Abstract
The biogenesis of the cytoskeletal proteins actin and tubulin involves interaction of nascent chains of each of the two proteins with the oligomeric protein prefoldin (PFD) and their subsequent transfer to the cytosolic chaperonin CCT (chaperonin containing TCP-1). Here we show by electron microscopy that eukaryotic PFD, which has a similar structure to its archaeal counterpart, interacts with unfolded actin along the tips of its projecting arms. In its PFD-bound state, actin seems to acquire a conformation similar to that adopted when it is bound to CCT. Three-dimensional reconstruction of the CCT:PFD complex based on cryoelectron microscopy reveals that PFD binds to each of the CCT rings in a unique conformation through two specific CCT subunits that are placed in a 1,4 arrangement. This defines the phasing of the CCT rings and suggests a handoff mechanism for PFD.
Keywords: actin/chaperonin/electron microscopy/prefoldin/protein folding
Introduction
Many proteins cannot reach their native conformation by themselves within the crowded environment of the cell. To do so, they need the help of one or more components of a large family of proteins termed molecular chaperones (reviewed in Bukau and Horwich, 1998; Bukau et al., 2000; Saibil, 2000; Frydman, 2001; Horwich et al., 2001). These proteins assist in productive folding by preventing the aggregation of an unfolded protein or by providing an appropriate environment in which the protein can reach the native state using the information encoded in its own amino acid sequence. Among chaperones, an essential role is played by the chaperonins, a group of proteins found in every known organism (Ellis, 1996). Chaperonins are toroidal complexes formed by the oligomerization of 60 kDa proteins. These complexes act on a large variety of proteins using a mechanism that involves the recognition of the unfolded polypeptide by hydrophobic residues at the entrance of the chaperonin’s central cavity. In the case of group I chaperonins (i.e. those present in eubacteria and endosymbiotic organelles), the polypeptide is discharged into the cavity as a result of interaction between the chaperonin and a small oligomer termed a co-chaperonin; there it has an opportunity to fold spontaneously. This general mechanism has been well characterized for GroEL, the chaperonin from Escherichia coli, and its co-chaperonin GroES (reviewed in Hartl, 1996; Buckle et al., 1997; Lorimer, 1997; Ranson et al., 1998; Grantcharova et al., 2001). It is generally supposed that a similar mechanism applies in the case of other chaperonins belonging to group I.
There is no homolog of GroES that operates in conjunction with group II chaperonins (i.e. those found in archaeabacteria and in the eukaryotic cytosol). Within this group, the cytosolic chaperonin CCT (chaperonin containing TCP-1; also termed c-cpn or TriC; Frydman et al., 1992; Gao et al., 1992; Lewis et al., 1992) is a toroidal complex formed by two rings each composed of eight different, albeit homologous subunits (reviewed in Willison, 1999, 2001). This chaperonin is essential for the productive folding of actins and tubulins in vitro and in vivo (Gao et al., 1992; Sternlicht et al., 1993; Chen et al., 1994; Vinh and Drubin, 1994). There is also evidence that CCT participates in the folding of Gα transducin (Farr et al., 1997), cyclin E (Won et al., 1998) and the Hippel–Landau tumor suppressor protein VHL (Feldman et al., 1999). The extent to which CCT might contribute to the facilitated folding of other newly synthesized cytosolic proteins (Thulasiraman et al., 1999; Feldman and Frydman, 2000; McCallum et al., 2000) is a matter of controversy (reviewed in Cowan, 2002). Nonetheless, it is likely that the heteromeric nature of CCT has functional significance: for example, recent studies point to the binding of unfolded actin and tubulin to specific CCT subunits (Llorca et al., 1999, 2000).
The folding mechanism of CCT has some features in common with that of GroEL–GroES, but there are also significant differences. Although both chaperonins undergo large conformational changes upon nucleotide binding, CCT lacks the concerted action of a co-chaperonin, although it has an extra domain that seals the cavity and mimics part of the co-chaperonin function (Klumpp et al., 1997). Moreover, CCT does not release its substrates into the cavity; rather, they remain bound to the chaperonin (Llorca et al., 2001a).
CCT does not function in isolation in the facilitated folding of actins and tubulins, but does so in conjunction with another molecular chaperone named prefoldin (PFD; also termed GimC; Geissler et al., 1998; Vainberg et al., 1998). PFD is a heterohexameric protein that exists in archaeabacterial and eukaryotic organisms and, like its partner CCT, has evolved from a simple oligomer in archaea to a more complex assembly in eukarya. In the case of archaeal PFD, the chaperone contains two different proteins (i.e. two α-subunits and four β-subunits). However, in eukaryotes, PFD exists as a more complex oligomer assembled from six different proteins, two of which are α-like and four of which are β-like (Vainberg et al., 1998; Leroux et al., 1999).
Recently, the structure of the archaeal PFD from Methanobacterium thermoautotrophicum has been obtained at atomic resolution (Siegert et al., 2000). The features of this molecule correlate well with its proposed function, i.e. the protection of relatively unfolded molecules pending their transfer to the chaperonin (Vainberg et al., 1998; Hansen et al., 1999). The structure of archaeal PFD resembles a jellyfish in that it is made up of a double β-barrel and six tentacles, each of them consisting of a coiled coil. The tips of the tentacles contain hydrophobic residues that are involved in the binding of unfolded proteins.
Here we show by electron microscopy that, at relatively low resolution, eukaryotic PFD possesses a similar structure to that of archaeal PFD, with the six arms protruding from the base of the oligomer. Three-dimensional reconstruction of the same oligomer complexed with an unfolded actin molecule reveals that the PFD-bound target protein has a defined conformation that crosses the PFD cavity and seems to interact with the tips of the chaperone. We also describe the three-dimensional reconstruction of a symmetric complex formed between CCT and one PFD oligomer bound to each of the CCT rings. This confirms the physical interaction between the two chaperones, and reveals that PFD interacts with two specific subunits in each of the CCT rings, placed in a 1,4 arrangement. The two PFD-binding subunits from one ring interact with the corresponding pair contained in the opposite ring through their equatorial domains, suggesting a model for the phasing of the two rings of CCT.
