP-2) bound to hnRNP A0 in LPS-stimulated macrophages, an interaction prevented by SB 203580. The LPS-induced stabilization of MIP-2 mRNA and production of MIP-2 protein were abolished when macrophages were incubated with SB 203580 plus PD 184352 (which inhibits the classical MAP kinase cascade). Our data suggest that LPS-induced binding of hnRNP A0 to AREs may contribute to the post-transcriptional regulation of specific mRNAs.
Keywords: AU-rich element/hnRNP/LPS/MAPKAP-K2/p38 MAP kinase
Introduction
Inflammation, a key process in the host defence system, is highly regulated in order to confine its action to the time and place where it is needed. Loss of control can lead to a number of diseases, including rheumatoid arthritis, neurodegenerative disorders and septic shock syndrome. Important mediators of inflammation are pro-inflammatory cytokines, such as tumour necrosis factor (TNF)-α, interleukin-6 (IL-6) and IL-8. The expression of these genes is regulated at both the transcriptional and post-transcriptional level. Thus, understanding how the expression of these proteins is controlled could be of great importance in overcoming the pathological effects of uncontrolled inflammation.
The presence of AU-rich sequences in the 3′-untranslated region (3′-UTR) of a number of inflammatory mediators and immediate early genes is critical for their post-transcriptional regulation (Grzybowska et al., 2001). These sequences are formed by the presence of one or more copies of the pentamer AUUUA (Xu et al., 1997), and bind to proteins involved in stabilizing or destabilizing these mRNAs. In the case of TNF-α, the AU-rich element (ARE) is known to be involved in repressing the translation of its mRNA (Kontoyiannis et al., 1999), while in the case of IL-6 and cyclooxygenase-2 (COX-2), it is critical for mRNA stability (Ridley et al., 1998; Winzen et al., 1999).
A number of proteins have been identified that bind to the TNF-α ARE. These include TIA-1 (Gueydan et al., 1999; Piecyk et al., 2000) and tristetraprolin (TTP) (Lai et al., 1999), which are thought to be negative regulators of TNF-α synthesis. Thus, in response to inflammatory stimuli that activate macrophages, such as lipopolysaccharide (LPS), intracellular signalling pathways are activated that carry the signal needed to activate inflammatory mediator production. LPS, acting via the toll-like receptor 4 (TLR4), stimulates several mitogen-activated protein (MAP) kinase cascades, as well as the pathway leading to activation of the transcription factor NF-κB (Lee et al., 2000; Zhu et al., 2000; Perera et al., 2001). A role for some of these pathways in the post-transcriptional regulation of TNF-α has already been reported. For example, the classical MAP kinase cascade appears to be involved in the nucleo-cytoplasmic transport of TNF-α mRNA via its ARE (Dumitru et al., 2000). The stress-activated protein kinase 2a (SAPK2a, also called p38) pathway also plays a key role in stimulating the translation of TNF-α, because SB 203580 and closely related pyridinyl imidazoles, which inhibit this protein kinase, suppress the synthesis of this cytokine (Lee et al., 1994). The effect of SAPK2a/p38 on TNF-α synthesis seems to be mediated via the activation of its downstream target MAPKAP-K2, because the LPS-induced production of TNF-α in vivo is decreased by 90% in mice that do not express MAPKAP-K2 (Kotlyarov et al., 1999). The production of IL-6 and interferon-γ is reduced similarly in MAPKAP-K2-deficient mice, although the effect on IL-6 appears to result from an increase in mRNA stability (Neininger et al., 2002). The stability of the mRNA encoding COX-2 is also decreased by SB 203580 (Ridley et al., 1998) and may involve MAPKAP-K2 (Lasa et al., 2000).
The mechanism by which MAPKAP-K2 controls the translation of TNF-α mRNA and the stability of the mRNA encoding IL-6 is unknown, but is dependent on the presence of the ARE (Kontoyiannis et al., 1999; Neininger et al., 2002) and on the catalytic activity of MAPKAP-K2 (Kotlyarov et al., 2002). A working hypothesis is that the mechanism involves the phosphorylation of one or more proteins that bind to the ARE. For this reason, we decided to identify the major proteins that bind to the TNF-α ARE and to investigate whether they were substrates for MAPKAP-K2. This study led us to focus on the heterogeneous nuclear ribonucleoproteins (hnRNPs). Here, we identify hnRNP A0 as a novel physiological substrate for MAPKAP-K2 and implicate this protein in the LPS-induced post-transcriptional regulation of TNF-α, COX-2 and macrophage inflammatory protein 2 (MIP-2).
Results
Identification of proteins that bind to the AU-rich element of the TNF-α mRNA
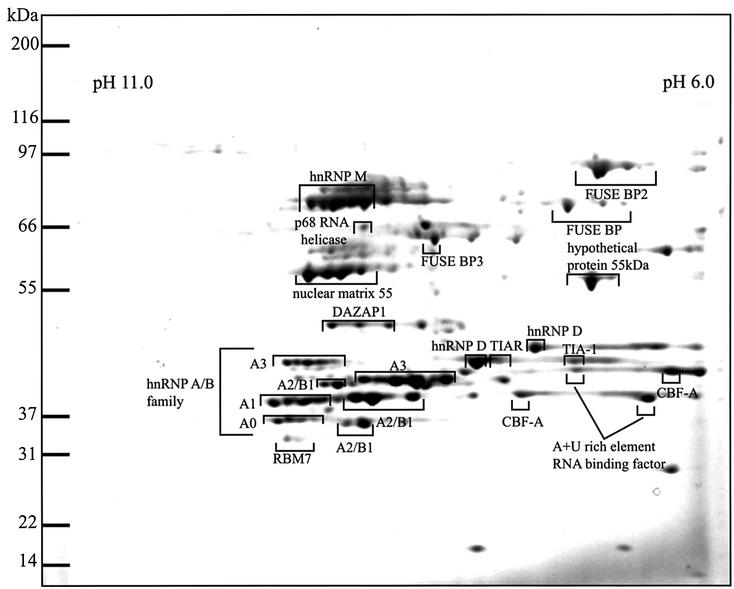
Extracts from RAW cells, a murine macrophage-like cell line, were subjected to affinity chromatography on magnetic beads coated with streptavidin to which biotinylated RNA oligonucleotides corresponding to the longer ARE of the TNF-α mRNA had been bound (see Materials and methods). Proteins that bound specifically were resolved on two-dimensional gels and stained with Sypro-Ruby (Figure 1). Each of the spots with pI values between 6 and 11 were excised, digested with trypsin and subjected to tryptic mass fingerprinting Those proteins that could be identified unequivocally are indicated (Table I; Figure 1). Proteins that could not be identified are unmarked. Many of the spots appearing as clusters with the same apparent mass but different net charge turned out to be different electrophoretic forms of the same protein and probably arise by post-translational modifications. Similarly, several spots with different molecular masses were identified as the same protein, presumably arising by alternative splicing of the same gene. Thus, the 60 major spots identified were accounted for by some 20 proteins. These included a few proteins already known to bind to AREs, such as TIA-1 (Gueydan et al., 1999), which is reported to be a negative regulator of TNF-α translation (Piecyk et al., 2000), hnRNP A0 (Myer and Steitz, 1995), hnRNP A1 (Hamilton et al., 1993) and hnRNP D (Loflin et al., 1999).
