Abstract
PEA-15 is a multifunctional protein that modulates signaling pathways which control cell proliferation and cell death. In particular, PEA-15 regulates the actions of the ERK MAP kinase cascade by binding to ERK and altering its subcellular localization. The three-dimensional structure of PEA-15 has been determined using NMR spectroscopy and its interaction with ERK defined by characterization of mutants that modulate ERK function. PEA-15 is composed of an N-terminal death effector domain (DED) and a C-terminal tail of irregular structure. NMR ‘footprinting’ and mutagenesis identified elements of both the DED and tail that are required for ERK binding. Comparison of the DED-binding surface for ERK2 with the death domain (DD)-binding surface of Drosophila Tube revealed an unexpected similarity between the interaction modes of the DD and DED motifs in these proteins. Despite a lack of functional or sequence similarity between PEA-15 and Tube, these proteins utilize a common surface of the structurally similar DD and DED to recognize functionally diverse targets.
Keywords: death domain/MAP kinase/NMR/three-dimensional structure
Introduction
The death effector domain (DED) is one of several small protein recognition modules that mediate the assembly of complexes required for signal transduction in programmed cell death. DEDs found in the adaptor protein FADD and the proforms of the initiator caspases, caspase-8 (FLICE, MACH) and caspase-10, play a pivotal role in the initiation of death receptor-mediated apoptosis, whereas DEDs in viral or cellular FLICE-inhibitory proteins (FLIPs) have the ability to block apoptosis (Ashkenazi and Dixit, 1998; Krammer, 2000). The DED, together with the structurally related death domain (DD) and caspase recruitment domain (CARD), are members of the death motif superfamily characterized by a conservedsix α-helix bundle structure (Aravind et al., 1999; Hofmann, 1999; Fesik, 2000). In addition to a common three-dimensional (3D) fold, these protein domains typically associate via homotypic interactions (DD–DD, DED– DED or CARD–CARD) with complementary domains in their binding partners. Surprisingly, no common protein interaction surface has been discernible for these structurally related motifs (Jeong et al., 1999; Qin et al., 1999; Xiao et al., 1999; Fesik, 2000).
DED-containing proteins are involved in other cellular signaling events besides the regulation of apoptosis. For example, the phosphoprotein enriched in astrocytes (PEA-15) activates the extracellular signal receptor-activated kinases (ERK1/2), members of the MAP kinase family (Ramos et al., 2000; Formstecher et al., 2001). PEA-15 is a small protein (15 kDa) that was first identified as an abundant phosphoprotein in brain astrocytes (Araujo et al., 1993) and subsequently was shown to be widely expressed in human tissues and highly conserved among mammals (Danziger et al., 1995; Estellés et al., 1996; Ramos et al., 1998). Several studies have shown that PEA-15 regulates multiple cellular functions including Fas- and tumor necrosis factor-α (TNF-α)-induced apoptosis (Condorelli et al., 1999; Estellés et al., 1999; Kitsberg et al., 1999) and phospholipase D expression (Zhang et al., 2000), and promotes resistance to insulin in type II diabetes (Condorelli et al., 1998).
PEA-15 has been recently demonstrated to regulate the subcellular localization of ERK1/2 and to control the biological outcomes of the ERK cascade (Formstecher et al., 2001). PEA-15 activates the ERK pathway in a Ras-dependent manner and binds specifically to ERK1/2 and not to the related MAP kinases p38 and JNK, or to other kinases in the ERK cascade (Ramos et al., 2000; Formstecher et al., 2001). The expression of PEA-15 in cells blocks ERK-dependent transcription and proliferation by preventing the translocation of ERK1/2 into the nucleus (Formstecher et al., 2001). Within the N-terminus of PEA-15, there is a predicted nuclear export sequence that could mediate the relocation of ERK to the cytoplasm, reminiscent of the MEK-dependent nuclear export of ERK (Fukuda et al., 1997; Adachi et al., 2000).
Death motifs usually are regarded as modules specialized for the recognition of complementary motifs in proteins that regulate apoptosis. However, the observation of an interaction between PEA-15 DED and ERK1/2 (Ramos et al., 2000; Formstecher et al., 2001) suggested a greater functional versatility for this structural motif. To examine this unusual death motif function, the 3D structure of PEA-15 was determined using NMR spectroscopy and its interaction with ERK2 established from an in vitro and in vivo analysis of PEA-15 mutant proteins. The structure of PEA-15 is comprised of a canonical N-terminal DED and an irregularly structured C-terminal tail. NMR ‘footprinting’ and mutagenesis of PEA-15 identified distinct regions in both the DED and C-terminal tail that are required for ERK2 binding. Remarkably, the ERK-binding surface of the PEA-15 DED is topologically similar to the Pelle-binding surface of Drosophila Tube DD (Xiao et al., 1999). Thus, despite the absence of functional or sequence similarity between PEA-15 and Tube, these proteins appear to utilize a common epitope to recognize structurally and functionally diverse targets.
Results
Three-dimensional structure of PEA-15
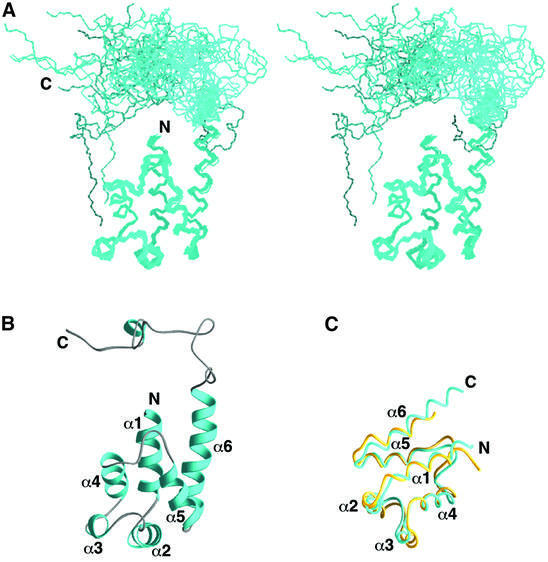
The structure of PEA-15 was determined from a total of 2937 NMR-derived restraints, including 2522 nuclear Overhauser effects (NOEs), 308 dihedral angle restraints and 107 3JNHα coupling constants (Figure 1). The 20 conformers exhibit good geometry, with no violations of distance restraints >0.5 Å and no dihedral angle violations >5° (Table I). The structure of the region corresponding to the DED is well defined by the NMR data. The atomic r.m.s.ds about the mean coordinate positions of the backbone atoms (N, Cα and C′) and all heavy atoms for residues 1–89 are 0.16 ± 0.02 and 0.62 ± 0.02 Å, respectively. The C-terminus of the protein does not adopt a regular structure in solution. This is evidenced by the absence of NOEs between non-sequential amino acids and by Cα and Cβ chemical shifts close to their random coil values. Residues 98–112 or 114–126 can be superimposed independently over the backbone atoms to <1.0 Å r.m.s.d.; however, no long-range NOEs were observed between the C-terminus and the DED to define their relative orientation.
Fig. 1. Three-dimensional structure of PEA-15. (A) Stereoview of the backbone atoms (N, Cα and C′) of 20 NMR conformers of PEA-15 superimposed over residues 1–97. (B) Ribbon representation of the averaged minimized structure of PEA-15. The orientation of the structure is as shown in (A). Residues 2–14, 17–27, 33–37, 42–51, 61–69 and 73–89 comprise helices α1, α2, α3, α4, α5 and α6, respectively. The helices are colored in cyan, and the loops and C-terminal tail in gray. (C) Backbone representation of the death effector domains of PEA-15 (cyan) and FADD (yellow) (Eberstadt et al., 1998; PDB accession code 1A1Z), superimposed over the Cα atoms of residues 1–83.
