Abstract
We have isolated the Xenopus p21-activated kinase 3 (XPak3) by virtue of its expression in the territory of primary neurogenesis in the developing embryo. XPak3, but not the other Pak variants, responds positively to X-Ngnr-1 and negatively to X-Notch-1. A constitutively active form of XPak3, generated by fusing a myristylation signal to the N-terminus (XPak3-myr), induces early cell cycle arrest at high concentrations, while ectopic expression of low amounts induces premature neuronal differentiation. Conversely, XPak3 loss of function achieved by use of an antisense morpholino oligonucleotide increases cell proliferation and inhibits neuronal differentiation; this phenotype is rescued by co-injection of XPak3-myr. We conclude that XPak3 is a novel member of the proneural pathway, functioning downstream of neurogenin to withdraw neuronally programmed cells from the mitotic cell cycle, thus allowing for their differentiation.
Keywords: cell cycle regulation/neurogenesis/Xenopus laevis/XPak3
Introduction
Early neural development in the vertebrate embryo is viewed as progressing in four major steps: competence, specification, commitment and differentiation. Experi mental observations made with amphibian embryos over the past century have led to a model for neural induction that, in its most simplistic version, relies primarily on the secretion of inhibitory signals from the Spemann organizer that counteract bone morphogenetic protein (BMP) signalling (reviewed in Hemmati-Brivanlou and Melton, 1997; Sasai and DeRobertis, 1997; Harland, 2000; Wilson and Edlund, 2001).
The gene network that leads to neuronal differentiation is governed by a basic helix–loop–helix (bHLH) transcription factor, referred to as neurogenin-related-1 (X-Ngnr-1) in Xenopus laevis (Ma et al., 1996). This protein activates a cascade of downstream events that includes the transcriptional activation of a set of genes equally encoding transcription regulators that are best defined as differentiation factors; these differentiation factors, such as X-NeuroD (Lee et al., 1995), X-MyT1 (Bellefroid et al., 1996) and Xebf3 (Pozzoli et al., 2001), are involved in the regulation of marker genes for differentiation, such as N-tubulin (Oschwald et al., 1991). This cascade of reactions, referred to as the proneural pathway, is antagonized by the neurogenic pathway that is governed by Delta/Notch signalling in a process known as lateral inhibition (Coffman et al., 1990; Chitnis et al., 1995; Chitnis and Kintner, 1996; Lamar et al., 2001). Therefore, only cells that express higher levels of proneural gene activity than their neighbours enter a neuronal fate (Van Doren et al., 1992). Interestingly, it has been demonstrated more recently that neuronal fate is promoted by neurogenin at the expense of glial fate and it has been proposed that Notch signalling may thus exert an instructive activity with respect to glial cell differentiation (Tomita et al., 2000; Sun et al., 2001).
Much less is known about the molecular mechanisms that link withdrawal from the mitotic cell cycle and the activity of cell fate determination/differentiation factors. In Xenopus, spatial and temporal patterns of cell cycle activities during early embryogenesis define the neural plate as an area of increased proliferative activity as compared with the non-neural ectoderm (Saka and Smith, 2001), raising the question as to how the developing neurons withdraw from the cell cycle in order to sustain neuronal differentiation. Early observations on Notch signalling had already indicated that deregulated Notch activity results in a prolonged proliferative phase within the anterior neural plate (Coffman et al., 1993). However, the molecular events responsible for this effect have not been elucidated. Rather, more recent studies have identified a regulatory circuit that is involved in proliferation control within the neural plate, but which does not seem to be linked directly to Notch signalling. XBF-1, an anterior neural plate-specific winged helix transcription factor, has been reported to promote proliferation of neuroectodermal cells at a high dose, while lower doses inhibit ectodermal proliferation and induce neural cell fate. The effect of XBF-1 on cell proliferation seems to be mediated by the cyclin-dependent kinase (Cdk) inhibitor p27XIC1, which is a direct target gene for XBF-1 (Hardcastle and Papalopulu, 2000). p27XIC1 previously had been demonstrated to exert a function in cell fate determination in the retina; overexpression of p27XIC1 in retinoblasts increases the number of Müller glial cells at the expense of bipolar neurons, while its inhibition reduces the number of Müller glial cells (Ohnuma et al., 1999). However, expression characteristics of both XBF-1 and p27XIC1 are not compatible with their functioning as cell cycle regulators in the context of primary neurogenesis. Thus, if and how neurogenin is linked to proliferation control still remains to be defined.