Fig. 1. Identification of proteins that bind to the TNF-α ARE. Untreated RAW cell extracts were incubated with a biotinylated RNA oligonucleotide corresponding to the AU-rich 3′-UTRs of the TNF-α mRNA (UUAUUAUUUAUUAUUUAUUUAUUAUUUAUUUAUUU). The ARE-binding proteins were resolved on a two-dimensional gel covering the range from pH 6 to pH 11 and stained with Sypro-Ruby. Proteins were identified by tryptic mass fingerprinting (Table I).
Table I. Identification of ARE-binding proteins isolated from murine RAW macrophages.
| Name of protein matched | Protein accession No. | Mass of protein (kDa) | % of tryptic peptides matched | Species matched to | Sequence coverage of peptides (%) |
|---|---|---|---|---|---|
| hnRNP M | 1710636 | 77.5 | 33 | Human | 43 |
| FUSE BP2 | 4504865 | 73.1 | 59 | Human | 32 |
| p68 RNA helicase | 6681157 | 69.3 | 13 | Mouse | 16 |
| FUSE BP | 4503801 | 67.5 | 45 | Human | 30 |
| FUSE BP3 | 1575609 | 63.9 | 43 | Human | 27 |
| Hypothetical protein 55 kDa | 7657015 | 55.2 | 43 | Human | 32 |
| Nuclear matrix protein 55 | 1895081 | 54.2 | 45 | Human | 42 |
| DAZAP1 | 8895708 | 43.1 | 15 | Mouse | 17 |
| TIAR | 6678349 | 43.3 | 56 | Mouse | 42 |
| TIA-1 | 14714709 | 41.5 | 27 | Mouse | 26 |
| RBM7 | 12837596 | 30.1 | 16 | Mouse | 29 |
| hnRNP D | 870749 | 38.4 | 25 | Human | 26 |
| AU-rich RNA-binding protein 1 | 6671602 | 29.9 | 25 | Mouse | 32 |
| hnRNPA3 | 1710627 | 39.7 | 20 | Human | 25 |
| hnRNPA2/B1 | 7949053 | 36.0 | 37 | Mouse | 56 |
| hnRNPA1 | 4504445 | 34.2 | 32 | Mouse | 54 |
| CBF-A (hnRNP A/B) | 6754222 | 30.9 | 18 | Mouse | 35 |
| hnRNP A0 | 12859567 | 30.5 | 36 | Mouse | 24 |
The masses of tryptic peptides were scanned against the Swiss-Prot, Genpep and NCBInr databases as described in Materials and methods.
The specificity of interaction of these proteins with the ARE was demonstrated by their failure to bind to a control RNA oligonucleotide of the same length that did not contain an ARE (data not shown). The pattern of proteins binding to the TNF-α ARE in LPS-stimulated and untreated RAW cells was similar (data not shown).
Phosphorylation of ARE-binding proteins by MAPKAP-K2
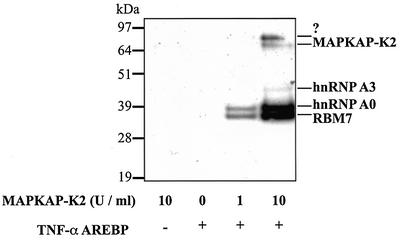
The proteins binding to the TNF-α ARE were incubated with Mg-[γ-32P]ATP in the absence or presence of MAPKAP-K2, and subjected to SDS–PAGE and autoradiography. These experiments revealed that two major bands were phosphorylated by MAPKAP-K2 with apparent Mrs of 38 and 36 kDa, together with two minor 32P-labelled bands of apparent Mrs 80 and 45 kDa (Figure 2). The 38 kDa 32P-labelled band was excised from the one-dimensional gel and identified as hnRNP A0 by tryptic mass fingerprinting. It also co-migrated with hnRNP A0 on two-dimensional gels (data not shown). Preliminary results suggest that the 36 kDa substrate is RNA-binding motif 7 protein (RBM7), but further experiments are needed to confirm this. The 90 kDa protein phosphorylated weakly by MAPKAP-K2 (Figure 2) has not yet been identified.
Fig. 2. MAPKAP-K2 phosphorylates two major proteins binding to the TNF-α ARE. Untreated RAW cell extracts were incubated with a biotin ylated RNA oligonucleotide corresponding to the TNF-α ARE. RNA-binding proteins were then phosphorylated in the absence or presence of 1 or 10 U/ml of MAPKAP-K2 and 20 nM [γ-32P]ATP (lanes 1–4). Proteins were then separated by SDS–PAGE, transferred to nitrocellulose membranes and autoradiographed.
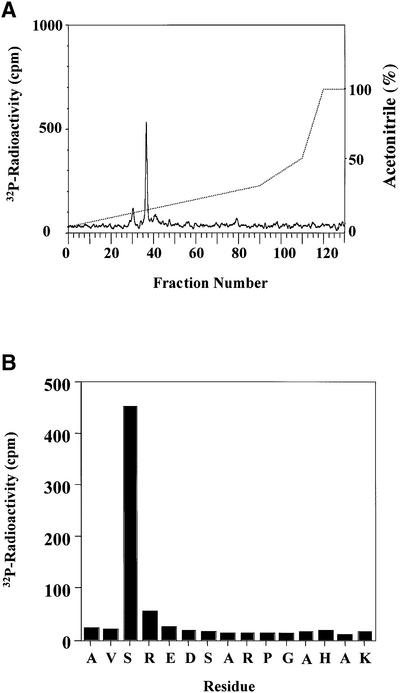
The human form of hnRNP A0 was cloned and expressed as a GST fusion protein. This protein could be phosphorylated to 0.7 mol/mol by incubation for 60 min with 150 U/ml MAPKAP-K2. The initial rate of phosphorylation was 3–5 times slower than that of heat shock protein 27 (HSP27), an established physiological substrate of MAPKAP-K2 (Eyers et al., 1999; Kotlyarov et al., 1999). Digestion with trypsin, followed by chromatography on a C18 column showed one major 32P-labelled peptide (Figure 3A). The mass of the peptide (m/z = 1631.8) corresponded to that of a monophosphorylated form of a tryptic peptide comprising residues 82–96. This provisional assignment was confirmed by Edman and solid phase sequencing, which also identified Ser84 as the site of phosphorylation (Figure 3B). An hnRNP A0 mutant in which Ser84 was changed to alanine could not be phosphorylated in vitro by MAPKAP-K2 (data not shown).