Table I. NMR structural statistics of PEA-15.
| R.m.s.ds from experimental distance restraints (Å) | |
| All (2522) | 0.067 ± 0.001 |
| Intraresidue (461) | 0.071 ± 0.003 |
| Sequential (|i – j| = 1) (623) | 0.062 ± 0.003 |
| Medium range (1<|i – j|≤5) (849) | 0.072 ± 0.002 |
| Long range (|i – j|>5) (503) | 0.067 ± 0.002 |
| Hydrogen bonds (86) | 0.008 ± 0.005 |
| R.m.s.ds from experimental torsion angle restraints | |
| Dihedral (°) (308)a | 0.23 ± 0.06 |
| 3JNHα coupling constants (Hz) (107) | 0.80 ± 0.02 |
| Deviations from idealized covalent geometry | |
| Bonds (Å) | 0.0041 ± 0.0001 |
| Angles (°) | 0.60 ± 0.01 |
| Impropers (°) | 0.56 ± 0.02 |
| Coordinate precision (Å)b | |
| Backbone (residues 1–89) | 0.16 ± 0.02 |
| All non-hydrogen atoms (residues 1–89) | 0.62 ± 0.02 |
| Quality factorsc | |
| % residues in most favorable region of Ramachandran plot (2380) | 84.2 |
| No. of bad contacts | 1.6 ± 1.1 |
| Prosa II Z-score | –7.9 ± 0.2 |
aThe torsion angle restraints comprised 121 φ, 95 ψ, 84 χ1 and 8 χ2.
bThe precision of the coordinates is defined as the average atomic r.m.s.d. between the 20 individual simulated annealing conformers and the mean coordinates for residues 1–89.
cPROCHECK_NMR (Laskowski et al., 1996) and Prosa II (Sippl, 1993) were used to assess the overall quality of the structures. There are no φ/ψ angles in the disallowed region of the Ramachandran plot, and the number of bad contacts (which provides a measure of the quality of non-bonded contacts) lies inside the range observed for ≤2 Å crystal structures. The Z-scores for random compact structures are distributed normally within a mean of 0 and a standard deviation of 1. The Z-scores for protein structures of 100–150 residues determined by X-ray crystallography are generally from –5 to –8 (Sánchez and Sâli, 1997). Thus, a Z-score of –7.9 is strongly indicative of a correct fold for the PEA-15 structure.
The 3D structure of PEA-15 consists of an N-terminal DED comprised of six antiparallel amphipathic α-helices closely packed around a central hydrophobic core, followed by a long C-terminal tail (Figures 1A and B). The C-terminal tail is irregular in structure, with the exception of residues 120–123, which appear to form a single turn of a 310-helix. The α-helices in the DED are connected by short loops, two of which contain β-turns (α2–α3 and α4–α5). The α-helices are arranged in a Greek key topology, with helices α1 and α2 being centrally located, α3 and α4 on one side and α5 and α6 on the other. This fold represents the core structure of the death motif superfamily that also includes the DD and CARD (Fesik, 2000).
The overall fold of the PEA-15 DED closely resembles the structure of the FADD DED (Eberstadt et al., 1998), the only other DED structure which has been determined to date. The two structures are very similar, with an r.m.s.d. of 1.8 Å for Cα atoms of residues 1–83, and differ primarily in the length of helix α6 (Figure 1C). Helix α6 of PEA-15 is seven residues longer than that of the FADD DED, and numerous NOEs were observed between residues in α1 and α6 of PEA-15 to define their relative orientation. The N-terminus of the FADD DED, on the other hand, is oriented away from α6 and the core of the protein as compared with PEA-15. PEA-15 also lacks the two hydrophobic patches observed on the surface of FADD DED. One of these patches in FADD consists of several residues in helix α2 that have been implicated in FADD’s apoptotic activity and interaction with the DEDs of procaspase-8 (Eberstadt et al., 1998). In contrast, many charged residues are present on the surface of the PEA-15 DED, suggesting that electrostatic interactions may mediate molecular recognition of ERK1/2.
PEA-15 DED and tail are required for ERK interaction
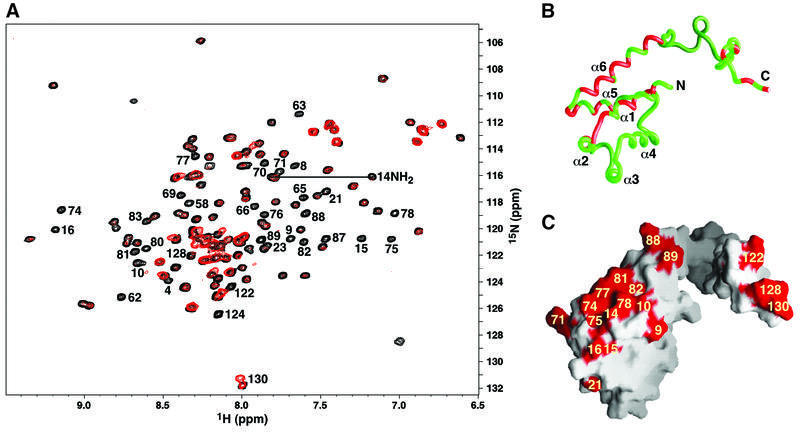
To explore the ERK-binding surface of PEA-15, complexes of 15N-labeled PEA-15 and unphosphorylated ERK2 were prepared, and the effect of ERK2 interaction on backbone N and HN chemical shifts (the NMR ‘footprint’) was examined. At the protein concentrations used in this study, unphosphorylated ERK2 is predicted to be dimeric (Khokhlatchev et al., 1998), resulting in a PEA-15–ERK2 complex of ∼115 kDa. As expected, the NMR spectrum of PEA-15 broadened away upon the addition of a stoichiometric amount of ERK2, consistent with the formation of a large, slowly tumbling complex in solution. At substoichiometric levels of ERK2, however, differential broadening of PEA-15 NMR resonances was observed, providing clues to where ERK2 may form direct contacts with PEA-15 (Figure 2A). Residues implicated in ERK binding were located in helices α1, α2, α5 and α6 of the DED, as well as in the C-terminal tail (Figure 2B and C).
Fig. 2. Identification of the ERK2-binding surface of PEA-15 by NMR ‘footprinting’. (A) Superposition of the 1H–15N HSQC spectra of PEA-15 in the free (black) and ERK2-bound form (red) at an ERK2:PEA-15 ratio of 0.7:1. Peaks are labeled for those backbone (NH) and/or side chain (Asn or Gln NH2) resonances that were completely broadened or displayed significant changes in chemical shift in the presence of ERK2. Mapping of the broadened resonances onto the (B) backbone structure and (C) molecular surface of PEA-15. Red shading corresponds to the residues labeled in (A). Helices α1, α2, α5 and α6, as well as several C-terminal residues, appear to contribute to the ERK2-binding surface of PEA-15.