Accordingly, we report here our findings on the p21-activated serine/threonine kinase 3 in X.laevis (XPak3). Paks serve as G-protein-activated downstream effectors that have been implicated in modulating the actin cyto skeleton, as well as activating the MAP kinase pathway (Daniels and Bokoch, 1999; Dan et al., 2001). We describe that XPak3 is expressed during primary neurogenesis in a pattern that is comparable with neuronal differentiation markers, such as N-tubulin, and that it is readily induced by ectopic neurogenin. We further show that ectopic activation of XPak3 in Xenopus embryos leads to cell cycle arrest and to premature neuronal differentiation in a dose-dependent manner. Conversely, inhibition of XPak3 by use of a specific morpholino antisense oligonucleotide results in increased cell proliferation, as well as in an inhibition of neuronal differentiation within the territory of XPak3 expression. On the basis of these findings, we propose that XPak3 regulates cell cycle withdrawal in the context of primary neurogenesis in Xenopus.
Results
Cloning and expression of XPak3
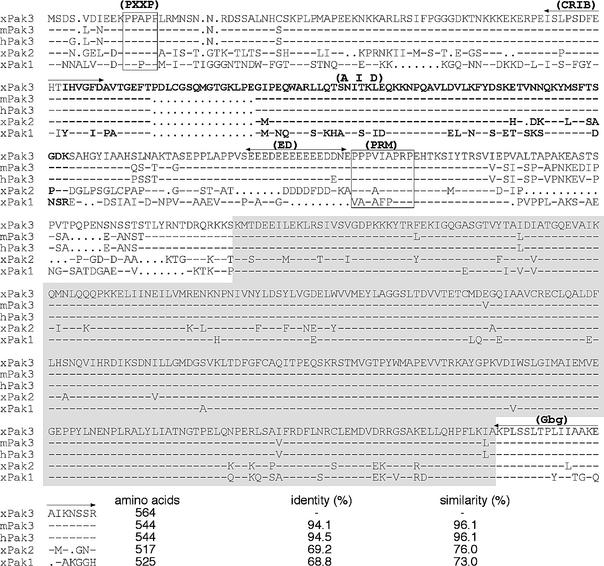
X-Ngnr-1, X-MyT1 and N-tubulin share a characteristic pattern of expression at the open neural plate stage during Xenopus embryogenesis that is defined by three longitudinal stripes on both sides lateral to the dorsal midline (Bellefroid et al., 1996). Since these genes are all part of the same intimately linked gene network, they define a prototype synexpression group (Niehrs and Pollet, 1999). In an attempt to define novel members of this same synexpression group, we have screened ∼1000 randomly selected clones from a tadpole stage head cDNA library by whole-mount in situ hybridization. Clone JS464 is one of several novel members of this synexpression group identified. Nucleotide sequence analysis reveals that it encodes the Xenopus orthologue of the mammalian p21-activated kinase 3 (XPak3). It is clearly distinct from the previously known XPak1 (Faure et al., 1997) and XPak2 (Cau et al., 2000), and most closely related to murine and human Pak3 (Figure 1). Characteristically, this family of kinases contains a conserved regulatory domain in the N-terminal portion, as well as a catalytic domain in the C-terminus (Manser et al., 1994). The proline-rich motif (PXXP), located at the N-terminal edge (Figure 1), has been reported to mediate Pak–Pix–G-protein complex anchorage to the plasma membrane through interaction with the Nck protein (Bokoch et al., 1996; Lu et al., 1997). The Cdc42/Rac1-interacting binding (CRIB) motif, covering the region from amino acid 70 to 88, includes a dimerization motif between amino acids 81 and 87 (Lei et al., 2000). Following these domains, XPak3 has a conserved 15 amino acid insertion. An autoinhibitory domain (AID), partially overlapping with CRIB (Figure 1), was found to inhibit Pak in trans (Frost et al., 1998). C-terminal to this domain, an acidic domain (ED) with an as yet unknown function is followed by a proline-rich motif (PRM) to which the guanine nucleotide exchange factor PIX binds in a G-protein-dependent manner (Daniels et al., 1999). At the C-terminus, XPak3 contains a G-β/γ subunit-binding domain (Gbg) (Leeuw et al., 1998).