Results
Three-dimensional structure of eukaryotic PFD
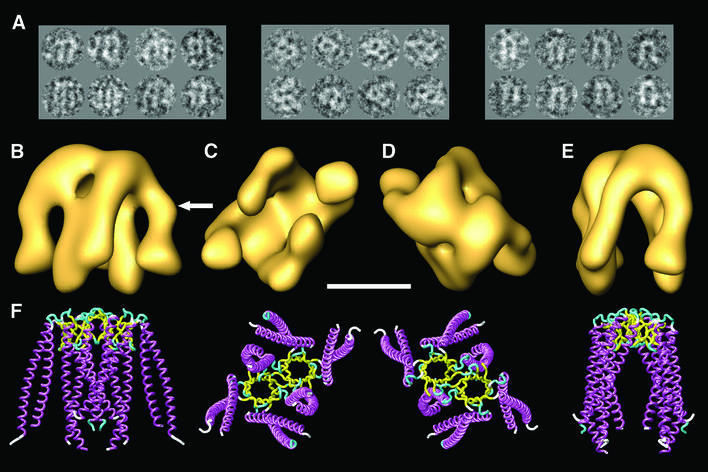
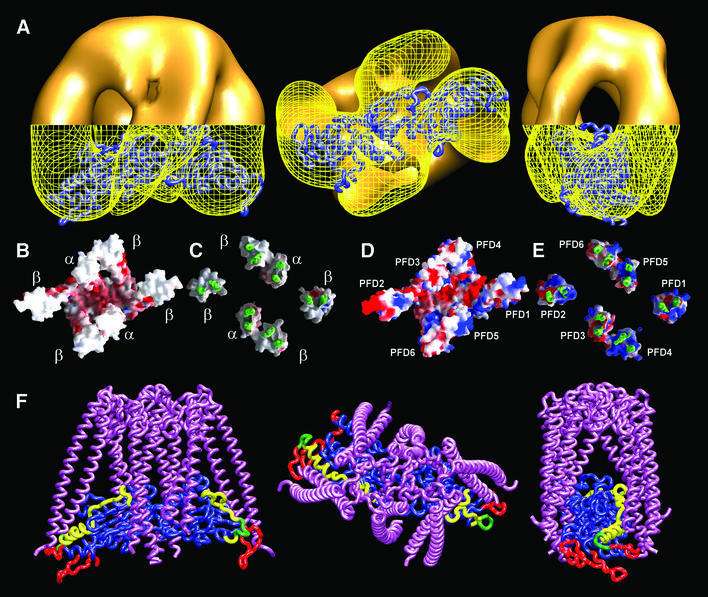
To ascertain the structural features of eukaryotic PFD, we examined its appearance in the electron microscope. Figure 1A shows a gallery of negatively stained particles of eukaryotic PFD. The gallery shows the three preferred orientations, which are orthogonal; this allows a comprehensive coverage of the whole space. No attempt was made to carry out microscopy of unstained specimens because of the very small size of the PFD molecule (∼90 kDa); indeed, the three-dimensional reconstruction was performed using the smallest molecule thus far reconstructed using single particle methodology. The results reveal features that are similar to the archaeal counterpart, although these appear at much lower resolution (compare the views of the three-dimensional reconstruction depicted in Figure 1B–E with the corresponding views of the atomic structure of archaeal PFD shown in Figure 1F). The same jellyfish-like appearance is observed, with the central tentacles protruding farther from the base of the structure than the peripheral ones. There are, however, some minor differences between the two structures, which were reproducible in each of the three independent reconstructions performed, the most prominent being a kink in two of the PFD tentacles (see arrow in Figure 1B). In the archaeal homolog, the two central tentacles belong to the two α-subunits and the four peripheral ones to the β-subunits (Fandrich et al., 2000; Siegert et al., 2000). The contacts observed between two of the peripheral tentacles and the two central ones are a result of the relatively low resolution obtained for the PFD molecule (24 Å). The identical effect is observed in the three-dimensional structure of the archaeal PFD when the resolution is limited to 20 Å (not shown), a higher resolution than that obtained in this reconstruction. The high degree of amino acid sequence similarity among archaeal and eukaryotic subunits (Vainberg et al., 1998; Leroux et al., 1999; Siegert et al., 2000) strongly suggests that the α-like subunits (PFD3/Gim2 and PFD5/Gim5) are located in the center of the structure, while the four β-like subunits (PFD1/Gim6, PFD2/Gim4, PFD4/Gim3 and PFD6/Gim1) are at the periphery. However, the arrangement of the latter has not been determined.
Fig. 1. Three-dimensional reconstruction of human PFD. (A) A gallery of negatively stained images similar to those used for the three-dimensional reconstruction of PFD. The three groups of images shown are archetypes of the three main views, which are orthogonal. (B–E) Several views of the three-dimensional reconstruction of human PFD. (F) Similar views of the atomic structure of the archaeal homolog of PFD (MtGimC from M.thermoautotrophicum; Siegert et al., 2000); PDB accession code 1FXK. The arrow points to a kink in the eukaryotic structure of PFD that is found reproducibly in all the three-dimensional reconstructions generated, and which is not present in the atomic structure of the archaeal PFD. Scale bar = 50 Å.
Three-dimensional structure of the PFD:actin complex
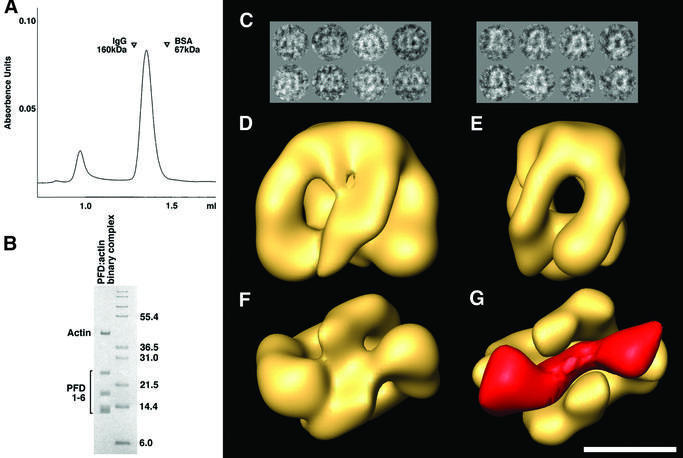
Genetic experiments in yeast have implicated actins and tubulins unambiguously as bona fide substrates for binding to eukaryotic PFD (Geissler et al., 1998; Vainberg et al., 1998). To generate PFD:actin complexes, we presented urea-unfolded actin to purified eukaryotic PFD. The resulting reaction products were isolated by gel filtration. A symmetrical peak emerged from the column with an apparent Mr of ∼120 kDa (Figure 2A). Analysis of this material by SDS–PAGE and inspection of the relative intensity of the Coomassie-stained bands corresponding to PFD and actin suggested that at least 30% of the PFD in this material was complexed with actin (Figure 2B). Similar results were obtained when the same biochemical material was examined by electron microscopy of negatively stained specimens, and the isolated particles subsequently were subjected to image classification methods. From these, 27% of the PFD particles were shown to contain a stain-excluding region at the level of the distal parts of the tentacles (a gallery of such particles is shown in Figure 2C). This region was not present in single particles used for the three-dimensional reconstruction of substrate-free PFD (Figure 1A). We conclude that eukaryotic PFD binds to unfolded or partially folded actin via the distal end of its tentacles. This conclusion was reinforced as a result of three-dimensional reconstruction of the PFD:actin complex by electron microscopy of randomly oriented, negatively stained specimens of the previously classified PFD:actin complexes (Figure 2D–F). The PFD:actin complex reconstructed volume revealed similar structural features to those described for substrate-free eukaryotic PFD, i.e. the same jellyfish-like appearance and the same kink in the two most peripheral tentacles. Such a kink is not present in the atomic structure of the archaeal PFD that was used as an initial model. The main difference consists of an almost cylindrical mass that traverses the PFD cavity of the PFD:actin complex and which interacts with the distal regions of the tentacles (Figure 2G). This mass, clearly representing the bound actin molecule, extends across the PFD molecule; the possible biological relevance of this observation is discussed below. The relatively low percentage of PFD:actin complexes obtained both biochemically and by image classification methods may be a result of dissociation during the isolation procedure, or there may have been a relatively low efficiency of formation of PFD binding-competent conformations formed upon dilution of the actin target protein from denaturant.