Fig. 3. Mapping of the site in hnRNP A0 phosphorylated by MAPKAP-K2. (A) Purified GST–hnRNP A0 was phosphorylated for 60 min in vitro with 150 U/ml of MAPKAP-K2 and 0.1 mM [γ-32P]ATP, and subjected to SDS–PAGE. The band corresponding to GST–hnRNP A0 was excised, digested with trypsin and the resulting peptides separated by reverse phase hydrophobic interaction chromatography on a C18 column equilibrated in 0.1% trifluoroacetic acid. 32P-radioactivity is shown by the full line, and the acetonitrile gradient by the broken line. (B) The mass of the major tryptic phosphopeptide from A (m/z = 1631.8) corresponds to that of a mono-phosphorylated form of a tryptic peptide comprising residues 82–96. The sequence of the peptide was confirmed by Edman sequencing and the site of phosphorylation was identified as the first serine in the peptide in a solid phase sequencing experiment. The methodology is detailed elsewhere (Stokoe et al., 1992).
hnRNP A0 is phosphorylated at Ser84 in response to LPS in RAW cells in an SB 203580-sensitive manner
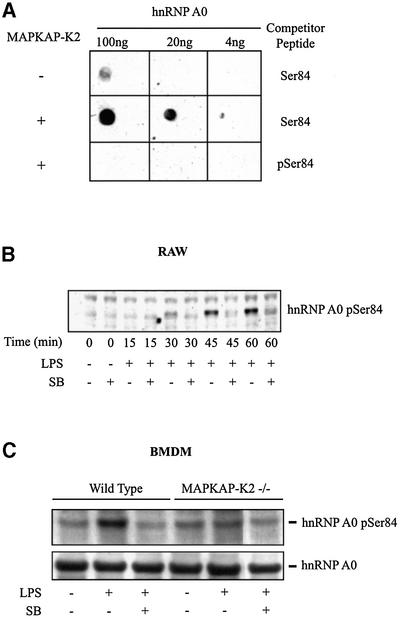
Antibodies were raised that recognize hnRNP A0 only when it is phosphorylated at Ser84 (Figure 4A) and used to investigate whether this protein was phosphorylated at this site in RAW macrophages. These experiments demonstrated that LPS did indeed induce the phosphorylation of a protein with the same apparent molecular mass as hnRNP A0 (Figure 4B), which was immunoprecipitated by an antibody that immunoprecipitates hnRNP A0 and co-migrated with hnRNP A0 on two-dimensional gels (data not shown). Moreover, phosphorylation was prevented by SB 203580, an inhibitor of SAPK2a/p38 (Figure 4B). Phosphorylation was unaffected by PD 184352 (1–10 µM; data not shown) which prevents activation of the classical MAP kinase cascade (Sebolt-Leopold et al., 1999) and the ERK5 pathway (Mody et al., 2001).
Fig. 4. hnRNP A0 is phosphorylated at Ser84 in LPS-stimulated macrophages. (A) GST–hnRNP A0 was phosphorylated for 60 min in vitro with 10 U/ml of MAPKAP K2 (+) and 0.1 mM ATP or left unphos phorylated (–). The proteins were spotted onto a nitrocellulose membrane in the amounts indicated. Blots were probed with the phospho-specific antibody that recognizes hnRNP A0 phosphorylated at Ser84 in the presence of the phosphorylated peptide antigen (pSer84) or the unphosphorylated Ser84 peptide (both at 10 µg/ml). (B) RAW cells were pre-treated or not for 15 min with 10 µM SB 203580 (SB) and either left untreated or stimulated with 50 ng/ml LPS for the times indicated. Lysates were separated by SDS–PAGE and, after transfer to nitrocellulose, the membranes were probed with the phospho-specific Ser84 antibody. (C) BMDMs from wild-type and MAPKAP-K2-deficient mice were pre-treated or not for 15 min with 10 µM SB 203580 (SB) and either left untreated or stimulated with 50 ng/ml LPS for 1 h. The cells were lysed and hnRNP A0 immunoprecipitated and immunoblotted as described in (B). The upper panel was probed with the Ser84 phospho-specific antibody and the lower panel with an antibody that recognizes the phosphorylated and dephosphorylated forms of hnRNP A0 equally well.
The LPS-induced phosphorylation of hnRNP A0 at Ser84 is greatly diminished in MAPKAP-K2-deficient mice
In order to investigate whether MAPKAP-K2, or another protein kinase activated by SAPK2a/p38, mediates the phosphorylation of hnRNP A0 at Ser84, we isolated bone marrow-derived macrophages (BMDMs) from wild-type and MAPKAP-K2-deficient mice. As in RAW cells, LPS induced the phosphorylation of a protein in BMDMs from wild-type mice that was recognized by the Ser84 phospho-specific antibody and was immunoprecipitated by an antibody that immunoprecipitates hnRNP A0 (Figure 4C). Phosphorylation was prevented by SB 203580, but not by PD 184352 (Figure 4C; data not shown). Importantly, the LPS-induced phosphorylation was greatly diminished in BMDMs from the MAPKAP-K2-deficient animals (Figure 4C). The residual phosphorylation observed in BMDM extracts from the deficient mice is also sensitive to SB 203580 (Figure 4C).
The mRNA encoding macrophage inflammatory protein 2 is bound by hnRNP A0
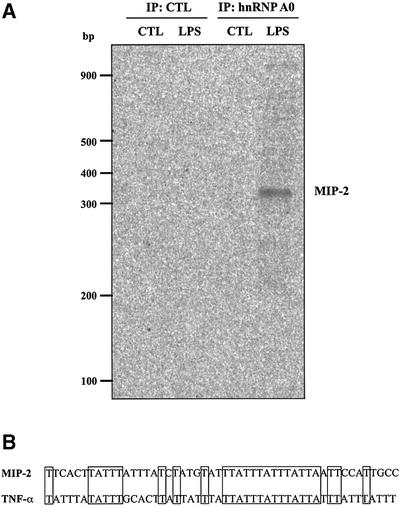
The finding that hnRNP A0 was a physiological substrate for MAPKAP-K2 raised the question of whether its phosphorylation played an important role in LPS-stimulated cytokine production. In order to investigate whether this was the case, we immunoprecipitated hnRNP A0 from RAW cell extracts and examined which mRNAs were bound to this protein (see Materials and methods). The method used was verified by the demonstration that TNF-α mRNA could be identified after immunoprecipitation of TIA-1 (data not shown), a protein known to bind to the ARE of the TNF-α mRNA (Piecyk et al., 2000). Using the same conditions, RNA bound to hnRNP A0 immunoprecipitates was amplified using random primer set 1 (see Materials and methods). This led to the amplification of one band that was present in the hnRNP A0 immunoprecipitated from extracts prepared from LPS-stimulated cells, but not from unstimulated cells or a control immunoprecipitation performed in the absence of the hnRNP A0 antibody (Figure 5A). This band was cut out, re-amplified, cloned and sequenced to determine its identity. The region retrieved was the 3′-UTR of MIP-2, which contains an ARE not dissimilar to that of TNF-α mRNA (Figure 5B). MIP-2 is a member of the CXC chemokine family and is involved in the recruitment of neutrophils to the site of inflammation (Nick et al., 2000).