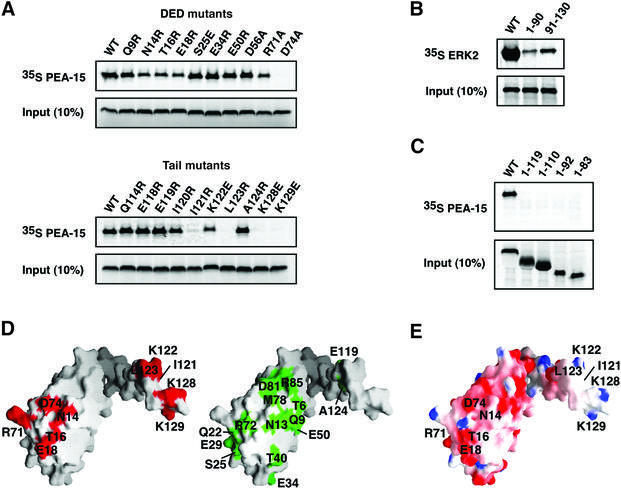
The segments of PEA-15 in which selective broadening was observed were functionally probed by site-directed mutagenesis and analyzed for binding to GST–ERK2 in an in vitro pull-down assay (Figure 3; Table II). Mutants were constructed in which surface-exposed residues were substantially altered in side chain length and/or charge, while minimizing the likelihood of altering the protein structure. Five out of 23 mutations introduced into the DED displayed either a reduced or complete loss of binding to ERK2 (Figure 3A; Table II). Three of these mutations were located in the α1–α2 loop (Asn14, Thr16 and Glu18) and the remaining two mutations in the α5–α6 loop (Arg71 and Asp74). Together, these five residues form a contiguous, predominantly negatively charged surface on the PEA-15 DED which defines part of the binding site for ERK2 (Figure 3D and E). In contrast, extensive mutagenesis of other regions of the DED had no effect on the interaction with ERK2 (Figure 3D; Table II).
Fig. 3. ERK2 recognition by PEA-15. (A) Effects of point mutations of PEA-15 on interaction with ERK2, determined by the binding activity of [35S]methionine-labeled wild-type (WT) or mutant PEA-15 to GST–ERK2 in a pull-down assay. The input lane represents 10% of the total protein added to the binding experiment. Mutants N14R, T16R, E18R, R71A and D74A in the DED, and I121R, K122E, L123R, K128E and K129E in the C-terminal tail had ≤50% ERK binding relative to wild-type PEA-15. (B) [35S]methionine-labeled ERK2 binding to GST–PEA-15, GST–PEA-15 DED (1–90) or GST–PEA-15 tail (91–130) in a pull-down assay. (C) Effects of C-terminal truncation of PEA-15 on the interaction with ERK2. The experiment was performed as described in (A). (D) Molecular surface representations of PEA-15 illustrating the amino acid residues that are important for ERK2 binding in red, while residues functionally insensitive to mutation are shown in green (compared with Table II). (E) Electrostatic potential surface representation of PEA-15. The color scale for the charge distribution extends from –8 kT/e– (red) to +8 kT/e– (blue), calculated using GRASP (Nicholls et al., 1991). The residues important for ERK2 binding deduced from (A) are labeled.
Table II. Mutational analysis of PEA-15.
| Mutationa | ERK2 bindingb | Locationc |
|---|---|---|
| T6R | ++ | α1 |
| Q9R | ++ | α1 |
| N13R | ++ | α1 |
| N14R | + | α1 |
| T16R | + | Loop(α1–α2) |
| E18R | + | α2 |
| Q22A | ++ | α2 |
| S25E | ++ | α2 |
| E29A | ++ | Loop(α2–α3) |
| E34R | ++ | α3 |
| T40R | ++ | Loop(α3–α4) |
| E50R | ++ | α4 |
| D56A | ++ | Loop(α4–α5) |
| S61R | ++ | α5 |
| E64A | ++ | α5 |
| H65A | ++ | α5 |
| R71A | + | Loop(α5–α6) |
| R72A | ++ | Loop(α5–α6) |
| D74A | – | α6 |
| T77A | ++ | α6 |
| M78R | ++ | α6 |
| D81A | ++ | α6 |
| R85A | ++ | α6 |
| Q114R | ++ | C-terminal tail |
| E118R | ++ | C-terminal tail |
| E119R | ++ | C-terminal tail |
| I120R | ++ | C-terminal tail |
| I121R | – | C-terminal tail |
| I121A | – | C-terminal tail |
| K122E | + | C-terminal tail |
| L123R | – | C-terminal tail |
| L123A | – | C-terminal tail |
| A124R | ++ | C-terminal tail |
| K128E | – | C-terminal tail |
| K128A | – | C-terminal tail |
| K129E | – | C-terminal tail |
| K129A | – | C-terminal tail |
aMutants were designed to introduce a significant change in the chemical properties of a surface-exposed residue without disruption of protein structure.
bBinding in a GST–ERK2 pull-down assay is defined as follows: ++, equivalent to wild-type; +, ≤50% wild-type; –, loss of binding.
cThe location of the mutation in the structure is indicated.
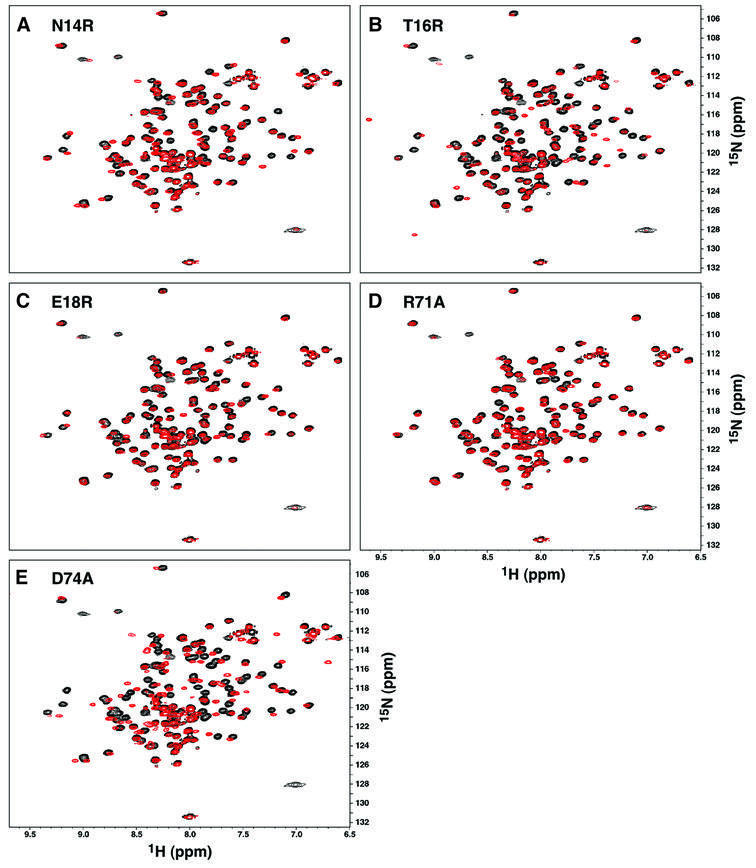
To assess the structural integrity of the five DED mutant proteins that demonstrated reduced binding to ERK, the 2D 1H-15N HSQC spectrum of each mutant was compared with that of wild-type PEA-15 (Figure 4). The sensitivity of the chemical shift to local environment permits the assessment of local and global changes in the structure of PEA-15 with amino acid resolution. Thus, the peak pattern and linewidth are diagnostic of the similarity in protein fold between the wild-type and mutant proteins. It is evident that the N14R, T16R, E18R and R71A mutant proteins are folded correctly, with only minor chemical shift perturbations observed for a few residues spatially proximal to the mutated site. The fifth mutant protein, D74A, displayed several differences in the N and HN chemical shifts compared with wild-type PEA-15 (Figure 4E). However, complete backbone chemical shift assignment of the D74A mutant revealed that neither significant local nor global structural changes had occurred. First, secondary chemical shift deviations for Cα and Cβ (Wishart et al., 1992) established that the wild-type and D74A proteins possessed identical helical segments, indicating that the secondary structure of D74A was unchanged by the mutation. Secondly, the absolute values of the Cα and Cβ chemical shifts are nearly identical between D74A and wild type, establishing that the global fold of the mutant protein is intact. These data strongly suggest that the D74A mutant was not significantly altered in structure. More probably, the removal of an exposed negative charge from a highly charged region of the protein (Figure 3E) altered the local electronic environment, resulting in the perturbation of several backbone N and HN resonances. Since all of the DED mutants were folded correctly, the loss of ERK binding observed for these mutants was most probably the consequence of a disruption in the interaction surface of PEA-15 for the enzyme.