Fig. 2. Generation of the PFD:actin complex and its three-dimensional reconstruction. (A) Isolation of PFD:actin complexes by gel filtration on Superdex 200. The locations of molecular mass markers run on the same column are shown. (B) Analysis by SDS–PAGE of material in the peak shown in (A) migrating with an apparent Mr of ∼100 kDa. Molecular mass markers (in kDa) are shown on the right. (C) A gallery of negatively stained images similar to those used for the three-dimensional reconstruction of the PFD:actin complex. The two main views are depicted. (D–F) Front, side and bottom views, respectively, of the three-dimensional reconstruction of the PFD:actin complex. (G) The volume of substrate-free PFD (solid yellow) placed in the same bottom view as the PFD:actin complex depicted in (F). The mass shown in red corresponds to the difference between the PFD:actin complex and substrate-free PFD, and is very suggestive of actin existing in an open conformation, traversing the PFD cavity and interacting with the tips of the tentacles. Scale bar = 50 Å.
Complexes between CCT and PFD
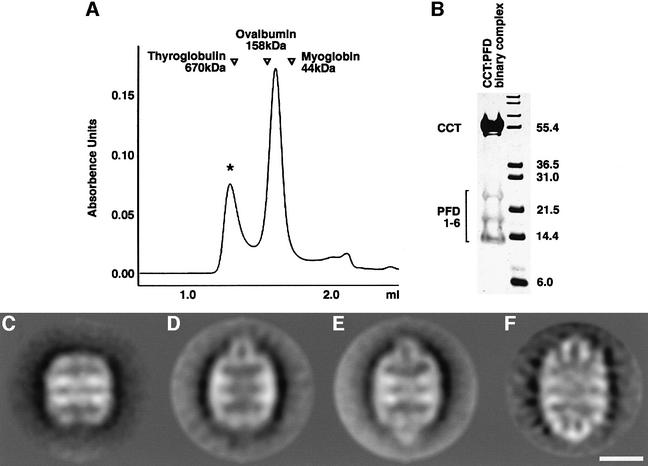
We co-incubated CCT and PFD and resolved the reaction products by gel filtration. Examination by SDS–PAGE of the content of the peak emerging from the column with an apparent Mr of ∼700 kDa (Figure 3A) showed it to contain peptides corresponding to both chaperones (Figure 3B). Upon examination in the electron microscope of the products of the co-incubation reaction, ∼30% of the CCT particles appeared with one or both of the rings capped by a small structure. These complexes never appeared when CCT preparations were visualized in the absence of PFD. Although the proportion of CCT:PFD complexes generated was quite low, we found that this could be increased by including a molecular crowding agent such as dextran-10 or dextran-60 (data not shown), presumably reflecting a relatively weak association between the two chaperones.
Fig. 3. Two-dimensional averages of CCT:PFD complexes. (A) Gel filtration on Superose 6 of the products resulting from co-incubation of CCT and PFD. The location of molecular size markers run on the same column under identical conditions is shown. (B) Analysis by SDS–PAGE of the products contained in the peak [marked with an asterisk in (A)] emerging from the column with an apparent Mr of ∼700 kDa. Note the presence of polypeptides characteristic of both CCT (in the size range around 55 kDa) and PFD (in the size range 14–25 kDa). (C) Average image obtained from negatively stained CCT particles in the absence of PFD (average image obtained from 350 particles; 27 Å resolution). (D) Average image obtained from negatively stained particles of CCT:PFD asymmetric complexes and (E) symmetric complexes (average images of 561 and 1128 particles; 27 and 24 Å resolution, respectively) (F) Average image of the CCT:PFD symmetric complex obtained from frozen-hydrated specimens (715 particles; 25 Å resolution). Scale bar = 100 Å.
To determine the shape of these complexes clearly, side views of negatively stained CCT particles were classified and homogeneous populations were averaged. As a result, together with the characteristic barrel shape of uncomplexed CCT particles (Figure 3C), two other CCT structures were observed in which an ‘inverted U-shape’ structure was seen to be interacting with the apical domain of one (Figure 3D) or both (Figure 3E and F) CCT rings. These three different classes of CCT particle have a size and shape that we ascribe to PFD-free CCT, asymmetric and symmetric CCT:PFD complexes, respectively, in which the PFD oligomer binds to CCT via the distal regions of its tentacles.
Three-dimensional structure of the CCT:PFD complex
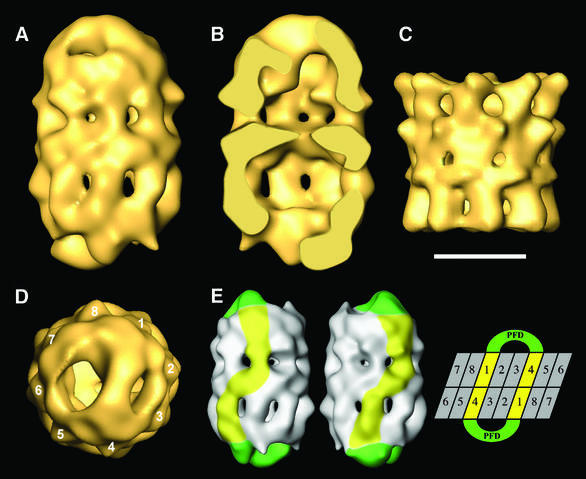
To study the interaction between CCT and PFD in more detail, a three-dimensional reconstruction of the CCT:PFD complex was carried out using unstained, frozen-hydrated preparations. After image classification and removal of the few CCT:PFD asymmetric particles and other particles with low correlation coefficients (see Materials and methods), a homogeneous population of 1385 CCT:PFD symmetric particles was obtained and used to generate a three-dimensional reconstruction of the CCT:PFD symmetric complex (Figure 4A and B). Compared with the three-dimensional reconstruction of apo-CCT (Llorca et al., 2000) (Figure 4C), the CCT:PFD complex shows some shared structural features such as the handedness of the particle and the ‘windows’ present at the level of the intermediate domain of each of the CCT monomers (Figure 4A and B). More important, however, are the structural differences, principal among which is a mass protruding from the apical domains of the two chaperonin rings. The most peripheral part of the protruding masses is rectangular in shape (Figure 4D), and is similar to the double β-barrel platform that links all six PFD subunits in the archaeal chaperone (Siegert et al., 2000). The peripheral tentacles interact with the CCT apical domains, and the central tentacles hang from the platform (see top ring in Figure 4B). A second difference lies in the upward movement of the apical domains of the chaperonin, induced by the interaction with PFD, also observed in the two-dimensional average images of the asymmetric and symmetric CCT:PFD complexes (Figure 3D–F), and which is much more evident here. Although the level of resolution obtained for the bound PFD molecules is low (30 Å), the volume reconstructed has important implications regarding the interaction between the two chaperones. Inspection of the way PFD interacts with CCT reveals that this occurs through two CCT subunits placed in a 1,4 arrangement (Figure 4D), in the same manner that unfolded actin has been observed to interact with CCT (Llorca et al., 1999, 2000). Only a single population of CCT:PFD symmetric complexes was obtained during the classification procedures leading to the three-dimensional reconstruction of the CCT:PFD complex. This implies that the interaction observed is unique, i.e. specific subunits of PFD interact with specific subunits of CCT. Indeed, careful observation of the CCT:PFD symmetric complex revealed that the two CCT subunits that interact with PFD in each ring are in contact with the other two PFD-interacting subunits from the other ring through their equatorial domains (see Figure 4E). This defines the phasing with respect to the subunits in the two rings of CCT (see diagram in Figure 4E).