Fig. 5. MIP-2 mRNA is bound by hnRNP A0 in extracts from LPS-stimulated cells. (A) RAW cells were left untreated or treated for 1 h with 50 ng/ml LPS and lysed. Then immunoprecipitation with (lanes 3–4) or without (lanes 1–2) the hnRNP A0 antibody was carried out from clarified extracts and the RNA bound identified using a differential display-based assay with random primer set 1 (see Materials and methods). CTL, control experiments. (B) Sequences of part of the 3′-UTR of murine MIP-2 mRNA (accession No. X53798) and TNF-α mRNA (accession No. X02611).
hnRNP A0 interaction with specific cytokine mRNAs is dependent on the SAPK2a/p38 pathway
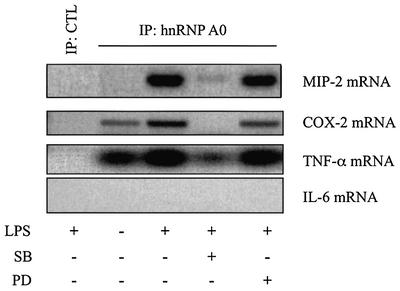
The identification of MIP-2 mRNA in hnRNP A0 immunoprecipitates was confirmed in a separate experiment using primers specific for the coding sequence of MIP-2 (Figure 6, lane 3). These experiments confirmed that this interaction only occurred in LPS-stimulated RAW cells and that it was prevented by SB 203580, but not by PD 184352 (Figure 6).
Fig. 6. SB 203580 prevents the LPS-induced interaction of hnRNP A0 with the mRNAs of MIP-2, COX-2 and TNF-α. RAW cells were pre-treated or not for 15 min with 10 µM SB 203580 (SB) or 1 µM PD 184352 (PD) and either left untreated or stimulated for 1 h with 50 ng/ml LPS. hnRNP A0 was immunoprecipitated as described in Figure 5 and mRNAs for MIP-2, TNF-α, COX-2 and IL-6 were amplified by RT–PCR with specific primers. CTL, control experiment.
Since MAPKAP-K2 has been implicated in the LPS-induced post-transcriptional regulation of COX-2, TNF-α and IL-6 (see Introduction), we used specific primers to examine whether any of these mRNAs could be amplified from the hnRNP A0 immunoprecipitates. These experiments revealed that COX-2 mRNA and TNF-α mRNA were also present in the hnRNP A0 immunoprecipitates, and that binding was markedly stimulated by LPS and inhibited by SB 203580. PD 184352 had no effect on the binding of TNF-α mRNA, but moderately decreased the binding of COX-2 mRNA. No IL-6 mRNA could be detected in hnRNP A0 immunoprecipitates even though it could be amplified from the extracts of LPS-stimulated cells in control experiments (Figure 6; data not shown).
The induction of MIP-2 protein and stability of the MIP-2 mRNA are prevented by inhibition of SAPK2a/p38 and the classical MAP kinase cascade
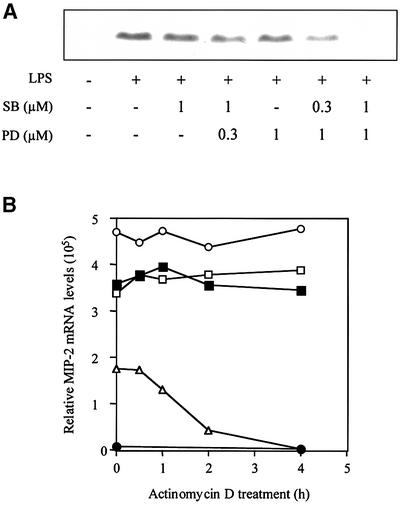
The MIP-2 protein is up-regulated following treatment with LPS (50 ng/ml). Induction can be observed after 1 h and is maximal after 2–4 h (data not shown). This up-regulation is partially inhibited by pre-treatment with SB 203580 or PD 184352, but blocked completely when both inhibitors are used at a concentration of 1.0 µM. At 1.0 µM SB 203580, half-maximal inhibition of MIP-2 induction by PD 184352 occurred at 0.3–0.5 µM, while at 1.0 µM PD 184352, half-maximal inhibition by SB 203580 occurred at 0.3 µM (Figure 7A). These concentrations are similar to those that block the LPS-induced activation of MAPKAP-K2 and the classical MAP kinase cascade, respectively (data not shown).
Fig. 7. The SAPK2a/p38 pathway and the classical MAP kinase cascade are both required for MIP-2 production at the mRNA and protein level. (A) RAW cells were pre-treated or not for 15 min with the indicated concentrations of SB 203580 (SB) and/or the indicated concentrations of PD 184352 (PD), and either left untreated or stimulated with 50 ng/ml LPS for 2 h. Lysates were separated by SDS–PAGE and, after transfer to nitrocellulose, the membranes were probed with a murine MIP-2-specific antibody. (B) RAW cells were treated for 15 min without (open and closed circles), with SB 203580 (closed squares), with PD 184352 (open squares) or SB 203580 plus PD 184352 (open triangles) and then for 1 h without (closed circles) or with (other symbols) LPS (time = 0 h). Transcription was then inhibited by the addition of actinomycin D and the level of MIP-2 RNA quantified at various times up to 4 h using an RNase protection assay.
In order to investigate the mechanism by which LPS stimulates the production of MIP-2, RAW cells were treated for 15 min with or without SB 203580 and/or PD 184352 and then for 1 h with LPS. This caused an enormous increase in the level of MIP-2 mRNA, which was only reduced by 25% in the presence of SB 203580 or PD 184352, but by 65% in the presence of both inhibitors (see time zero in Figure 7B). This indicates that at least one other signal transduction pathway (resistant to SB 203580 plus PD 184352) is required for maximal induction of MIP-2 mRNA, probably at the level of gene transcription.
One hour after stimulation with LPS, transcription was inhibited by the addition of actinomycin D, and the level of MIP-2 RNA was quantified at various times using an RNase protection assay (Figure 7). Pre-treatment with either SB 203580 or PD 184352 had little effect on mRNA stability in the 4 h subsequent to the addition of actinomycin D. In contrast, pre-treatment with SB 203580 plus PD 184352 drastically decreased MIP-2 mRNA stability, the half-life decreasing to <2 h (Figure 7).