Fig. 1. XPak3 sequence analysis. The XPak3 protein sequence, as predicted from the cDNA, was aligned with the murine (m) and human (h) orthologues, as well as with the two other Xenopus variants, XPak1 and XPak2 (Bagrodia et al., 1995; Faure et al., 1997; Allen et al., 1998; Cau et al., 2000). Dashes represent identical amino acids while dots indicate the absence of amino acids. The kinase domain is shaded in grey. The Cdc42/Rac1-interacting binding (CRIB) domain is indicated and partially overlaps with the autoinhibitory domain (AID) indicated in bold. The proline-rich motifs (PXXP and PRM), specific for Nck and Pix interactions, are boxed. The G-β/γ-binding domain (Gbg) is conserved at the C-terminal edge. The percentages of sequence identity and similarity are indicated.
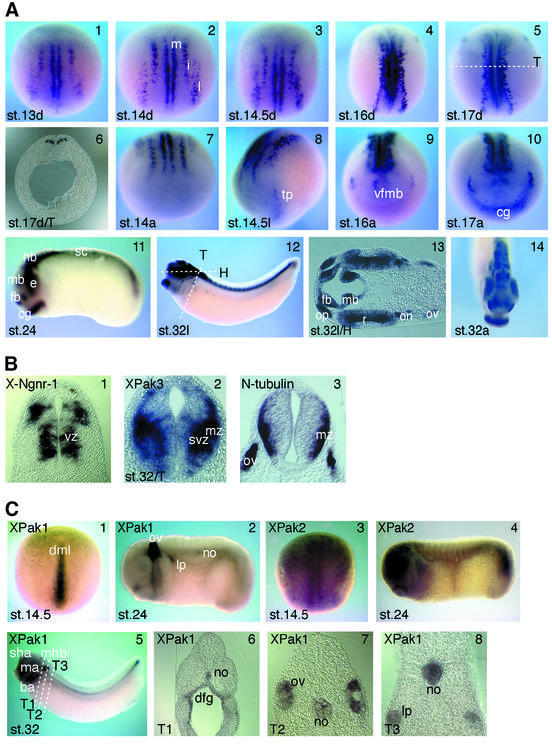
Embryonic expression of XPak3 was analysed using RT–PCR and whole-mount in situ hybridization. The first method reveals the presence of XPak3 transcripts from the egg to tailbud stage embryos (not shown). However, by whole-mount in situ hybridization, XPak3 transcripts are first detected at late gastrula/early neurula stages of development within the posterior neuroectoderm (Figure 2A). Cells expressing XPak3 fall into a pattern that is reminiscent of the neuron-specific N-tubulin gene, whose expression marks the territories of primary neurons (Chitnis et al., 1995). XPak3 expression is restricted to three groups of cells arranged in a bilaterally symmetrical pattern on either side of the dorsal midline of the posterior neural plate. The same pattern of expression is maintained in later stage neurula embryos, as the lateral stripes of XPak3-expressing cells extend along and converge towards the dorsal midline. Expression of XPak3 in the anterior neural plate starts at midneurulation (stage 14.5) in the area of the tri geminal placodes (Figure 2A, panel 8). At embryonic stage 16, a second group of XPak3-expressing cells appears more centrally in a site corresponding to the presumptive brain (Figure 2A, panel 9). In contrast to the known marker genes of neurogenesis, XPak3 is strongly expressed in the cement gland, starting at embryonic stage 17 (Figure 2A, panel 10). During tailbud and tadpole stages of development, XPak3 transcripts are found in the entire central nervous system. In the neural tube, X-Ngnr-1 and XPak3 exhibit predominantly complementary patterns of expression. While XPak3 is localized in the subventricular and marginal zones, X-Ngnr-1 expression defines the proliferating cells of the ventricular zone. Furthermore, the XPak3 expression territory includes the N-tubulin-expressing, differentiated cells of the outer margin (Figure 2B). Thus, the pattern of XPak3 expression in the neural tube already suggests that this kinase may have a function downstream of X-Ngnr-1 and upstream of N-tubulin.