Fig. 4. Analysis of the structural integrity of the PEA-15 DED mutant proteins. (A–E) Superpositions of the 1H–15N HSQC spectra of wild-type (black) and mutant (red) PEA-15 proteins N14R, T16R, E18R, R71A and D74A.
Regions outside the DED of PEA-15 have previously been implicated in ERK2 binding, as overexpression of the PEA-15 DED alone did not activate ERK (Ramos et al., 2000; Formstecher et al., 2001). NMR ‘footprinting’ also suggested the involvement of the C-terminal tail in binding to ERK2 (Figure 2). Furthermore, neither the DED nor C-terminal tail of PEA-15 bound ERK well in isolation (Figure 3B), and deletion of only 11 amino acids from the tail was sufficient to disrupt interaction with ERK (Figure 3C). Site-directed mutagenesis within the tail revealed that Ile121, Lys122, Leu123 (all in the 310-helix) in addition to Lys128 and Lys129 were important for ERK interaction (Figure 3A; Table II). The irregular structure of the tail and its lack of interaction with the DED suggests that none of the tail mutants would have affected the integrity of the protein. Although two hydrophobic residues were changed to arginine, which could have disrupted the putative 310-helix (I121R and L123R), mutation of these residues to alanine displayed an identical phenotype to that of the arginine mutants (Table II). Thus, the functional defect observed for these mutants was not likely to be a consequence of the introduction of a conformational change in the tail. These results demonstrate that electrostatic interactions involving residues in the DED as well as hydrophobic and charged residues in the tail are critical for PEA-15 binding to ERK2.
Functional characterization of PEA-15 mutants
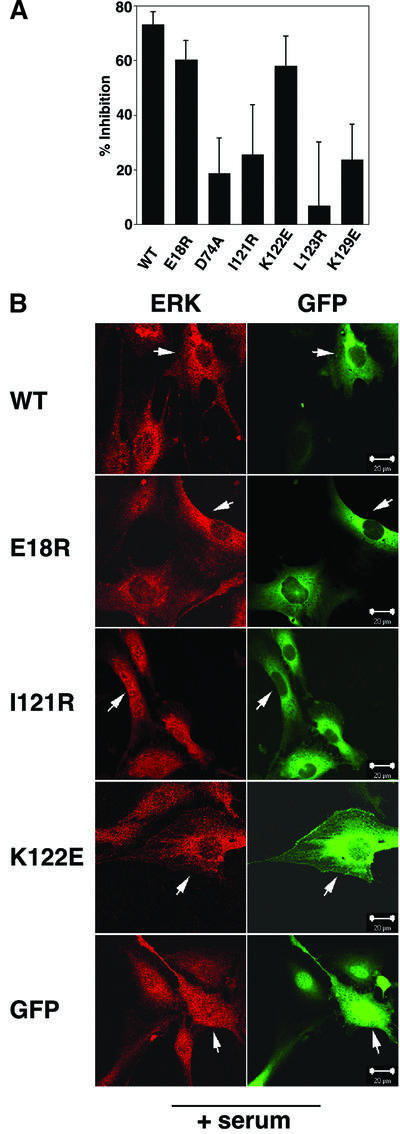
To correlate the in vitro ERK binding deficiency of the PEA-15 mutants with their functional activity in vivo, the ability of wild-type or mutant PEA-15 to inhibit ERK nuclear signaling was assessed (Figure 5A). Wild-type PEA-15 markedly attenuated the ERK-stimulated transcriptional activation of Elk-1 (Formstecher et al., 2001), whereas mutants of PEA-15 that lacked the capacity to bind ERK2 in vitro (Figure 3A) also displayed re duced inhibition of Elk-1-dependent transcription in vivo (Figure 5A). Mutations in the DED, such as E18R, were clearly less effective at reversing the inhibition of transcription compared with mutations in the C-terminal tail (e.g. I121R, L123R and K129E). These results correlate well with the residual ERK-binding activity of the DED and tail mutants observed in the pull-down assay (Figure 3A). Thus, the capacity of PEA-15 to bind ERK correlates with its ability to block the phosphorylation and transcriptional activity of ERK nuclear substrates.
Fig. 5. In vivo analysis of PEA-15 mutants. (A) Elk-1-dependent transcriptional activity was measured in serum-stimulated CHO-K1 cell lysates transfected with expression plasmids encoding either wild-type PEA-15, mutant PEA-15 or control vector. The data are from an experiment done in triplicate and expressed as percentage inhibition of transcription relative to a vector-only control. (B) NIH 3T3 cells expressing either wild-type or mutant GFP–PEA-15 were serum starved for 16–20 h and then stimulated with 20% serum for 3 h and stained for ERK. The proteins were imaged by immunofluorescence in confocal microscopy. PEA-15 mutants such as I121R that are unable to bind ERK2 in vitro do not prevent ERK translocation to the nucleus upon serum stimulation (arrows), contrary to wild-type GFP–PEA-15 (arrows). The scale bar represents 20 µm.
The effects of PEA-15 on transcription were associated with an alteration in the nuclear translocation of ERK (Figure 5B). Mutations that completely abrogated ERK binding (e.g. I121R) failed to block ERK nuclear translocation. A similar outcome was shown previously for the D74A mutant in the DED (Formstecher et al., 2001). In contrast, mutants that retained some ability to bind ERK2 in vitro, such as E18R and K122E, also substantially retained ERK in the cytoplasm. Thus, the ERK-binding activity of PEA-15 is required for its biological effects on ERK localization.
A common binding surface in the death motif mediates diverse protein–protein interactions
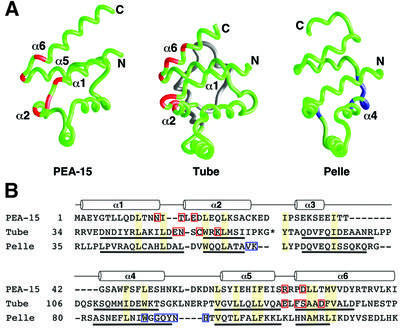
A comparison of the ERK-binding surface of PEA-15 with that observed for Tube DD in the crystal structure of the Tube–Pelle DD complex (Xiao et al., 1999) revealed an unexpected similarity in the binding motifs of Tube and PEA-15 (Figure 6). The interaction between Tube and the serine/threonine kinase Pelle through their N-terminal DDs is required for the activation of dorsal–ventral patterning genes during Drosophila embryogenesis (Belvin and Anderson, 1996). The molecular determinants by which Tube recognizes Pelle include a compact surface within the Tube DD as well as a distinct set of interactions formed by its irregularly structured C-terminal tail (Xiao et al., 1999). These elements of Tube contact adjacent sites on the Pelle DD, forming an almost continuous interaction surface. The similarity to the PEA-15–ERK2 interaction resides in the interaction surface of Tube DD, formed by the α1–α2 and α5–α6 loops and α6 (Figure 6). Mutations introduced into the these regions of PEA-15 were functionally deficient for ERK2 binding, thereby restoring ERK2 nuclear translocation and consequent stimulation of transcription. Mutants within the corresponding loops and helix of Tube DD rendered the protein inactive (Xiao et al., 1999), indicative of an important role for these regions in the interaction between the Tube and Pelle DDs. Since the targets of Tube and PEA-15 are structurally and functionally unrelated, the similarity between the DD and DED surfaces implicated in protein–protein interaction is suggestive of a conserved binding epitope inherent to the common fold of the DD and DED of these proteins. While the irregularly structured C-terminal tails of Tube and PEA-15 both appear to play critical roles in the recognition of their binding partners, a correspondence between their respective roles cannot be drawn in the absence of the 3D structure of the PEA-15–ERK2 complex.