Discussion
Elucidating the mechanisms by which LPS stimulates the production of inflammatory mediators involves understanding how the mRNAs encoding these proteins are regulated at the post-transcriptional level. Numerous studies have shown how the ARE found in the 3′-UTR of many of these mRNAs is critical in controlling their stability and translation (Kontoyiannis et al., 1999; Dumitru et al., 2000; Lasa et al., 2000; Neininger et al., 2002). However, the mechanism by which this is accomplished in macrophages remains to be elucidated. In order to understand this phenomenon better, we initially characterized the proteins that bind to the TNF-α ARE in macrophages using a proteomic-based approach. Interestingly, some of the proteins known to bind to the ARE (hnRNP A0, hnRNP A1, hnRNP D and TIA-1) and to be involved in TNF-α production (TIA-1) were identified in our analysis, although others (such as TTP) were not. Most of the proteins also bound to the c-fos ARE as well as the AREs in the mRNAs encoding COX-2 and granulocyte–macrophage colony-stimulating factor (GM-CSF).
Most of the proteins we characterized contain RNA recognition motifs (RRMs) present in 1–3 copies per protein, but the KH-type RNA-binding domain was found in the FUSEBP family. A subset of the RNA-binding proteins, which included members of the hnRNP A/B family, also possessed an arginine/glycine-rich domain (RGG) that is thought to be involved in protein–protein interaction as well as RNA binding (Krecic and Swanson, 1999). The largest group of proteins (∼50%) belonged to the hnRNP family, known to shuttle between the nucleus and the cytoplasm and to be involved in many aspects of RNA processing. The members of this group are also known to exist in multiple forms, resulting from both alternative splicing and post-translational modifications (Krecic and Swanson, 1999), which is in accordance with our observations (Figure 1). This gives the cell an impressive pool of regulatory proteins with which to control the different steps of RNA processing. Other proteins found are better known for their involvement in splicing (FUSEBP, nuclear matrix protein 55), but might also be part of RNA–protein complexes shuttling between the nucleus and the cytoplasm. Our identification of most of the proteins binding specifically to the ARE should facilitate further experiments aimed at understanding the nature of the protein complexes that bind to the ARE.
The post-translational modifications present in ARE-binding proteins (AREBPs) may be involved in the LPS-mediated post-transcriptional control of inflammatory mediators. Since signalling through MAPKAP-K2 appears to be crucial for the production of TNF-α, IL-6, interferon-γ and COX-2 (see Introduction), we investigated whether MAPKAP-K2 was able to phosphorylate any of the AREBPs. Two proteins were phosphorylated strongly by MAPKAP-K2 in vitro, one of which was identified as hnRNP A0 (Figure 2). The phosphorylation site was mapped to Ser84 within a consensus phosphorylation sequence for MAPKAP-K2 found only in hnRNP A0 and not in other hnRNP A/B. For example, although the equivalent serine (Ser113) is present in hnRNP A3, it is not phosphorylated by MAPKAP-K2 in vitro (data not shown), probably because of the lack of a large hydrophobic residue five amino acids N-terminal to the site of phosphorylation (Stokoe et al., 1993).
A phospho-specific antibody was developed against Ser84 of hnRNP A0. With this antibody, the LPS-stimulated phosphorylation of Ser84 was observed in both RAW cells and BMDMs. This phosphorylation was sensitive to the SAPK2a/p38 inhibitor SB 203580, unaffected by PD 184352, which blocks the classical MAPK pathway, or wortmannin, which inhibits class I phosphatidylinositol 3-kinases (data not shown), and greatly decreased in BMDMs from MAPKAP-K2-deficient mice (Figure 4C). Although these results suggest that MAPKAP-K2 is the major protein kinase phosphorylating hnRNP A0 at Ser84 in vivo, it has been reported that the level of SAPK2a/p38 protein is diminished in primary macrophages from MAPKAP-K2-deficient mice for reasons that are not yet understood (Kotlyarov et al., 2002). It could therefore be argued that the decrease in SAPK2a/p38 activity in BMDMs reduces the activity of other protein kinases that are switched on by SAPK2a/p38 and that it is these enzymes, rather than MAPKAP-K2, that phosphorylate Ser84 in vivo. Indeed, we have found that hnRNP A0 can also be phosphorylated at Ser84 by two such enzymes, MAPKAP-K3 and p38-regulated and activated kinase (PRAK, also called MAPKAP-K5) (McLaughlin et al., 1996; New et al., 1998) in vitro (data not shown).
In order to address this problem, we studied the LPS-induced phosphorylation of the transcription factor CREB, which is known to be catalysed by mitogen and stress-activated proteins kinases 1 and 2 (MSK1 and MSK2; Caivano and Cohen, 2000; Wiggin et al., 2002), two closely related protein kinases that are activated by SAPK2a/p38 (Deak et al., 1998). In the presence of PD 184352 to prevent the activation of MSK1/MSK2 by the MAP kinases of the classical MAP kinase cascade (Deak et al., 1998), we found that the LPS-induced phosphorylation of CREB was not decreased in BMDMs from MAPKAP-K2-deficient mice (data not shown). This indicates that, although the level of SAPK2a/p38 protein is reduced in MAPKAP-K2-deficient BMDMs, its activity towards at least one of its substrates (MSK1/MSK2) is not impaired.
The residual phosphorylation of hnRNP A0 at Ser84 in BMDMs from MAPKAP-K2-deficient mice was also sensitive to SB 203580 (Figure 4C). This suggests that the weak phosphorylation of Ser84 observed under these conditions may be mediated MAPKAP-K3 and/or PRAK.
In order to investigate which mRNAs bind specifically to hnRNP A0, we immunoprecipitated this protein from macrophage extracts. These studies revealed that the mRNAs encoding TNF-α, COX-2 and MIP-2 could all be detected in the immunoprecipitates. Strikingly, the mRNA encoding MIP-2 could only be detected after immunoprecipitation of hnRNP A0 from extracts of LPS-stimulated cells, but not if the cells were pre-incubated with SB 203580 prior to stimulation with LPS (Figure 6), even though the level of MIP-2 mRNA in the macrophage extracts was only decreased slightly by SB 203580 (Figure 7B). The amount of mRNA encoding TNF-α and COX-2 was also stimulated by LPS and suppressed by SB 203580. The simplest explanation for these findings is that the phosphorylation of hnRNP A0 at Ser84 by MAPKAP-K2 enhances binding to the AREs of these mRNAs or allows hnRNP A0 to displace another protein(s) from the AREs.
The LPS-induced stabilization of the MIP-2 mRNA was unaffected by the inhibition of SAPK2a/p38 (Figure 7), even though the interaction of hnRNP A0 with the MIP-2 mRNA was blocked (Figure 6). Our working hypothesis is that at least one other phosphorylation event is sufficient for MIP-2 stabilization, because the LPS-induced stabilization of MIP-2 mRNA is only blocked by SB 203580 in combination with PD 184352 (Figure 7), an inhibitor of a distinct signal transduction pathway. This situation is likely to apply to the post-transcriptional regulation of other inflammatory mediators, since inhibition of both the classical MAP kinase cascade and SAPK2a/p38 is also required to prevent the LPS-induced production of the mRNA and protein encoding COX-2 and IL-1 in RAW cells (Caivano and Cohen, 2000).