Fig. 2. Differential expression patterns of XPak genes during Xenopus development. (A) Spatial and temporal expression of XPak3 was analysed at different stages (st.) of neurogenesis using whole-mount in situ hybridization. (1–5) XPak3 transcripts are found in three bilateral stripes, lateral (l), intermediate (i) and medial (m), in the posterior neural plate. Embryos are shown in a dorsal (d) view with anterior up. (6) In a transverse (T) section of a stage 17 embryo at the level shown in panel 5 (white dotted line), XPak3 transcripts are detectable in the deep layer of the neuroectoderm in regions corresponding to the medial, intermediate and lateral stripes. Panels 7–10 show the expression in the anterior neural plate; (8) XPak3-expressing cells first appear in the trigeminal placode (tp), then (9) in a centrally located, semicircular array corresponding to a ventral area within the prospective fore-/midbrain where neurons first develop (vfmb) and (10) in the cement gland (cg). (11–14) At later stages of neurogenesis, XPak3 is expressed in the forebrain (fb), midbrain (mb), hindbrain (hb), retina (r), optic nerve (on), olfactory placode (op), otic vesicle (ov) and cement gland (cg): (11 and 12) lateral view (l), (13) horizontal (H) section of the head region at the level indicated in panel 12, (14) anterior view (a). (B) Differential expression of neuronal marker genes in the neural tube of stage 32 embryos. (1) X-Ngnr-1 is expressed in the ventricular zone (vz), (3) N-tubulin in the marginal zone (mz) and (2) XPak3 in the marginal and subventricular zone (svz). The level of the transverse (T) sections is shown in (A) panel 12. (C) Expression patterns of XPak1 and XPak2. (1) XPak1 transcripts are present in the dorsal midline (dml) and (2 and 5–8) later in the otic vesicle, mandibular arch (ma), branchial arch (ba), midbrain–hindbrain boundary (mhb), stomodeal–hypophyseal anlage (sha), lateral placode (lp), notochord (no) and dorsal foregut (dfg). (3) Early on, XPak2 is expressed ubiquitously in the neural plate and (4) later the expression becomes stronger in the brain, eye and tailbud. Embryos are shown in (1 and 3) dorsal view with anterior up and (2, 4 and 5) lateral view. (6–8) Levels of the transverse (T) sections (T1–3) are indicated in panel 5.
Other members of the Xenopus Pak gene family are also expressed differentially during development, but in patterns clearly distinct from that of XPak3. In neurula stage embryos, XPak1 is expressed mainly in the notochord (Figure 2C, panel 1). During later phases of embryogenesis, XPak1 expression is maintained in the notochord but also prominently detected in tissues derived from dorsolateral cranial placodes (Figure 2C, panels 2 and 5–8; see also Islam et al., 2000). XPak2 is expressed more ubiquitously in both early and late stages of Xenopus embryogenesis (Figure 2C, panels 3 and 4).