Fig. 4. Three-dimensional reconstruction of the CCT:PFD symmetric complex. (A) Side view of the CCT:PFD symmetric complex. (B) A section along the longitudinal axis of the same volume. (C) Side view of the three-dimensional reconstruction of apo-CCT (Llorca et al, 2000). (D) Top view of the three-dimensional reconstruction showing the 1,4 interaction between PFD and the CCT subunits. (E) A scheme showing the interaction of the two PFD oligomers with the CCT subunits of the two rings. The two volumes on the left represent two opposing views of the CCT:PFD complex. The PFD oligomers are highlighted in green and the CCT subunits interacting with PFD are shown in yellow. The topology of the CCT subunits depicted on the right is for illustrative purposes only, and the numbers have the sole purpose of discriminating among the subunits. Scale bar = 100 Å in (A–D).
Discussion
Interaction between PFD and unfolded actin
PFD is a molecular chaperone that interacts with certain non-native proteins and protects them from aggregation pending their transfer to chaperonin for the final step(s) in their folding (Geissler et al., 1998; Vainberg et al., 1998). PFD can therefore legitimately be considered as a co-chaperonin, although its role seems to be quite different from that played by classical co-chaperonins such as GroES, which acts by capping the GroEL cylinder.
It has been shown that hetero-complexes formed by co-assembly of subunits of archaeal and yeast origin can partially complement yeast strains lacking some β-like subunits (Gim1; Leroux et al., 1999). This in itself suggests that all PFD complexes share a similar three-dimensional structure. Indeed, the three-dimensional reconstruction of human PFD generated by electron microscopy uncovers similar basic structural features to those present in its archaeal counterpart (Figure 1B–E), i.e. a platform from which six tentacles hang. The shape of the molecule and the role of this chaperone in preventing the aggregation of nascent proteins strongly suggest that the tentacles of the oligomer serve to embrace and thus protect the unfolded substrate from unwanted interactions. Indeed, deletion experiments show the tips of the tentacles to be required for substrate binding (Siegert et al., 2000). Our three-dimensional reconstruction of the complex between human PFD and denatured actin is consistent with these results, with the unfolded actin located along the interior of the PFD cavity, bound to the tips of the tentacles (Figure 2D–F). The PFD-bound, unfolded actin has a rod-like shape (Figure 2G) very suggestive of the structure that is thought to be recognized by and bound to CCT (Llorca et al., 1999, 2000). Docking of this molecule into the volume of the actin complexed to PFD (Figure 5A) is very good, and strengthens the notion that unfolded actin may have acquired this quasi-native conformation before interacting with CCT. This conclusion is consistent with data showing that denatured actin is capable of spontaneously acquiring a large portion of its native tertiary structure (Schüler et al., 2000).
Fig. 5. Model of the interaction between denatured actin and PFD. (A) Several views of the three-dimensional reconstruction of the PFD:actin complex in which the atomic model of unfolded actin generated upon its docking into the three-dimensional reconstruction of actin bound to nucleotide-free CCT (Llorca et al., 1999, 2000) has been fitted into the actin part of the PFD:actin complex. The envelope of the tips of the PFD:actin complex has been drawn as a yellow grid to allow visualization of the atomic model of unfolded actin. (B and C) Bottom and top views, respectively, of the tips of the tentacles of the atomic structure of the archaeal PFD from M.thermoautotrophicum (Siegert et al., 2000). (D and E) The same views of the structural model generated with human PFD subunits. In these images, the α and β archaeal subunits were used as templates, and the human α- and β-like subunits were placed in the center and outer positions, respectively. The β-like subunits are placed randomly. Human PFD was modeled using the homology modeling programs Swiss-PdbViewer and Swiss-Model (Guex and Peitsch, 1997; Schwede et al., 2000). In (C) and (E), the base of the structure and part of the tentacles have been removed to allow visualization of the outer surface of the tentacle tips. Surfaces (negatively charged residues in red, positively charged residues in blue) were generated using GRASP (Nicholls et al., 1991). (F) Several views of the modeled interaction between unfolded actin and PFD. The atomic structure of the archaeal PFD and the atomic model of unfolded actin shown in (A) were used as structures for the modeling of this interaction. Actin residues putatively involved in the interaction of PFD with CCT are highlighted as follows: residues involved in the interaction with CCT (Llorca et al., 2000, 2001b) (red); residues involved in PFD binding (Rommelaere et al., 2001) (yellow); residues involved in both functions (green).
Given the structural data presented here, we conclude that the interaction between unfolded actin and PFD must be of a specific nature and must be governed by contacts between particular domains in both the chaperone and its substrate. Compared with the paucity of positive and negatively charged amino acids in the interior of the tentacle tips of archaeal PFD (Figure 5B), the same regions of the modeled eukaryotic PFD are covered by charged residues (Figure 5D). Using truncation and mutagenesis experiments, Rommelaere et al. (2001) have identified two regions in actin and tubulin that are required for their interaction with PFD. Interestingly, in the atomic model of quasi-folded actin bound to CCT (Llorca et al., 2000), these two actin PFD-binding regions [site I (residues 60–79) and site II (residues 170–198)] are located on the surface and face the PFD tentacles (yellow and green regions in Figure 5F). These observations reinforce the idea that unfolded actin interacts with PFD in a quasi-folded conformation that requires the protection of the chaperone. PFD has been shown to bind unfolded actin after the synthesis of the first ∼145 amino acids (Hansen et al., 1999). Importantly, these 145 residues form the N-terminal domain of actin (Kabsch et al., 1990). Similarly, PFD interacts with unfolded β-tubulin after the synthesis of the first ∼250 amino acids, slightly longer than the region that constitutes its N-terminal domain (Nogales et al., 1998). In both actin and tubulin, the two fragments that are necessary for PFD interaction encompass the strongest PFD-binding domains described by Rommelaere et al. (2001). Taken together, these results suggest that the nascent ribosome-associated actin chain binds to PFD via PFD-binding site I, once the N-terminal domain has folded (or at least attained a quasi-folded conformation). This PFD:actin complex persists until completion of actin synthesis, followed by folding of the second, C-terminal domain, which binds to PFD via PFD-binding site II. The complete actin molecule bound to PFD may thus attain a conformation in which the two domains are folded or almost folded (Figure 5F), a conformation that is well suited for its transfer to CCT (see below).