Fig. 6. Comparison of the PEA-15 DED, Tube DD and Pelle DD binding epitopes. (A) Backbone representations of the death motifs of PEA-15, Tube and Pelle (Xiao et al., 1999; PDB accession code 1D2Z) with residues important for binding their target proteins highlighted in red for PEA-15 and Tube and in blue for Pelle. Tube contains an insertion of two short α-helices between helices α2 and α3 (shown in gray) that is not part of the core structure of the DD. (B) Structure-based sequence alignment of mouse PEA-15, Drosophila melanogaster Tube DD and D.melanogaster Pelle DD created from pairwise superpositions calculated in DALI (Holm and Sander, 1993). The positions of helices are indicated by cylinders for PEA-15, and the helices for Tube and Pelle are underlined. Conserved residues which form part of the hydrophobic core in each protein are shaded yellow. The asterisk in the Tube sequence indicates a two-helix insertion (Xiao et al., 1999) that has been omitted for clarity. Sites corresponding to mutations that are deleterious to function are highlighted with red boxes for PEA-15 (this study). Interfacial residues observed in the Tube–Pelle DD complex structure are indicated by red boxes for Tube DD and blue boxes for Pelle DD (Xiao et al., 1999).
Discussion
The death motif superfamily, composed of the DD, DED and CARD families, has emerged as the primary mediator of protein–protein interactions required for the transmission and regulation of apoptotic signals (Aravind et al., 1999; Hofmann, 1999). Despite low sequence identity between the death motif subfamilies, they have been shown to share a common 3D fold containing six antiparallel α-helices (Fesik, 2000). In addition to a common fold, each of these subfamilies typically interacts with other proteins through homotypic interactions in which DD–DD, DED–DED and CARD–CARD contacts are formed. The elucidation of the molecular basis and specificity of death motif interactions is central to understanding the apoptotic process. Two recent X-ray crystal structures of heterodimer complexes formed by the CARDs of Apaf-1 and procaspase-9 (Qin et al., 1999) and the DDs of Drosophila proteins Tube and Pelle (Xiao et al., 1999) have provided valuable insights into the association mechanisms of death motifs. However, the residues that form the interface are dramatically different between these two heterodimer structures, and also differ from the sur faces suggested by mutagenesis to mediate other death motif interactions (Tartaglia et al., 1993; Huang et al., 1996; Eberstadt et al., 1998; Jeong et al., 1999; Telliez et al., 2000; McDonald et al., 2001). The apparent absence of a conserved interaction surface within this common protein fold has led to the suggestion that death motifs may associate with one another by a variety of mechanisms (Xiao et al., 1999; Fesik, 2000).
To date, the interaction between a death motif superfamily member and a heterotypic binding partner has not been described. PEA-15 is unusual given that it has been suggested to interact with other death motif proteins (FADD and procaspase-8) via homotypic interactions (Condorelli et al., 1999; Kitsberg et al., 1999) and to interact with structurally unrelated proteins such as ERK MAP kinases through a previously unknown mechanism (Ramos et al., 2000; Formstecher et al., 2001). Elucidation of the 3D structure of PEA-15 in conjunction with functional mutagenesis has revealed the molecular determinants by which PEA-15 recognizes ERK. Previous studies had established the importance of helix α6 of the DED for PEA-15 function (Ramos et al., 1998, 2000; Formstecher et al., 2001), and in this study the ERK-binding epitope was defined further to include the α1–α2 and α5–α6 loops (Figure 3). A number of mutations to surface-exposed residues in other regions of the PEA-15 DED did not disrupt the binding to ERK2 (Table II), indicating that the interaction interface within the DED is confined to this face of the motif. Essential determinants for ERK interaction also reside in the C-terminal tail of PEA-15, although the structural relationship between the DED- and tail-binding determinants has not been determined from the structure of PEA-15 alone in solution. The effects of the PEA-15 mutations on ERK binding in vitro correlated well with in vivo functional analysis. Although mutants in the DED and C-terminal tail displayed differential effects on PEA-15 function, these regions of PEA-15 are not functionally independent, suggesting that they may contact adjacent binding surfaces on ERK.
Recently, several regions of MAP kinases have been implicated in mediating interactions with their substrates, activators and regulators. For example, a docking groove has been identified in the C-terminal domain that appears to function as a general binding site (Tanoue et al., 2000, 2001; Chang et al., 2002), and may be recognized by many proteins that contain a conserved docking site sequence comprised of basic and hydrophobic motifs (Sharrocks et al., 2000). This docking groove contains an acidic patch termed the CD domain (Tanoue et al., 2000) and adjacent hydrophobic patches (Chang et al., 2002) to accommodate the basic and hydrophobic motifs of the docking site peptides. PEA-15, however, does not contain a recognizable docking site sequence and was found instead to bind the αG helix and MAP kinase insert region of ERK (J.M.Hill, F.-L.Chou, J.-C.Hsieh, J.W.Ramos, M.H.Ginsberg and M.H.Werner, unpublished data). An essential role for this region has also been demonstrated recently for the interaction of ERK2 with the MAP/ERK kinases MEK1 and MEK2 (Robinson et al., 2002).
Unexpectedly, the location of the ERK-binding epitope on the PEA-15 DED is similar to that observed for Drosophila Tube DD in the crystal structure of the Tube and Pelle DD heterodimer (Xiao et al., 1999). Both the PEA-15 DED and Tube DD utilize residues in the α1–α2 loop, α5–α6 loop and helix α6 for interaction with their binding partners (Figure 6). Particularly intriguing is the similarity between the homotypic DD interaction of Tube–Pelle and the heterotypic interaction implied by functional mutagenesis of PEA-15. This observation seems inconsistent with the hypothesis of diverse association mechanisms for the DD and DED motifs (Xiao et al., 1999; Fesik, 2000), and suggests that the binding epitopes may be structurally conserved. Certainly, the similarity between Tube and PEA-15 is not complete given that contacts made by the C-terminal tail and DD of Tube form a nearly continuous binding surface on Pelle DD, while the precise role of the PEA-15 tail and its relative orientation with respect to PEA-15 DED are not yet known.