XPak3 expression is positively regulated by X-Ngnr-1 and negatively by Notch signalling
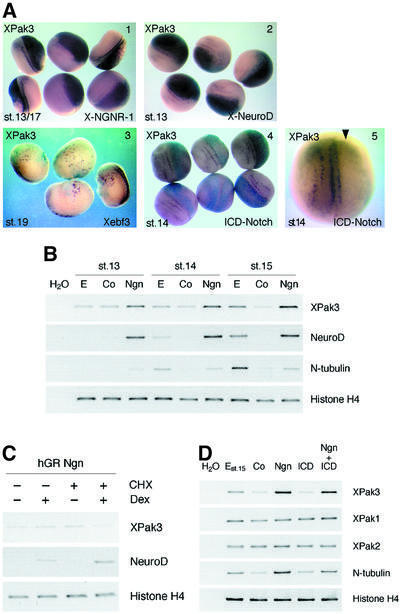
As a member of the X-Ngnr-1/X-MyT1/N-tubulin synexpression group, XPak3 expression is likely to be responsive to proneural and neurogenic genes. To address this question directly, embryos were injected at the two-cell stage with synthetic X-Ngnr-1 mRNA, along with LacZ RNA as a tracer, and analysed at the neural plate stage for XPak3 expression, as well as with X-gal staining to reveal the distribution of the injected RNAs. The same type of analysis was carried out with other proneural regulators such as X-NeuroD, functioning downstream of X-Ngnr-1 (Lee et al., 1995; Ma et al., 1996), and Xebf3, functioning downstream of NeuroD (Pozzoli et al., 2001). The results obtained (Figure 3A) reveal that overexpression of X-Ngnr-1 or X-NeuroD both strongly activate ectopic fields of XPak3 expression. In contrast, Xebf3 overexpression induces merely a scattered pattern of a few ectopic, XPak3-expressing cells, similar to the effects that have been described for Xebf3 on N-tubulin expression (Pozzoli et al., 2001). Based on these results, and on the spatial correlation of X-Ngnr-1 and XPak3 expression described above, we conclude that XPak3 is part of the proneural gene cascade, functioning downstream of X-Ngnr-1, NeuroD and Xebf3.
Fig. 3. Regulation of XPak3 expression in both embryos and animal caps. (A) XPak3 expression is positively regulated by proneural genes and negatively regulated by X-Notch-1. Xenopus albino embryos were injected into one cell at the two-cell stage with synthetic RNA encoding X-Ngnr-1, X-NeuroD, Xebf3 and ICD-Notch, along with LacZ RNA as tracer. Embryos were fixed at the neurula stage, then stained with X-gal (light blue) and analysed by whole-mount in situ hybridization for XPak3 expression (purple). (1) X-Ngnr-1 and (2) X-NeuroD injection results in strong ectopic expression of XPak3 (100 and 96%, n = 76 and 64, respectively). (3) Xebf3 is sufficient to turn on ectopically the expression of XPak3 but in a weak and scattered manner (62%, n = 48). (4 and 5) The activated form of X-Notch-1, ICD-Notch, blocks XPak3 expression (100%, n = 56). (B) X-Ngnr-1 is sufficient to activate XPak3 expression in animal caps. Pigmented embryos were injected with 50 pg of X-Ngnr-1 mRNA. Animal caps were isolated and cultivated to the indicated stages. Total RNAs were prepared from the caps and analysed by RT–PCR for XPak3 expression, using NeuroD, N-tubulin and histone H4 as controls. For each stage, E (embryos), Co (control caps) and Ngn (neurogenin-injected caps) are shown. (C) XPak3 is not induced directly by X-Ngnr-1. Embryos were injected into two blastomeres of the two-cell stage with hGR Ngn. Cycloheximide (CHX) treatment was from stage 11.5 followed by 2.5 h treatment with or without dexamethasone (Dex). Total RNAs were prepared from these caps and analysed by RT–PCR for XPak3, NeuroD and histone H4. (D) XPak3, but not XPak1 and XPak2, is activated by X-Ngnr-1 in animal cap explants. Pigmented embryos were injected with X-Ngnr-1 RNA and/or ICD-Notch RNA and allowed to grow up to stage 9. Animal caps were isolated and cultivated to stage 15. Total RNAs were prepared from the caps and analysed by RT–PCR for the genes indicated. From left to right, the columns represent: water control (H2O), uninjected control embryos stage 15 (E), uninjected control caps (Co), X-Ngnr-1-injected caps (Ngn), ICD-Notch-injected caps (ICD) and X-Ngnr-1/ICD-Notch-co-injected caps (Ngn + ICD).