Interaction between PFD and CCT
It has been shown previously that productive actin folding is accelerated at least 5-fold in the presence of PFD (Siegers et al., 1999). In this reaction, PFD transfers the unfolded substrate to CCT through a mechanism that does not involve the release of the substrate into solution, but occurs via a physical interaction between PFD and CCT (Vainberg et al., 1998). The interaction between PFD and CCT is probably transient in vivo. Here we used gel filtration and electron microscopy to identify and visualize complexes formed between eukaryotic PFD and CCT (Figures 3 and 4).
Two types of CCT:PFD complexes were identified, one asymmetric with a PFD oligomer bound to one of the CCT rings, and a more preponderant symmetric complex with a PFD oligomer bound to each of the two CCT rings. This is surprising, in that the existence of CCT:PFD symmetric complexes contrasts with previous data in which actin or tubulin was found bound to only one of the CCT rings, regardless of the amount of unfolded substrate presented to the chaperonin (Llorca et al., 1999, 2000, 2001a). It is possible that in the presence of target protein, PFD interacts with CCT in such a way as to give rise to an inter-ring signal that prevents a second substrate molecule from binding to the opposite CCT ring. Alternatively, it is possible that only in the presence of PFD, their natural carrier, do actin or tubulin molecules interact with both rings of CCT. Examination of stable CCT:PFD:actin complexes would distinguish between these possibilities. Thus far, however, all attempts to form such ternary complexes have been unsuccessful, probably because of their very transient existence.
The two-dimensional average images of the CCT:PFD complexes clearly showed that the interaction between the two chaperones occurs at the level of the outer regions of the PFD tentacles and the inner surface of the apical domains of the chaperonin (Figure 3). The interaction with each of the CCT rings differs in orientation with respect to the longitudinal axis of the chaperonin (see Figure 3F). This observation was confirmed in the three-dimensional reconstruction of the CCT:PFD symmetric complexes generated by cryoelectron microscopy of frozen-hydrated specimens (Figure 4). The volume generated revealed, besides the two PFD oligomers bound to the apical domains of each CCT ring, an upward movement of the apical domains of the chaperonin that is induced by the binding of the chaperone.
Three-dimensional reconstruction of the CCT:PFD complex also showed that the PFD oligomers interact in each ring with two CCT subunits placed in a 1,4 arrangement. This mode of interaction is identical to that found in complexes between CCT and unfolded actin. In the latter case, the interaction is not only geometry dependent but also subunit specific, with the unfolded actin binding through the tip of the small, N-terminal domain to the CCTδ subunit and through the tip of the large, C-terminal domain to either the CCTβ or CCTε subunit (Llorca et al., 1999). Because each of the six PFD subunits is different, it is not difficult to envisage an interaction between CCT and PFD that is also subunit specific. This notion is reinforced by the presence of a large number of charged residues in the exterior of the tentacle tips of eukaryotic PFD (Figure 5E) that are absent in the archaeal counterpart (Figure 5C). Two other observations support this idea further. First, the three-dimensional reconstruction of the CCT:PFD complex corresponds to the only population of symmetric CCT:PFD complexes detected during the classification procedure, which clearly suggests that the way in which the two PFD oligomers interact with CCT is unique. Since it has been established that the subunits within CCT are arranged with a defined geometry (Liou and Willison, 1997), it follows that interactions between the PFD oligomer and the two rings of the chaperonin occur through the same two CCT subunits. Secondly, the two PFD-interacting CCT subunits from a given ring are in contact with the corresponding pair in the opposite ring through their equatorial domains (as described in Figure 4E). Thus, one of the two CCT subunits involved in PFD binding (which we arbitrarily define as CCT1) interacts indirectly with the other PFD-binding subunit within the same CCT ring (i.e. CCT4) via PFD itself, and directly with CCT4 from the other ring through their equatorial domains. These kinds of interaction could generate intra- and/or inter-ring signaling and thereby contribute to the functional cycle of CCT, in the same way that binding of GroES to a given GroEL ring generates a set of signals that influence the opposite GroEL ring (Rye et al., 1997). This analysis also leads to a model for the phasing of the subunits in the two rings of CCT (diagram in Figure 4E).
A model for the interaction between actin, PFD and CCT
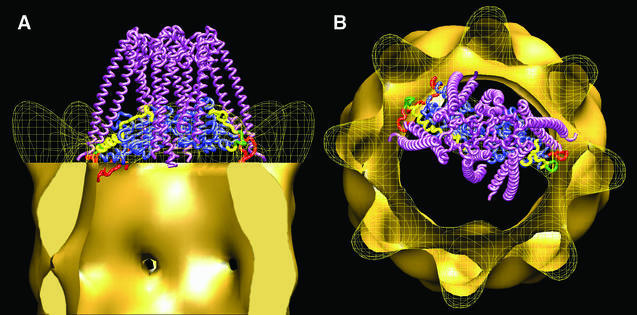
A model depicting the interaction between denatured actin, PFD and CCT is presented in Figure 6. This model, which uses the atomic model of open actin docked into the three-dimensional reconstruction of the CCT:actin complex (Llorca et al., 2000), and the atomic structure of archaeal PFD (Siegert et al., 2000) placed in such a way that it fits the density mass of the PFD oligomer within the three-dimensional reconstruction of the CCT:PFD complexes, is completely consistent with the two-dimensional average images and the three-dimensional reconstruction of the CCT:PFD complexes shown in Figures 3 and 4. According to this model, the PFD:actin complex interacts with specific CCT subunits and presents the CCT-binding regions of partially unfolded actin to the apical domains of specific CCT subunits (red and green regions in Figure 6). It is tempting to speculate that the CCT subunits involved in the binding of PFD and partially unfolded actin are the same. If this is the case, the PFD:actin complex would interact with CCTδ and CCTβ or with CCTδ and CCTε. As stated above, CCT and PFD interact in a unique way, so that only one of the two possible modes of interaction described for actin is possible in this case. Taken together, the data presented here suggest a mechanism whereby PFD recognizes and binds actin in an open, quasi-folded conformation via the tips of its tentacles. The PFD:actin complex thus formed binds to CCT, and PFD is then displaced by the CCT binding of the actin it is carrying. CCT then stabilizes this intermediate actin conformation, and folding proceeds to completion following conformational changes promoted by the cytosolic chaperonin upon nucleotide binding.