The functional importance of the α1–α2 and α5–α6 loops in the DED has been extended recently to include two proteins associated with cell death signaling—the viral FLICE inhibitory protein MC159 (Garvey et al., 2002) and the death adaptor protein FADD (Thomas et al., 2002). The α5–α6 loop contains a conserved RxDLφ motif (x = any amino acid, φ = any hydrophobic residue) that is found in many DED-containing proteins (Figure 7). For example, the DEDs of cellular FLIP, most of the viral FLIPs, the FADD DED, one of two DEDs in procaspase-8 and the nuclear targeted proteins DEDD (Stegh et al., 1998) and DEDD2/FLAME-3 (Roth et al., 2002; Zhan et al., 2002) all contain the motif. Garvey et al. (2002) demonstrated that the α1–α2 and α5–α6 loops of both DEDs in MC159 are important for its ability to inhibit apoptosis via interaction with FADD and procaspase-8. Reverse two-hybrid analysis also suggested a functional role for the α5–α6 loop of FADD DED during assembly of the CD95 (Fas, APO-1) death-inducing signaling complex (Thomas et al., 2002). These results indicate that the similarities between Tube DD and PEA-15 DED may extend to a number of proteins which harbor these motifs, irrespective of their physiological diversity. Thus, a common binding surface in the DED and DD may govern protein–protein interactions with their binding partners.
Fig. 7. Sequence alignment of death effector domains. Listed are the DEDs of mouse PEA-15 (Q62048), human FADD (Q13158), human caspase-8/FLICE (Q14790), human cellular FLIP (c-FLIP) (O15519), molluscum contagiosum virus (MCV) MC159 protein (Q98325), equine herpesvirus 2 (EHV-2) E8 protein (Q66674), human DEDD (NP_127491) and human DEDD2/FLAME3 (NP_579874). Swiss-Prot accession numbers are indicated in parentheses. The positions of helices are indicated by cylinders for PEA-15, and the helices for FADD (Eberstadt et al., 1998) are underlined. Highly conserved hydrophobic residues are shaded yellow and the conserved RxDLφ motif (x = any amino acid, φ = any hydrophobic residue) at the beginning of helix α6 is indicated by a box.
Materials and methods
Protein expression and purification
Full-length PEA-15 was cloned into a pQE-9 vector (Qiagen), which produces the recombinant protein with a hexahistidine (His6) sequence at the N-terminus. Uniformly 15N- and 15N/13C-labeled proteins were overexpressed in Escherichia coli BL21 cells by adaptive control fermentation (Werner et al., 2001) in minimal medium containing 15NH4Cl and/or [13C]glucose as the sole nitrogen and carbon sources, respectively. Cells were grown at 37°C to an optical density (A600 nm) of ∼3.0 and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. The cells were suspended in 50 mM Tris buffer pH 8.0, 1 M NaCl, 30 mM imidazole and 10 mM benzamidine hydrochloride, lysed with a French press, and centrifuged. The protein was isolated by Ni2+ affinity chromatography and subsequently purified to homogeneity by anion-exchange chromatography (MonoQ HR 16/10, Amersham Pharmacia Biotech). NMR samples contained 1 mM protein in 10 mM sodium phosphate buffer pH 7.0, 1 mM dithiothreitol (DTT) and 50 µM NaN3.
Selected point mutations (N14R, T16R, E18R, R71A and D74A) were introduced into PEA-15 contained in pQE-9 using the QuikChange site-directed mutagenesis kit (Stratagene). The resulting mutations were confirmed by DNA sequencing. 15N-labeled PEA-15 mutant proteins were produced on a small scale (400 ml) in shake flasks and purified using Ni2+ affinity chromatography. 2D 1H–15N HSQC spectra of the mutant proteins were obtained using the sample conditions described for the wild-type protein, except that the protein concentration was ∼0.3 mM.
Differential exchange broadening of PEA-15–ERK2 complexes was carried out with unphosphorylated ERK2 that was expressed and purified as described (Zhang et al., 1993). Both PEA-15 and ERK2 were dialyzed into 10 mM sodium phosphate buffer pH 7.0, 150 mM NaCl, 1 mM DTT and 50 µM NaN3 prior to complex formation. Samples of 0.2 mM 15N-labeled PEA-15 and substoichiometic amounts of ERK2 (0.3, 0.5 and 0.7 molar equivalents) were prepared and resonances monitored by 1H–15N correlation spectroscopy.
NMR spectroscopy
All NMR experiments were acquired at 25°C on either a Bruker DMX500 or DMX600 spectrometer equipped with a z-shielded gradient triple resonance probe. The backbone and side chain 1H, 13C and 15N resonances of the protein were assigned using CBCA(CO)NH, HNCACB, HBHA(CO)NH, C(CO)NH, H(CCO)NH and 3D HCCH-TOCSY experiments (Clore and Gronenborn, 1998). 3JNHα, 3JNHβ, 3JCγN and 3JCγCO coupling constants were measured by quantitative J correlation spectroscopy (Vuister et al., 1999). NOEs involving protons of the protein were obtained from 3D 15N-separated, 3D 13C-separated and 4D 13C/13C-separated NOE spectra (mixing times of 110, 120 and 90 ms, respectively). NMR spectra were processed with NMRPipe/NMRDraw (Delaglio et al., 1995) and analyzed using PIPP and STAPP (Garrett et al., 1991).
Structure calculations
NOEs within the protein were grouped into four distance ranges, 1.8–2.7, 1.8–3.3, 1.8–5.0 and 1.8–6.0 Å, corresponding to strong, medium, weak and very weak intensities. Distances involving methyl protons, aromatic ring protons and non-stereospecifically assigned methylene protons were represented as a (Σr–6)–1/6 sum (Nilges, 1993). A total of 121 φ, 84 χ1 and eight χ2 angle restraints were derived from an analysis of 3JNHα, 3JNHβ, 3JCγN and 3JCγCO coupling constants (Vuister et al., 1999), and 95 ψ angle restraints were determined by chemical shift database analysis using the program TALOS (Cornilescu et al., 1999). The minimum range employed for φ, ψ, χ1 and χ2 torsion angle restraints was 30°. Protein backbone hydrogen bonding restraints (rNH–O = 1.5–2.8 Å, rN–O = 2.4–3.5 Å) within areas of α-helical structure derived from an analysis of 13Cα chemical shifts and characteristic Hα-NHi + 3 and Hα-Hβi + 3 NOE patterns were introduced during the final stages of refinement. A single turn of 310-helix was concluded to exist for residues 120–123 based on consecutive Hα-NHi + 2 and Hα-Hβi + 3 NOEs (Wüthrich, 1986), but no hydrogen bonding restraints were included. The structures were calculated with the program X-PLOR 3.843 (Brünger, 1992) adapted to incorporate pseudo-potentials for 3JNHα coupling constants (Garrett et al., 1994), secondary 13Cα and 13Cβ chemical shifts (Kuszewski et al., 1995) and a conformational database potential (Kuszewski and Clore, 2000). There were no hydrogen-bonding, electrostatic or 6–12 Lennard–Jones empirical potential energy terms in the target function. The final ensemble of 20 NMR conformers was selected on the basis of lowest energy and least number of restraint violations; these structures had no distance restraint violations >0.5 Å and no dihedral angle violations >5°. Structure quality was assessed with PROCHECK_NMR (Laskowski et al., 1996) and ProsaII (Sippl, 1993). Structures were displayed and analyzed using GRASP (Nicholls et al., 1991) and Ribbons (Carsons, 1987).