In contrast to MIP-2, COX-2 and IL-6, where post-transcriptional regulation occurs mostly at the level of mRNA stabilization, the LPS-induced production of TNF-α is thought to be due mainly to the release of inhibition of mRNA translation, resulting from the interaction of proteins with the ARE (Kontoyiannis et al., 1999; Neininger et al., 2002). Moreover, this is largely suppressed by inhibition of SAPK2a/p38 alone (Lee et al., 1994) and in the MAPKAP-K2-deficient mice (Kotlyarov et al., 1999). This raises the possibility that the MAPKAP-K2-catalysed phosphorylation of hnRNP A0 and/or the other major substrate for this protein kinase that we have detected in the ARE-binding fraction (Figure 2) may play a key role in stimulating translation of the TNF-α message. Interfering with endogenous hnRNP A0 either with small interfering RNA (siRNA) or by replacement of wild-type hnRNP A0 with the Ser84Ala mutant will be important to establish whether the phosphorylation of this site is critical for cytokine production.
Gene regulation through post-transcriptional mechanisms is likely to play an important role in controlling many steps of inflammation and other physiological processes. In order to understand this mechanism better, it will be important to comprehend the dynamics of protein complex formation on specific mRNAs and how they are regulated by signal transduction pathways. In this study, we have addressed this question by, on the one hand, purifying many AREBPs bound to a prototypical ARE and, on the other hand, by identifying specific mRNAs bound by one AREBP. It will be interesting to find out if different members of the hnRNP A/B family bind the same or distinct mRNAs. Moreover, as we have observed for MIP-2 and COX-2, a network of signal transduction pathways clearly operates to control gene regulation at different levels.
Materials and methods
Materials
[γ-32P]ATP, [α-33P]dATP, PreScission protease and materials for two-dimensional gel analysis were obtained from Amersham Pharmacia Biotech (Little Chalfont, UK), except for the pre-cast SDS–polyacrylamide gels, running buffer, transfer buffer and colloidal blue staining kit which were obtained from Invitrogen (Groningen, The Netherlands). Unlabelled ATP, Pefabloc, dithiothreitol (DTT) and ‘complete protease inhibitor set EDTA-free’ were from Roche Molecular Biochemicals (Lewes, UK), cell culture media from Bio-Whittaker (Wokingham, UK) and SB 203580 from Calbiochem (Nottingham, UK). Maxiscript T7 kit, Retroscript kit, RPA III kit, RNase-free buffers and solutions were purchased from Ambion (Abingdon, UK). Streptavidin-coated magnetic beads and MPC-1 magnet were from Dynal (Bromborough, UK). The access RT–PCR kit and RNAsin were from Promega (Southampton, UK). The RNAimage kit for differential display was purchased from GenHunter Corporation (Kimbolton, UK). The RNAeasy Mini and Midi kit were purchased from Qiagen (Crawley, UK). Sypro-Orange and Sypro-Ruby gel and blot stains were from Molecular Probes (Leiden, The Netherlands). Other chemicals were purchased from Merck (Poole, UK) or Sigma-Aldrich (Poole, UK).
Antibodies
Phospho-specific antibodies were affinity purified from antisera on phosphopeptide antigen–Sepharose columns, followed by passage through an unphosphorylated peptide–Sepharose column. The flowthrough fractions were collected and used for experimentation. The antibodies were used at 0.1–1.0 µg/ml in the presence of 10 µg/ml of the unphosphorylated peptide antigen. The antibody that recognizes hnRNP A0 phosphorylated at Ser84 was raised against the peptide clkravs*redsarpga (synthesized by Dr Graham Blumberg, University of Bristol, UK), where the phosphorylated serine is represented by S*. The peptide was conjugated separately to both bovine serum albumin and keyhole limpet haemocyanin via the N-terminal cysteine before injection into sheep at Diagnostics Scotland (Edinburgh, UK). The antibody that recognizes hnRNP A0 was raised by injecting GST–hnRNP A0 and purifying the antisera through an affinity column followed by passage down a GST column. These antibodies were provided by Drs Jane Leitch and Chris Armstrong (Division of Signal Transduction Therapy, University of Dundee). Antibodies against mouse TNF-α and mouse MIP-2 were purchased from R&D systems (Abingdon, UK). The antibody against mouse TIA-1 was purchased from BD Biosciences (Oxford, UK). Rabbit anti-sheep IgG antibodies and goat anti-rabbit IgG, both conjugated to peroxidase, were obtained from Perbio Science Ltd. (Tattenhall, UK).
Plasmids
Image clone 755060 was amplified using primers 5′_HA_Bam (ggatccgccaccatgtacccatacgatgtgccagattacgccgagaattctcagttgtgtaagctgttc) and 3′_Bam (ggatccttagaaggagctgcctccatagccaccccc) with the GC-Rich PCR System (Roche) following the manufacturer’s instructions, which produced a single product of 963 bp. These primers introduced BamHI sites at both ends along with a Kozak sequence and 5′ haemagglutinin (HA) tag. The PCR product was cloned into pCR2.1 TOPO (Invitrogen) and fully sequenced. pCR2.1A0 was digested with BamHI and cloned into the same site in pGEX6P-1 (Amersham Pharmacia Biotech) to produce bacterially expressed GST/HA tag hnRNP A0.
Proteins
All recombinant proteins used were expressed as GST fusion proteins in Escherichia coli strain BL21. Where indicated, the GST was cleaved using the PreScission protease. MAPKAP-K2 was activated by incubation with GST–ERK2 (extracellular signal-regulated kinase-2) and the activator removed by chromatography on S-Sepharose; alternatively, MAPKAP-K2 was activated with GST–SAPK2a/p38 and used in the presence of 10 µM SB 203580. These enzymes were prepared in the Division of Signal Transduction Therapy, Dundee. HSP27 was a generous gift from Professor Roy Quinlan, University of Durham, UK. One unit of MAPKAP-K2 activity was that amount that catalysed the incorporation of 1 nmol of phosphate into the peptide substrate KKLNRTLSVA in 1 min under standard assay conditions (Stokoe et al., 1993).
Cell culture
RAW264 macrophages were maintained in a 95% air, 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% (v/v) heat-inactivated fetal calf serum (FCS), 100 U/ml penicillin and 100 µg/ml streptomycin. The day before stimulation, the macrophages were plated at a density of 2 × 106 cells/6 cm plate. BMDMs were isolated by flushing mice femurs with RPMI-1640, passed through a cell strainer (70 µM; Falcon), centrifuged and re-suspended in RPMI-1640 containing 10% (v/v) heat-inactivated FCS, 30% L929 conditioned media, 100 U/ml penicillin and 100 µg/ml streptomycin. They were plated on 9 cm bacteriological plastic and left to grow for a week. One day prior to plating, the cells were washed with fresh media.