Fig. 6. Model of the interaction between unfolded actin, PFD and CCT. (A and B) Side view and top views, respectively, of the proposed ternary interaction between actin, PFD and CCT. The PFD:actin complex shown in Figure 5 interacts with specific CCT subunits through the tips of some of the PFD subunits and presents the actin molecule to the CCT subunits that interact with this molecule (Llorca et al., 1999). The actin molecule has been fitted into the actin part of the three-dimensional reconstruction of the CCT:actin complex used to generate the model (Llorca et al., 2000). The color code of the actin molecule is as in Figure 5.
Materials and methods
Purification of CCT and eukaryotic PFD
CCT was purified from soluble extracts of bovine testis (Gao et al., 1994) by a modification of the procedure of Norcum (1996). Briefly, an ammonium sulfate cut (30–50%) was prepared and redissolved in buffer [50 mM HEPES pH 7.2, 5 mM MgAc, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF)]. This material was centrifuged over sucrose cushions containing 1.0 M sucrose, and proteins migrating into this sucrose layer were purified further by cation exchange chromatography on a column of heparin– agarose as described (Norcum, 1996). Fractions emerging from this column were identified by SDS–PAGE as containing CCT by the characteristic appearance of a cluster of bands in the 55–62 kDa range. These fractions were pooled and purified further first by affinity selection on a column of agarose-bound ATP and then by gel filtration on a Superose 6 column as described (Gao et al., 1992). Material thus obtained was stripped of bound target protein by incubation for 1 h at 30°C in folding buffer (Tian et al., 1995) containing 1 mM ATP and a 5- to 10-fold molar excess of purified Hsp60SR1 (Nielsen and Cowan, 1998). This mutant form of the mitochondrial chaperonin was included as a trap for the capture of cycling target proteins (Weissman et al., 1995; Farr et al., 1997). CCT was re-isolated from this reaction mixture by cation exchange chromatography on a column of agarose-bound heparin. This material was judged by Coomassie Blue staining of SDS gels to be minimally 98% pure, and (in contrast to CCT that had not been thus treated) contained barely detectable amounts of actin or tubulin target proteins as judged by western blotting with anti-actin or anti-tubulin monoclonal antisera (data not shown).
Bovine PFD was purified from the same ammonium sulfate cut prepared from a soluble bovine testis tissue extract as described above, using the chromatographic dimensions described (Vainberg et al., 1998). Human PFD was assembled from individual subunits of PFD produced individually as soluble recombinant proteins in E.coli (C.T.Simons, S.A.Lewis, F.Bartolini and N.J.Cowan, unpublished data). This human recombinant PFD behaved like purified bovine PFD in all in vitro binding and folding assays.
Formation of PFD:actin and CCT:PFD complexes
PFD:actin binary complexes were formed by rapid dilution from 8 M urea of denatured, purified β-actin into 60 µl of folding buffer (Tian et al., 1995) containing purified bovine PFD (1.1 mg/ml) and incubating for 1 h on ice. The final concentration of actin in these reactions was 10 µM. The reaction mixture was applied to a 2 ml column of Superdex 200 run in 20 mM Tris–HCl pH 7.3, 0.15 M NaCl, 1 mM EGTA, 1 mM DTT, using an Amersham Pharmacia Smart system. PFD:actin complexes made in this way yield native actin when incubated with CCT and ATP (Vainberg et al., 1998).
CCT:PFD complexes were generated by co-incubating purified bovine CCT and recombinant human PFD at 30°C at a molar ratio of 1:10 for 10 min. This material was applied to a 2 ml Superose 6 gel filtration column run in 20 mM MES pH 6.8, 20 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT. Fractions emerging from the column were analyzed by SDS–PAGE.
Electron microscopy
For cryoelectron microscopy, 5 µl aliquots of a solution containing CCT:PFD complexes were applied to glow-discharged carbon grids for 1 min, blotted for 5 s and frozen rapidly in liquid ethane at –180°C. Images were recorded at 0°-tilt under minimum dose conditions in a JEOL 1200EX-II electron microscope equipped with a Gatan cold stage operated at 120 kV and recorded on Kodak SO-163 film at 60 000× nominal magnification and ∼2.0 µm underfocus. For electron microscopy of negatively stained samples, aliquots were applied to glow-discharged carbon grids for 1 min and then stained for 1 min with 2% uranyl acetate. Images were recorded at 20°-tilt, in the same way as for cryoelectron microscopy experiments.
Image processing, two-dimensional averaging and three-dimensional reconstruction
Micrographs were digitized in a Zeiss SCAI scanner with a sampling window corresponding to 2.4 Å/pixel for PFD and the PFD:actin complex, and 3.5 Å/px for the CCT:PFD complexes. For two-dimensional classification and averaging, images were selected, aligned using a free-pattern algorithm (Penczek et al., 1992) and classified using a self-organizing map algorithm as described by Marabini et al. (1998). The resolution of the average images was estimated by the spectral signal to noise ratio (SSNR) method (Unser et al., 1987) and average images subsequently were filtered to the resolution obtained.
Three-dimensional reconstructions of PFD and PFD:actin complexes were generated respectively from 971 and 5026 negatively stained, randomly oriented particles. The orientations of particles were determined using the angular refinement algorithms provided by SPIDER (Frank, 1996). As a first model for the iterative procedure, the atomic structure of the archaeal PFD from M.thermoautotrophicum (Siegert et al., 2000) with the resolution limited at 35 Å was used to determine the rough orientation of the particles. All subsequent volumes were generated with the eukaryotic particles using ART (algebraic reconstruction techniques) with blobs (Marabini et al., 1998). No symmetry was applied to any of the volumes generated during the iterative procedure. In the case of the PFD:actin complexes, the following strategy was used to separate the actin-containing PFD particles from the substrate-free ones. First, all the particles were refined against the atomic structure of the archaeal PFD from M.thermoautotrophicum (Siegert et al., 2000) with the resolution limited at 35 Å. Then the particles were classified, attending to the angles provided by the refinement procedure. In this way, two main populations (accounting for 90% of the total number of particles) were obtained: those having an ‘M’ shape (70% of the total number of particles) and those having a ‘U’ shape (20% of the total number of particles) (see the two galleries of Figure 2C). These two populations were subjected independently to a second classification procedure, using neuronal networks (Marabini and Carazo, 1994), to separate the actin-free particles from the PFD:actin complexes. This procedure yielded 1379 PFD:actin particles classified as such (27% of the total number of particles, a value very similar to that obtained in biochemical experiments) that were selected to perform the refinement using the standard iterative procedure. In this case, and although the volume used in the first iteration was the same as that utilized for the reconstruction of substrate-free PFD, the volume generated showed an additional mass crossing the PFD cavity. This clearly indicates that the procedure followed does not introduce a bias towards the first volume used. The volume was refined several times until no further improvement was obtained. In the three-dimensional reconstruction of both PFD and PFD:actin complexes, some particles were discarded using the correlation coefficients of each particle with the model. By performing this process iteratively, final reconstructions were made of homogeneous populations. The final resolution was estimated with the 0.5 criterion for the Fourier shell correlation coefficient between two independent reconstructions using the ‘Bsoft’ software package (Heymann, 2001). The values obtained, 24 Å for PFD and 26 Å for the PFD:actin complex, were used to low-pass filter the volumes.