Mutagenesis and GST pull-down assays
Full-length PEA-15 (residues 1–130) and several C-terminal truncation mutants of PEA-15 encoding residues 1–119, 1–110, 1–92 and 1–83 were amplified by PCR and subcloned into a pET-21a vector (Novagen). Specific amino acid changes were introduced into full-length PEA-15 contained in pET-21a using the QuikChange site-directed mutagenesis kit (Stratagene). The resulting mutations were confirmed by DNA sequencing. A complete list of the mutants constructed is given in Table II. Wild-type and mutant proteins were expressed in vitro using the TNT T7 Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]methionine. Rat wild-type ERK2 was overproduced as a GST fusion protein in E.coli BL21 cells using a pGEX-4T-1 vector (Amersham Pharmacia Biotech) and batch purified using glutathione–agarose beads (Sigma). 35S-labeled wild-type and mutant PEA-15 proteins were incubated with GST–ERK2 in 150 µl of 50 mM HEPES pH 7.4, 50 mM NaCl, 5 mM EDTA, 1% NP-40 and 10% glycerol. Samples were incubated for 2 h at 4°C and then washed three times with 0.5 ml of the same buffer. The bound proteins were separated on 16% Tris-Tricine gels and visualized by phosphoimaging.
Full-length PEA-15, DED (residues 1–90) and C-terminus (residues 91–130) were overproduced as GST fusion proteins in E.coli BL21 cells using a pGEX-4T-1 vector (Amersham Pharmacia Biotech) and batch purified using glutathione–agarose beads (Sigma). ERK2 contained in a NpT7-5 vector (Zhang et al., 1993) was expressed in vitro using the TNT T7 Coupled Reticulocyte Lysate System in the presence of [35S]methionine. The pull-down assays were performed as described above.
Transcription assays
Transcription assays were done using the Pathdetect trans-reporting kit according to the manufacturer’s instructions (Stratagene). Briefly, CHO-K1 cells were co-transfected with GAL4-Elk-1 (50 ng), GAL4-luciferase (500 ng) and PEA-15, wild-type or mutant constructs in pcDNA3 vector (200 ng) or vector alone (200 ng). GAL4-Elk-1 contains the DNA-binding domain of GAL4 fused to the trans-activation domain of Elk-1. GAL4-luciferase has the luciferase reporter gene under the control of a synthetic promoter containing five tandem repeats of the yeast GAL4-binding sites. Cells were allowed to grow until 90% confluent and then transfected with Lipofectamine 2000 (Invitrogen Life Technologies) according to the manufacturer’s protocol. Cells were serum starved for 16–20 h in Dulbecco’s modified Eagle’s medium (DMEM) containing 0.5% fetal bovine serum (FBS) and then stimulated with 20% serum for 30 min. Elk-1 transcription was measured by expression of active luciferase, for 10 µg of total protein from cell lysates, using the Promega Luciferase Assay System according to the manufacturer’s instructions.
Immunofluorescence microscopy
NIH 3T3 cells, grown on coverslips, were transfected with 1 µg of an expression plasmid encoding wild-type or mutant PEA-15 fused to GFP, or with vector alone using Lipofectamine Plus (Invitrogen Life Technologies) according to the manufacturer’s protocol. The transfected cells were serum starved for 16–20 h before stimulation with 20% serum for 3 h. The cells were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 10 min. They were then permeabilized with 0.2% Triton X-100 in PBS for 10 min. Cells were incubated with anti-ERK1/2 antibodies (Santa Cruz Biotechnologies) overnight in PBS containing 3% bovine serum albumin (BSA) and 0.2% Triton X-100. ERK staining was detected using Texas red secondary antibody (Molecular Probes). Coverslips were mounted using Fluoromount-G (Southern Biotechnology Associates) and viewed under a Zeiss Axiovert microscope to detect GFP or ERK localization.
Coordinates
The coordinates of the 20 PEA-15 NMR structures have been deposited in the RCSB protein data bank with accession number 1N3K.
Acknowledgments
Acknowledgements
We thank Melanie Cobb for providing cDNA of ERK2. This work was supported by fellowships awarded by the Human Frontier Science Program (LT0537), the NHMRC of Australia (997045) and the Norman and Rosita Winston Foundation to J.M.H., and by a National Science Foundation Grant (MCB-0095074) to M.H.W. M.H.W. is a Distinguished Young Scholar of the W.M.Keck Foundation.
References
- Adachi M., Fukuda,M. and Nishida,E. (2000) Nuclear export of MAP kinase (ERK) involves a MAP kinase kinase (MEK)-dependent active transport mechanism. J. Cell Biol., 148, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo H., Danziger,N., Cordier,J., Glowinski,J. and Chneiweiss,H. (1993) Characterization of PEA-15, a major substrate for protein kinase C in astrocytes. J. Biol. Chem., 268, 5911–5920. [PubMed] [Google Scholar]
- Aravind L., Dixit,V.M. and Koonin,E.V. (1999) The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci., 24, 47–53. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A. and Dixit,V.M. (1998) Death receptors: signaling and modulation. Science, 281, 1305–1308. [DOI] [PubMed] [Google Scholar]
- Belvin M.P. and Anderson,K.V. (1996). A conserved signaling pathway: the Drosophila toll–dorsal pathway. Annu. Rev. Cell. Dev. Biol., 12, 393–416. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Carsons M.J. (1987) Ribbon models of macromolecules. J. Mol. Graph., 5, 103–106. [Google Scholar]
- Chang C., Xu,B., Akella,R., Cobb,M.H. and Goldsmith,E.J. (2002) Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell, 9, 1241–1249. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1998) Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol., 16, 22–34. [DOI] [PubMed] [Google Scholar]
- Condorelli G. et al. (1998) PED/PEA-15 gene controls glucose transport and is overexpressed in type 2 diabetes mellitus. EMBO J., 17, 3858–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G. et al. (1999) PED/PEA-15: an anti-apoptotic molecule that regulates Fas/TNFR1-induced apoptosis. Oncogene, 18, 4409–4415. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Danziger N., Yokoyama,M., Jay,T., Cordier,J., Glowinski,J. and Chneiweiss,H. (1995) Cellular expression, developmental regulation and phylogenetic conservation of PEA-15, the astrocytic major phosphoprotein and protein kinase C substrate. J. Neurochem., 64, 1016–1025. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Eberstadt M., Huang,B., Chen,Z., Meadows,R.P., Ng,S.-C., Zheng,L., Lenardo,M.J. and Fesik,S.W. (1998) NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature, 392, 941–945. [DOI] [PubMed] [Google Scholar]
- Estellés A., Yokoyama,M., Nothias,F., Vincent,J.-D., Glowinski,J., Vernier,P. and Chneiweiss,H. (1996) The major astrocytic phospho protein PEA-15 is encoded by two mRNAs conserved on their full length in mouse and human. J. Biol. Chem., 271, 14800–14806. [DOI] [PubMed] [Google Scholar]
- Estellés A., Charlton,C.A. and Blau,H.M. (1999) The phosphoprotein protein PEA-15 inhibits Fas- but increases TNF-R1-mediated caspase-8 activity and apoptosis. Dev. Biol., 216, 16–28. [DOI] [PubMed] [Google Scholar]
- Fesik S.W. (2000) Insights into programmed cell death through structural biology. Cell, 103, 273–282. [DOI] [PubMed] [Google Scholar]
- Formstecher E. et al. (2001) PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase. Dev. Cell, 1, 239–250. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Gotoh,Y. and Nishida,E. (1997) Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleo cytoplasmic transport of MAP kinase. EMBO J., 16, 1901–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett D.S., Powers,R., Gronenborn,A.M. and Clore,G.M. (1991) A common sense approach to peak picking in two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson., 95, 214–220. [DOI] [PubMed] [Google Scholar]