Cell lysis
The cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in buffer A [50 mM Tris–HCl pH 7.4, 150 mM KCl, 0.1 mM EDTA, 1% (w/v) NP-40, 4 mM DTT, 20 mM NaF, 40 mM sodium β-glycerophosphate, 2 mM sodium orthovanadate, 1 mM Pefabloc, complete EDTA-free protease inhibitors]. The lysates were centrifuged for 10 min at 13 000 g and the supernatants removed, and used immediately or frozen in liquid nitrogen and stored at –80°C.
RNA affinity chromatography
Streptavidin-coated magnetic beads were made RNase free and bound to different biotinylated RNA oligonucleotides (Xeragon) according to the manufacturer’s instructions (Dynal). Lysates from RAW cells were incubated for 30 min at 4°C with a biotinylated RNA oligonucleotide corresponding to the AU-rich region of TNF-α (UUAUUAUUUAUUA UUUAUUUAUUAUUUAUUUAUUU), c-fos (GUUUUUAAUUUAU UUAUUAAGAUGGAUUcU) or an irrelevant RNA (AAGCUUGGG CUGCAGGUCGACUCUAGAGGA) in binding solution. This consisted of equal volumes of buffer A and buffer B [50 mM KCl, 10 mM MgCl2, 10% (v/v) glycerol, 2 mg/ml heparin, 2 mg/ml tRNA, 50 U/ml of RNAsin]. The beads were washed extensively in buffer C [25 mM Tris–HCl pH 7.4, 100 mM NaCl, 5 mM MgCl2, 5% (v/v) glycerol], and the AREBPs were eluted in one of two ways. For two-dimensional gel analysis, the AREBPs were eluted directly in the re-hydration solution (8 M urea, 2 M thiourea, 2% CHAPS, 3 mM TCEP), while for analysis of phosphorylation the AREBPs were eluted with 1.5 M NaCl in buffer C.
Two-dimensional gel analysis
AREBP were resolved on IPG 6–11 strips for the first dimension using the IPGphor isoelectric focusing system (Amersham). This pH range was selected because most of the proteins that were retained by the RNA affinity column had pI values within that region due to their affinity for RNA (data not shown). The second dimension was performed on 4–12% ZOOM gels from Invitrogen using standard electrophoresis technique. The gels were stained with Sypro-Ruby and the image captured using a Fuji Imager with LAS1000IR software.
Phosphorylation of AREBPs
The mixture of AREBPs from the RNA affinity column was diluted 10-fold in buffer D [50 mM Tris–HCl pH 7.5, 0.1% (v/v) β-mercaptoethanol, 0.1 mM sodium orthovanadate, 5 mM sodium β-glycerophosphate] and activated MAPKAP-K2 added to a final concentration of 1 or 10 U/ml. The reactions (0.03 ml) were initiated by the addition of 15 mM MgCl2 and γ-32P-labelled ATP (2.5 × 106 c.p.m.) at the concentrations specified in the figure legends, and incubated for 7.5 min at 30°C. Purified hnRNP A0 and HSP27 were phosphorylated at a final concentration of 1 µM.
Identification of RNA bound to immunoprecipitated hnRNP A0
Cells were lysed in 10 mM Tris–HCl pH 7.6, 1 mM potassium acetate, 1.5 mM magnesium acetate, 0.5 % (w/v) NP-40, 2 mM DTT, 1000 U/ml of RNAsin, 1 mM Pefabloc, complete EDTA-free protease inhibitors. The lysates were centrifuged for 10 min at 13 000 g and the supernatants removed and used immediately for immunoprecipitation. This was carried out using the Seize-X immunoprecipitation kit from Pierce according to the manufacturer’s instructions. Briefly, 25 µg of hnRNP A0 antibody was cross-linked with disuccinimidyl suberate (DSS) to protein G–Sepharose. Lysates were diluted with an equal volume of 8 mM Na2PO4, 2 mM KH2P04, 140 mM NaCl, 10 mM KCl pH 7.4 and incubated for 1 h at 4°C in the presence of the bound antibody. They were then washed four times with 25 mM Tris–HCl pH 7.2 containing 150 mM NaCl, eluted with Immunopure Elution buffer (Pierce, pH 2.8) and neutralized by the addition of 10 µl of 1 M Tris base to 190 µl of eluate. RNA in the eluate was extracted using the RNAeasy Kit (Qiagen).
For the identification of unknown RNA species present in hnRNP A0 immunoprecipitates, differential display was carried out using the GenHunter RNAimage kit number 4. The 3′ primer used for reverse transcription was a mix of the three primers H-T11G, H-T11A and H-T11C. The PCR was then carried out with these primers in the presence of [α-33P]dATP (3000 Ci/mmol) and a mix of the following four 5′ primers, H-AP25, H-AP26, H-AP27 and H-AP28 (termed random primer set 1). The mixture was resolved on 6% denaturing polyacrylamide gels and specific bands were cut out and re-amplified before being cloned into pCR2.1 TOPO (Invitrogen) and sequenced. The identification was confirmed by RT–PCR using specific primers for identified genes (see below).
RNase protection assay and RT–PCR
Relative levels of MIP-2 RNA were quantified by RNase protection with the RPA III kit (Ambion). The MIP-2 probe was synthesized with the following primers (5′, ATGGCCCCTCCCACCTGCC; 3′, TCAGTTAG CCTTGCCTTTGTTCAGTATCTTTTG; 303 bp product) and the cyclophilin probe using the pTRI template (Ambion). RNA levels were determined with a phosphoimager and normalized to cyclophilin control. In some cases, RNA was amplified using the Access RT–PCR kit from Promega with specific primers for TNF-α, COX-2 and IL-6 from Ambion and those for MIP-2 as described above for synthesis of the RPA probe.
Mass spectrometry
Tryptic peptides were analysed on a Perseptive Biosystems (Framingham, MA) Elite STR matrix-assisted laser desorption time of flight (MALDI-TOF) mass spectrometer with saturated α-cyanocinnamic acid as the matrix. The mass spectrum was acquired in the reflector mode and was mass calibrated internally. The tryptic peptide ions obtained were scanned against the Swiss-Prot and Genpep databases using the MS-FIT program of Protein Prospector. Where there was an ambiguity in the identity of a protein sample, LC-MS/MS was performed. The tryptic digest was injected onto a 0.075 × 100 mm PepMap C18 capillary column, equilibrated with 0.1% formic acid in water, attached to a LC-Packings Ultimate HPLC system (Dionex UK; Camberley, UK). The column was developed with a discontinuous acetonitrile gradient at 0.2 µl/min and the column was interfaced to a Q-TOF2 mass spectrometer (Micro mass, Manchester, UK). The peptide ions generated by the electrospray interface were fragmented automatically using machine-defined collision voltages. The resultant peak lists were searched using the Sonar search engine (Genomic Solutions, Ann Arbor, MI) against the NCBInr database.