As a component of the proneural gene network, XPak3 is predicted to be subject to lateral inhibition. To test this, we examined XPak3 expression in ICD-Notch-injected embryos (Figure 3A, panels 4 and 5). The results obtained demonstrate that activation of Notch signalling represses XPak3 expression, strongly indicating that the scattered pattern of XPak3 expression is generated by lateral inhibition. We further tested the regulation of XPak3 transcription by use of the animal cap system (Figure 3B–D). A low level of XPak3, that may reflect the maternal contribution, is detected in naive control caps. Injection of X-Ngnr-1 is found to be sufficient to activate XPak3 transcription. However, unlike with NeuroD, which is also induced by X-Ngnr-1, the inductive effect on XPak3 expression increases with time (from the equivalent of stage 13 to the equivalent of stage 15) (Figure 3B), resembling the temporal increase of N-tubulin expression (Koyano-Nakagawa et al., 1999) and indicating that XPak3, just like N-tubulin, is not a direct target of X-Ngnr-1. Indeed, use of an inducible version of neurogenin in the presence of a protein synthesis inhibitor reveals that, in contrast to NeuroD, XPak3 cannot be activated directly (Figure 3C). We also tested if Notch signalling is able to repress XPak3 in the presence of ectopic X-Ngnr-1, as has been found to be the case for other downstream regulators of the proneural network, such as X-MyT1 (Bellefroid et al., 1996). The experiment reveals that ICD-Notch is unable to repress X-Ngnr-1-induced XPak3 gene transcription in the animal cap system (Figure 3D). The same results were obtained in animal caps neuralized by Noggin (data not shown). The other Pak variants, XPak1 and XPak2, respond neither to X-Ngnr-1 nor to lateral inhibition (Figure 3D). Thus, we conclude that XPak3, but not other Pak variants, is an indirect downstream target of X-Ngnr-1 and that its expression is most likely to be negatively regulated by Notch signalling via inhibition of X-Ngnr-1 gene transcription.