Three-dimensional reconstruction of the CCT:PFD symmetric complex was carried out from 1625 side views whose orientations were determined using angular refinement algorithms provided by SPIDER. As a preliminary three-dimensional reference model for this procedure, a volume was generated by applying 8-fold symmetry to the frozen-hydrated average image of the side view of the CCT:PFD symmetric complex. This initial refinement yielded a first volume that is asymmetric (due to the noisy nature of the cryoimages), and this asymmetric volume was then used as the first model for the iterative refinement procedure. No symmetrization was applied to any of the volumes obtained during the iterative procedure or to the final volume shown in this work. Volumes were again generated using ART with blobs. The resolution obtained (30 Å) was used to low-pass filter the volume. Visualization of the volumes was carried out using AMIRA (http://amira.zeb.de).
Modeling of PFD:actin and CCT:PFD:actin complexes
To model the PFD:actin complex, the atomic structure of actin (Kabsch et al., 1990) opened to fit the three-dimensional reconstruction of actin bound to nucleotide-free CCT (Llorca et al., 2000) was docked visually using ‘O’ software (Jones et al., 1991) into the actin part of the three-dimensional reconstruction of the PFD:actin complex. In the case of the CCT:PFD:actin complex, the atomic structure of PFD from the archaea M.thermoautotrophicum (Siegert et al., 2000) was docked visually into the three-dimensional reconstruction of the CCT:actin complex. Visualization of the structures was carried out using VMD (Humphrey et al., 1996).
Acknowledgments
Acknowledgements
We thank Dr David L.Stokes for advice and helpful discussions. This work was partially supported by grants from the CICYT, FEDER, CAM (J.M.V.), the NIH (N.J.C.) and DGICYT (J.L.C.). J.B. is a postdoctoral fellow from the Comunidad Autónoma de Madrid (CAM).
References
- Buckle A.M., Zahn,R. and Fersht,A.R. (1997) A structural model for GroEL-polypeptide recognition. Proc. Natl Acad. Sci. USA, 94, 3571–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B. and Horwich,A.L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell, 92, 351–380. [DOI] [PubMed] [Google Scholar]
- Bukau B., Deuerling,E., Pfund,C. and Craig,E.A. (2000) Getting newly synthesized proteins into shape. Cell, 101, 119–122. [DOI] [PubMed] [Google Scholar]
- Chen X., Sullivan,D.S. and Huffaker,T.C. (1994) Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc. Natl Acad. Sci. USA, 91, 9111–9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan N.J.and.Lewis.,S.A. (2002) Type II chaperonins, prefoldin and the tubulin-specific chaperones. Adv. Protein Chem., 59, 73–104. [DOI] [PubMed] [Google Scholar]
- Ellis R.J. (1996) The Chaperonins. Academic Press, San Diego, CA.
- Fandrich M., Tito,M.A., Leroux,M.R., Rostom,A.A., Hartl,F.U., Dobson,C.M. and Robinson,C.V. (2000) Observation of the noncovalent assembly and disassembly pathways of the chaperone complex MtGimC by mass spectrometry. Proc. Natl Acad. Sci. USA, 97, 14151–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr G.W., Scharl,E.C., Schumacher,R.J., Sondek,S. and Horwich,A.L. (1997) Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell, 89, 927–937. [DOI] [PubMed] [Google Scholar]
- Feldman D.E. and Frydman,J. (2000) Protein folding in vivo: the importance of molecular chaperones. Curr. Opin. Struct. Biol., 10, 26–33. [DOI] [PubMed] [Google Scholar]
- Feldman D.E., Thulasiraman,V., Ferreyra,R.G. and Frydman,J. (1999) Formation of the VHL–elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol. Cell, 4, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Frank J. (1996) Three-dimensional Electron Microscopy of Macro molecular Assemblies. Academic Press, San Diego, CA, pp. 182–246.
- Frydman J. (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem., 70, 603–647. [DOI] [PubMed] [Google Scholar]
- Frydman J., Nimmesgern,E., Erdjument-Bromage,H., Wall,J.S., Tempst,P. and Hartl,F.U. (1992) Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J., 11, 4767–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Thomas,J.O., Chow,R.L., Lee,G.H. and Cowan,N.J. (1992) A cytoplasmic chaperonin that catalyzes β-actin folding. Cell, 69, 1043–1050. [DOI] [PubMed] [Google Scholar]
- Gao Y., Melki,R., Walden,P.D., Lewis,S.A., Ampe,C., Rommelaere,H., Vandekerckhove,J. and Cowan,N.J. (1994) A novel cochaperonin that modulates the ATPase activity of cytoplasmic chaperonin. J. Cell Biol., 125, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S., Siegers,K. and Schiebel,E. (1998) A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J., 17, 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantcharova V., Alm,E.J., Baker,D. and Horwich,A.L. (2001) Mechanisms of protein folding. Curr. Opin. Struct. Biol., 11, 70–82. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hansen W.J., Cowan,N.J. and Welch,W.J. (1999) Prefoldin–nascent chain complexes in the folding of cytoskeletal proteins. J. Cell Biol., 145, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U. (1996) Molecular chaperones in cellular protein folding. Nature, 381, 571–579. [DOI] [PubMed] [Google Scholar]
- Heymann J.B. (2001) Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol., 133, 156–169. [DOI] [PubMed] [Google Scholar]
- Horwich A.L., Fenton,W.A. and Rapoport,T.A. (2001) Protein folding taking shape: workshop on molecular chaperones. EMBO Rep., 2, 1068–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke,A. and Schulten,K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz,H.G., Suck,D., Pai,E.F. and Holmes,K.C. (1990) Atomic structure of the actin:DNase I complex. Nature, 347, 37–44. [DOI] [PubMed] [Google Scholar]
- Klumpp M., Baumeister,W. and Essen,L.O. (1997) Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin. Cell, 91, 263–270. [DOI] [PubMed] [Google Scholar]
- Leroux M.R., Fandrich,M., Klunker,D., Siegers,K., Lupas,A.N., Brown,J.R., Schiebel,E., Dobson,C.M. and Hartl,F.U. (1999) MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J., 18, 6730–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis V.A., Hynes,G.M., Zheng,D., Saibil,H. and Willison,K. (1992) T-complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol. Nature, 358, 249–252. [DOI] [PubMed] [Google Scholar]