Acknowledgments
Acknowledgements
We thank the Protein Production Team of the Division of Signal Transduction Therapy at Dundee for providing activated MAPKAP-K2, and the Dundee Transgenic Mouse Facility for maintaining the colonies of MAPKAP-K2-deficient mice. S.R. is supported by a postdoctoral fellowship from the Canadian Institute of Health Research. The work was supported by the UK Medical Research Council, the Royal Society, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, NovoNordisk and Pfizer.
References
- Caivano M. and Cohen,P. (2000) Role of mitogen-activated protein kinase cascades in mediating lipopolysaccharide-stimulated induction of cyclooxygenase-2 and IL-1β in RAW264 macrophages. J. Immunol., 164, 3018–3025. [DOI] [PubMed] [Google Scholar]
- Deak M., Clifton,A.D., Lucocq,L.M. and Alessi,D.R. (1998) Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38 and may mediate activation of CREB. EMBO J., 17, 4426–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru C.D. et al. (2000) TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell, 103, 1071–1083. [DOI] [PubMed] [Google Scholar]
- Eyers P.A., van den,I.P., Quinlan,R.A., Goedert,M. and Cohen,P. (1999) Use of a drug-resistant mutant of stress-activated protein kinase 2a/p38 to validate the in vivo specificity of SB 203580. FEBS Lett., 451, 191–196. [DOI] [PubMed] [Google Scholar]
- Grzybowska E.A., Wilczynska,A. and Siedlecki,J.A. (2001) Regulatory functions of 3′UTRs. Biochem. Biophys. Res. Commun., 288, 291–295. [DOI] [PubMed] [Google Scholar]
- Gueydan C., Droogmans,L., Chalon,P., Huez,G., Caput,D. and Kruys,V. (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem., 274, 2322–2326. [DOI] [PubMed] [Google Scholar]
- Hamilton B.J., Nagy,E., Malter,J.S., Arrick,B.A. and Rigby,W.F. (1993) Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem., 268, 8881–8887. [PubMed] [Google Scholar]
- Kontoyiannis D., Pasparakis,M., Pizarro,T.T., Cominelli,F. and Kollias,G. (1999) Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity, 10, 387–398. [DOI] [PubMed] [Google Scholar]
- Kotlyarov A., Neininger,A., Schubert,C., Eckert,R., Birchmeier,C., Volk,H.D. and Gaestel,M. (1999) MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nat. Cell. Biol., 1, 94–97. [DOI] [PubMed] [Google Scholar]
- Kotlyarov A., Yannoni,Y., Fritz,S., Laass,K., Telliez,J.B., Pitman,D., Lin,L.L. and Gaestel,M. (2002) Distinct cellular functions of MK2. Mol. Cell. Biol., 22, 4827–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa M., Mahtani,K.R., Finch,A., Brewer,G., Saklatvala,J. and Clark,A.R. (2000) Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol., 20, 4265–44274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Mira-Arbibe,L. and Ulevitch,R.J. (2000) TAK1 regulates multiple protein kinase cascades activated by bacterial lipo polysaccharide. J. Leukoc. Biol., 68, 909–915. [PubMed] [Google Scholar]
- Lee J.C. et al. (1994) A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature, 372, 739–746. [DOI] [PubMed] [Google Scholar]
- Loflin P., Chen,C.Y. and Shyu,A.B. (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev., 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M.M., Kumar,S., McDonnell,P.C., Van Horn,S., Lee,J.C., Livi,G.P. and Young,P.R. (1996) Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J. Biol. Chem., 271, 8488–8492. [DOI] [PubMed] [Google Scholar]
- Mody N., Leitch,J., Armstrong,C., Dixon,J. and Cohen,P. (2001) Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett., 502, 21–24. [DOI] [PubMed] [Google Scholar]
- Myer V.E. and Steitz,J.A. (1995) Isolation and characterization of a novel, low abundance hnRNP protein: A0. RNA, 1, 171–182. [PMC free article] [PubMed] [Google Scholar]
- Neininger A., Kontoyiannis,D., Kotlyarov,A., Winzen,R., Eckert,R., Volk,H.D., Holtmann,H., Kollias,G. and Gaestel,M. (2002) MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem., 277, 3065–3068. [DOI] [PubMed] [Google Scholar]
- New L., Jiang,Y., Zhao,M., Liu,K., Zhu,W., Flood,L.J., Kato,Y., Parry,G.C. and Han,J. (1998) PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J., 17, 3372–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick J.A., Young,S.K., Brown,K.K., Avdi,N.J., Arndt,P.G., Suratt,B.T., Janes,M.S., Henson,P.M. and Worthen,G.S. (2000) Role of p38 mitogen-activated protein kinase in a murine model of pulmonary inflammation. J. Immunol., 164, 2151–2159. [DOI] [PubMed] [Google Scholar]
- Perera P.Y., Mayadas,T.N., Takeuchi,O., Akira,S., Zaks-Zilberman,M., Goyert,S.M. and Vogel,S.N. (2001) CD11b/CD18 acts in concert with CD14 and Toll-like receptor (TLR) 4 to elicit full lipopolysaccharide and taxol-inducible gene expression. J. Immunol., 166, 574–581. [DOI] [PubMed] [Google Scholar]
- Piecyk M. et al. (2000) TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J., 19, 4154–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley S.H., Dean,J.L., Sarsfield,S.J., Brook,M., Clark,A.R. and Saklatvala,J. (1998) A p38 MAP kinase inhibitor regulates stability of interleukin-1-induced cyclooxygenase-2 mRNA. FEBS Lett., 439, 75–80. [DOI] [PubMed] [Google Scholar]
- Sebolt-Leopold J.S. et al. (1999) Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med., 5, 810–816. [DOI] [PubMed] [Google Scholar]
- Stokoe D., Campbell,D.G., Nakielny,S., Hidaka,H., Leevers,S.J., Marshall,C. and Cohen,P. (1992) MAPKAP kinase-2; a novel protein kinase activated by mitogen-activated protein kinase. EMBO J., 11, 3985–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D., Caudwell,B., Cohen,P.T. and Cohen,P. (1993) The substrate specificity and structure of mitogen-activated protein (MAP) kinase-activated protein kinase-2. Biochem. J., 296, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggin G.R., Soloaga,A., Foster,J.M., Murray-Tait,V., Cohen,P. and Arthur,J.S. (2002) MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol., 22, 2871–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzen R. et al. (1999) The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J., 18, 4969–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Chen,C.Y. and Shyu,A.B. (1997) Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol., 17, 4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Downey,J.S., Gu,J., Di Padova,F., Gram,H. and Han,J. (2000) Regulation of TNF expression by multiple mitogen-activated protein kinase pathways. J. Immunol., 164, 6349–6358. [DOI] [PubMed] [Google Scholar]