Modulation of XPak3 activities affects neuronal differentiation
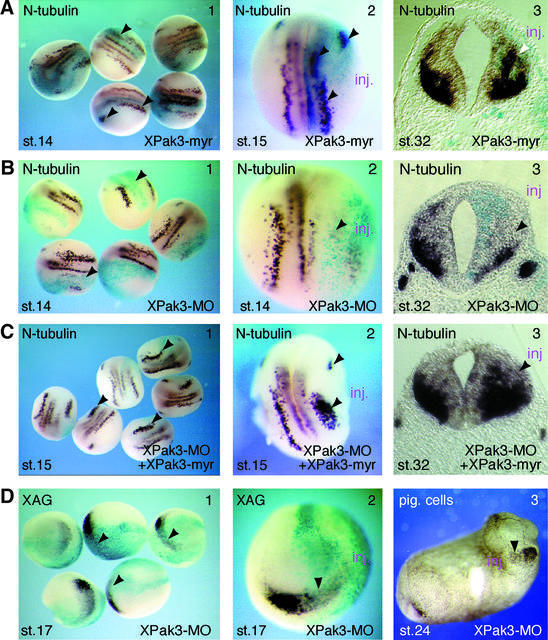
The expression pattern of XPak3, as well as its mode of transcription regulation, suggest that XPak3 may play an active role during neuronal differentiation in Xenopus embryos. However, misexpression of XPak3 by means of mRNA injection had obvious effects neither on the development of the embryos nor on the expression of N-tubulin (data not shown). Previous studies on p21-activated protein kinases had demonstrated that recruitment of human Pak1 to the plasma membrane leads to up-regulation of its kinase activity (Lu and Mayer, 1999). Similarly, in Drosophila, localization of the XPak3 homologue to the cell membrane by artificial myristylation was found to be sufficient to activate its kinase function constitutively (Hing et al., 1999). Accordingly, we generated a membrane-anchored version of XPak3, termed XPak3-myr, by fusion of a myristylation signal to the N-terminus of the wild-type protein. Microinjection of low doses of the mRNA encoding this construct has no overt effect on the morphology of the embryos, but results in specific consequences for the expression of neuronal marker genes. As shown in Figure 4A, panels 1 and 2, microinjection of up to 10 pg of XPak3-myr mRNA enhances N-tubulin staining in the domains where neurons normally form in early neurula (stage 14–15) embryos. Transverse sections of a microinjected tadpole (stage 32) embryo reveal that overexpression of XPak3-myr expands/shifts the N-tubulin expression domain towards the ventricular zone at the expense of the expression in the marginal zone (Figure 4A, panel 3), indicating premature neuronal differentiation. Importantly, XPak3-myr misexpression has no overt effect on the regulation of proneural factors such as X-Ngnr-1 (not shown), indicating that the above phenotype is not the result of feedback regulation.
Fig. 4. XPak3 activation induces premature neuronal differentiation. (A–C) Modulation of XPak3 activity influences neuronal differentiation. Xenopus albino embryos were injected into one blastomere at the two-cell stage with (A) 10 pg of XPak3-myr RNA, (B) 2.5 pmol of XPak3-MO and (C) 2.5 pmol of XPak3-MO/12.5 pg XPak3-myr RNA, all with LacZ RNA as tracer. Embryos were fixed at neurula and tailbud stages, then stained with X-gal (light blue) and analysed for neuronal differentiation, as marked by N-tubulin expression. (A3, B3 and C3) Transverse sections of stage 32 embryos were prepared at the level of the hindbrain. (A1 and 2) XPak3-myr injection results in increased N-tubulin expression within the trigeminal placode and within the territories of primary neurons, as indicated by arrowheads (71%, n = 46). (A3) In the neural tube, XPak3-myr injections expand/shift the expression domain of N-tubulin from the marginal zone towards the ventricular zone (white arrowhead). (B1–3) XPak3-MO injections strongly inhibit N-tubulin expression within both the trigeminal placode and the territories of primary neurons (100%, n = 92), and within the neural tube. (C1 and 2) Co-injection of XPak3-MO and XPak3-myr RNA results in rescue of N-tubulin expression in a pattern that is not identical to, but reminiscent of the normal one (84%, n = 73). (C3) In the neural tube, co-injection increases/expands the expression domain of N-tubulin similarly to XPak3-myr injection (see A, panel 3). (D) XPak3 is necessary for cement gland formation. Embryos were injected into one blastomere of two-cell stage embryos with 2.5 pmol of XPak3-MO and LacZ RNA as tracer. One batch of these embryos was fixed at stage 17, stained for β-galactosidase activity (light blue) and bleached. The other batch was fixed at stage 24. (1 and 2) By whole-mount in situ hybridization, XAG is found to be suppressed, as marked by arrowheads (100%, n = 71). (3) In stage 24 embryos, pigmented cement gland cells are missing on the injected side.