- Garrett D.S., Kuszewski,J., Hancock,T.J., Lodi,P.J., Vuister,G.W., Gronenborn,A.M. and Clore,G.M. (1994) The impact of direct refinement against three-bond HN–CαH coupling constants on protein structure determination by NMR. J. Magn. Reson. B, 104, 99–103. [DOI] [PubMed] [Google Scholar]
- Garvey T.L., Bertin,J., Siegel,R.M., Wang,G., Lenardo,M.J. and Cohen,J.I. (2002) Binding of FADD and caspase-8 to molluscum contagiosum virus MC159 v-FLIP is not sufficient for its antiapoptotic function. J. Virol., 76, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. (1999) The modular nature of apoptotic signaling proteins. Cell. Mol. Life Sci., 55, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Huang B., Eberstadt,M., Olejniczak,E.T., Meadows,R.P. and Fesik,S.W. (1996) NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature, 384, 638–641. [DOI] [PubMed] [Google Scholar]
- Jeong E.J., Bang,S., Lee,T.H., Park,Y.I., Sim,W.S. and Kim,K.S. (1999) The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. J. Biol. Chem., 274, 16337–16342. [DOI] [PubMed] [Google Scholar]
- Khokhlatchev A.V., Canagarajah,B., Wilsbacher,J., Robinson,M., Atkinson,M., Goldsmith,E. and Cobb,M.H. (1998) Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell, 93, 605–615. [DOI] [PubMed] [Google Scholar]
- Kitsberg D. et al. (1999) Knock-out of the neural death effector domain protein PEA-15 demonstrates that its expression protects astrocytes from TNF-α induced apoptosis. J. Neurosci., 19, 8244–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer P.H. (2000) CD95’s deadly mission in the immune system. Nature, 407, 789–795. [DOI] [PubMed] [Google Scholar]
- Kuszewski J. and Clore,G.M. (2000) Sources of and solutions to problems in the refinement of protein NMR structures against torsion angle potentials of mean force. J. Magn. Reson., 146, 249–254. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Qin,J., Gronenborn,A.M. and Clore,G.M. (1995) The impact of direct refinement against 13Cα and 13Cβ chemical shifts on protein structure determination by NMR. J. Magn. Reson. B, 106, 92–96. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK_NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- McDonald E.R. III, Chui,P.C., Martelli,P.F., Dicker,D.T. and El-Deiry,W.S. (2001) Death domain mutagenesis of KILLER/DR5 reveals residues critical for apoptotic signaling. J. Biol. Chem., 276, 14939–14945. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nilges M. (1993) A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins, 17, 297–309. [DOI] [PubMed] [Google Scholar]
- Qin H., Srinivasa,S.M., Wu,G., Fernandes-Alnemri,T., Alnemri,E.S. and Shi,Y. (1999) Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature, 399, 549–557. [DOI] [PubMed] [Google Scholar]
- Ramos J.W., Kojima,T.K., Hughes,P.E., Fenczik,C.A. and Ginsberg,M.H. (1998) The death effector domain of PEA-15 is involved in its regulation of integrin activation. J. Biol. Chem., 273, 33897–33900. [DOI] [PubMed] [Google Scholar]
- Ramos J.W., Hughes,P.E., Renshaw,M.W., Schwartz,M.A., Formstecher, E., Chneiweiss,H. and Ginsberg,M.H. (2000) Death effector domain protein PEA-15 potentiates Ras activation of extracellular signal receptor-activated kinase by an adhesion-independent mechanism. Mol. Biol. Cell, 11, 2863–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson F.L., Whitehurst,A.W., Raman,M. and Cobb,M.H. (2002) Identification of novel point mutations in ERK2 that selectively disrupt binding to MEK1. J. Biol. Chem., 277, 14844–14852. [DOI] [PubMed] [Google Scholar]
- Roth W., Stenner-Liewen,F., Pawlowski,K., Godzik,A. and Reed,J.C. (2002) Identification and characterization of DEDD2, a death effector domain-containing protein. J. Biol. Chem., 277, 7501–7508. [DOI] [PubMed] [Google Scholar]
- Sánchez R. and Sâli,A. (1997) Evaluation of comparative protein structure modeling by MODELLER-3. Proteins, 1, 50–58. [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D., Yang,S.-H. and Galanis,A. (2000) Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci., 25, 448–453. [DOI] [PubMed] [Google Scholar]
- Sippl M.J. (1993) Recognition of errors in three-dimensional structures of proteins. Proteins, 17, 355–362. [DOI] [PubMed] [Google Scholar]
- Stegh A.H., Schickling,O., Ehret,A., Scaffidi,C., Peterhänsel,C., Hofmann,T.G., Grummt,I., Krammer,P.H. and Peter,M.E. (1998) DEDD, a novel death effector domain-containing protein, targeted to the nucleolus. EMBO J., 17, 5974–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T., Adachi,M., Moriguchi,T. and Nishida,E. (2000) A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol., 2, 110–116. [DOI] [PubMed] [Google Scholar]
- Tanoue T., Maeda,R., Adachi,M. and Nishida,E. (2001) Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J., 20, 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia L.A., Ayres,T.M., Wong,G.H. and Goeddel,D.V. (1993) A novel domain within the 55 kd TNF receptor signals cell death. Cell, 74, 845–853. [DOI] [PubMed] [Google Scholar]
- Telliez J.B., Xu,G.Y., Woronicz,J.D., Hsu,S., Wu,J.L. and Lin,L. (2000) Mutational analysis and NMR studies of the death domain of the tumor necrosis factor receptor-1. J. Mol. Biol., 300, 1323–1333. [DOI] [PubMed] [Google Scholar]
- Thomas L.R., Stillman,D.J. and Thorburn,A. (2002) Regulation of Fas-associated death domain interactions by the death effector domain identified by a modified reverse two-hybrid screen. J. Biol. Chem., 277, 34343–34348. [DOI] [PubMed] [Google Scholar]
- Vuister G.W., Tessari,M., Karini-Nejad,Y. and Whitehead,B. (1999) Pulse sequences for measuring coupling constants. In Krishna,N.R. and Berliner,L.J. (eds), Biological Magnetic Resonance 16. Kluwer Press, New York, NY, pp. 195–259.
- Werner M.H., Gupta,V., Lambert,L.J. and Nagata,T. (2001) Uniform 13C/15N-labeling of DNA by tandem repeat amplification. Methods Enzymol., 338, 283–304. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Sykes,B.D. and Richards,F.M. (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry, 31, 1647–1651. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. (1986) NOE observable 1H–1H distances in proteins. NMR of Proteins and Nucleic Acids. John Wiley & Sons, New York, NY, 117–129.
- Xiao T., Towb,P., Wasserman,S.A. and Sprang,S.R. (1999) Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell, 99, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Hegde,R., Srinivasula,S.M., Fernandes-Alnemri,T. and Alnemri,E.S. (2002) Death effector domain-containing proteins DEDD and FLAME-3 form nuclear complexes with the TFIII102 subunit of human transcription factor IIIC. Cell Death Differ., 9, 439–447. [DOI] [PubMed] [Google Scholar]
- Zhang F., Robbins,D.J., Cobb,M.H. and Goldsmith,E.J. (1993) Crystallization and preliminary X-ray studies of extracellular signal-regulated kinase-2/MAP kinase with an incorporated His-tag. J. Mol. Biol., 233, 550–552. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Redina,O., Altshuller,Y.M., Yamazaki,M., Ramos,J., Chneiweiss,H., Kanaho,Y. and Frohman,M.A. (2000) Regulation of expression of phospholipase D1 and D2 by PEA-15, a novel protein that interacts with them. J. Biol. Chem., 275, 35224–35232. [DOI] [PubMed] [Google Scholar]