- Liou A.K. and Willison,K.R. (1997) Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J., 16, 4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O., McCormack,E.A., Hynes,G., Grantham,J., Cordell,J., Carrascosa,J.L., Willison,K.R., Fernandez,J.J. and Valpuesta,J.M. (1999) Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature, 402, 693–696. [DOI] [PubMed] [Google Scholar]
- Llorca O., Martin-Benito,J., Ritco-Vonsovici,M., Grantham,J., Hynes,G.M., Willison,K.R., Carrascosa,J.L. and Valpuesta,J.M. (2000) Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J., 19, 5971–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O., Martin-Benito,J., Gomez-Puertas,P., Ritco-Vonsovici,M., Willison,K.R., Carrascosa,J.L. and Valpuesta,J.M. (2001a) Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J. Struct. Biol., 135, 205–218. [DOI] [PubMed] [Google Scholar]
- Llorca O., Martin-Benito,J., Grantham,J., Ritco-Vonsovici,M., Willison,K.R., Carrascosa,J.L. and Valpuesta,J.M. (2001b) The ‘sequential allosteric ring’ mechanism in the eukaryotic chaperonin-assisted folding of actin and tubulin. EMBO J., 20, 4065–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer G. (1997) Protein folding. Folding with a two-stroke motor. Nature, 388, 720–721, 723. [DOI] [PubMed] [Google Scholar]
- Marabini R. and Carazo,J.M. (1994) Pattern recognition and classification of images of biological macromolecules using artificial neural networks. Biophys. J., 66, 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini R., Herman,G.T. and Carazo,J.M. (1998) 3D reconstruction in electron microscopy using ART with smooth spherically symmetric volume elements (blobs). Ultramicroscopy, 72, 53–65. [DOI] [PubMed] [Google Scholar]
- McCallum C.D., Do,H., Johnson,A.E. and Frydman,J. (2000) The interaction of the chaperonin tailless complex polypeptide 1 (TCP1) ring complex (TRiC) with ribosome-bound nascent chains examined using photo-cross-linking. J. Cell Biol., 149, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nielsen K.L. and Cowan,N.J. (1998) A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol. Cell, 2, 93–99. [DOI] [PubMed] [Google Scholar]
- Nogales E., Wolf,S.G. and Downing,K.H. (1998) Structure of the αβ tubulin dimer by electron crystallography [published erratum appears in Nature, 1998, 393, 191]. Nature, 391, 199–203. [DOI] [PubMed] [Google Scholar]
- Norcum M.T. (1996) Novel isolation method and structural stability of a eukaryotic chaperonin: the TCP-1 ring complex from rabbit reticulocytes. Protein Sci., 5, 1366–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P., Radermacher,M. and Frank,J. (1992) Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy, 40, 33–53. [PubMed] [Google Scholar]
- Ranson N.A., White,H.E. and Saibil,H.R. (1998) Chaperonins. Biochem. J., 333, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere H., De Neve,M., Neirynck,K., Peelaers,D., Waterschoot,D., Goethals,M., Fraeyman,N., Vandekerckhove,J. and Ampe,C. (2001) Prefoldin recognition motifs in the nonhomologous proteins of the actin and tubulin families. J. Biol. Chem., 276, 41023–41028. [DOI] [PubMed] [Google Scholar]
- Rye H.S., Burston,S.G., Fenton,W.A., Beechem,J.M., Xu,Z., Sigler,P.B. and Horwich,A.L. (1997) Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature, 388, 792–798. [DOI] [PubMed] [Google Scholar]
- Saibil H. (2000) Molecular chaperones: containers and surfaces for folding, stabilising or unfolding proteins. Curr. Opin. Struct. Biol., 10, 251–258. [DOI] [PubMed] [Google Scholar]
- Schüler H., Lindberg,U., Schutt,C.E. and Karlsson,R. (2000) Thermal unfolding of G-actin monitored with the DNase I-inhibition assay. Eur. J. Biochem., 267, 476–486. [DOI] [PubMed] [Google Scholar]
- Schwede T., Diemand,A., Guex,N. and Peitsch,M.C. (2000) Protein structure computing in the genomic era. Res. Microbiol., 151, 107–112. [DOI] [PubMed] [Google Scholar]
- Siegers K., Waldmann,T., Leroux,M.R., Grein,K., Shevchenko,A., Schiebel,E. and Hartl,F.U. (1999) Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin–GimC system. EMBO J., 18, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R., Leroux,M.R., Scheufler,C., Hartl,F.U. and Moarefi,I. (2000) Structure of the molecular chaperone prefoldin. Unique interaction of multiple coiled coil tentacles with unfolded proteins Cell, 103, 621–632. [DOI] [PubMed] [Google Scholar]
- Sternlicht H., Farr,G.W., Sternlicht,M.L., Driscoll,J.K., Willison,K. and Yaffe,M.B. (1993) The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc. Natl Acad. Sci. USA, 90, 9422–9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasiraman V., Yang,C.F. and Frydman,J. (1999) In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J., 18, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Vainberg,I.E., Tap,W.D., Lewis,S.A. and Cowan,N.J. (1995) Specificity in chaperonin-mediated protein folding. Nature, 375, 250–253. [DOI] [PubMed] [Google Scholar]
- Unser M., Trus,B.L. and Steven,A.C. (1987) A new resolution criterion based on spectral signal-to-noise ratios. Ultramicroscopy, 23, 39–51. [DOI] [PubMed] [Google Scholar]
- Vainberg I.E., Lewis,S.A., Rommelaere,H., Ampe,C., Vandekerckhove, J., Klein,H.L. and Cowan,N.J. (1998) Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell, 93, 863–873. [DOI] [PubMed] [Google Scholar]
- Vinh D.B. and Drubin,D.G. (1994) A yeast TCP-1-like protein is required for actin function in vivo. Proc. Natl Acad. Sci. USA, 91, 9116–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman J.S., Hohl,C.M., Kovalenko,O., Kashi,Y., Chen,S., Braig,K., Saibil,H.R., Fenton,W.A. and Horwich,A.L. (1995) Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell, 83, 577–587. [DOI] [PubMed] [Google Scholar]
- Willison K.R. (1999) Composition and function of the eukaryotic cytosolic chaperonin containing TCP1. In Bukau,B. (ed.), Molecular Chaperones and Folding Catalysts. Harwood Academic Publishers, Amsterdam, The Netherlands, pp. 555–571.
- Willison K.R. (2001) The roles of cytosolic chaperonin, CCT, in normal eukaryotic cell growth. In Lund,P. (ed.), Molecular Chaperones: Frontiers in Molecular Biology. Oxford University Press, Oxford, England, pp. 90–118.
- Won K.A., Schumacher,R.J., Farr,G.W., Horwich,A.L. and Reed,S.I. (1998) Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol. Cell. Biol., 18, 7584–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]