Conversely, inhibition of XPak3 expression using a morpholino antisense oligonucleotide (XPak3-MO) correlates with an inhibition of neuronal differentiation, as marked by the suppression of N-tubulin gene transcription both in the open neural plate and in the neural tube (Figure 4B). Importantly, co-injection of both the XPak3-MO and XPak3-myr (but not XPak3) mRNA is sufficient to rescue the expression of the N-tubulin gene in a pattern that is not identical to, but reminiscent of the normal one (Figure 4C, panels 1 and 2). Furthermore, an increased/expanded expression of N-tubulin in the neural tube (Figure 4C, panel 3) indicates that this rescue experiment correlates with the gain-of-function effect reported above (Figure 4A, panel 3). XPak3 expression is also prominent in the cement gland; suppression of XPak3 expression leads to an inhibition of cement gland formation, as evident from the marked reduction of the cement gland marker XAG in neurula (stage 17) embryos (Figure 4D, panels 1 and 2), as well as from the reduced pigment of later (stage 24) embryos (Figure 4D, panel 3). Taken together, our findings indicate that ectopic activation of XPak3 correlates with premature/increased neuronal differentiation, whereas XPak3 loss of function results in an inhibition of neuronal differentiation.
XPak3 inhibits cell proliferation
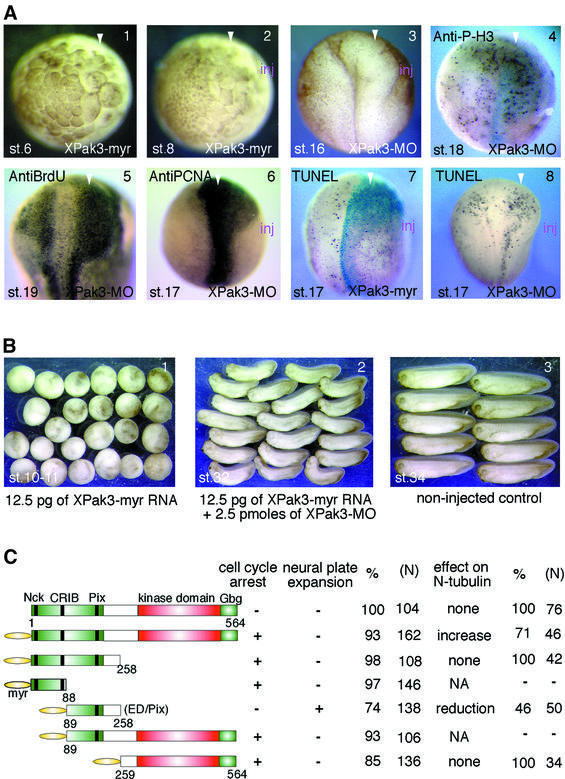
Microinjection of XPak3-myr mRNA leads to cell cycle arrest in a dose-dependent manner; high amounts (>50 pg) induce developmental arrest prior to mid-blastula transition (MBT), while intermediate amounts give a similar phenotype after MBT (Figure 5A, panels 1 and 2); low amounts (<10 pg) produce no morphological abnormality (Figure 4A, panels 1 and 2). Conversely, microinjection of XPak3-MO results in an expansion of the anterior neural plate, which may reflect increased proliferative activity (Figure 5A, panel 3). In order to characterize these effects further, morpholino-injected embryos were stained for bromodeoxyuridine (BrdU) incorporation, proliferating cell nuclear antigen (PCNA) expression and histone H3 phosphorylation. XPak3-MO-injected embryos show significantly increased staining within the expanded area of the neural plate in all three assays (Figure 5A, panels 4–6), providing further evidence for increased proliferation being responsible for the morphological effect observed. We further analysed if increasing or decreasing XPak3 activity would influence apoptosis in Xenopus embryos using the TUNEL assay (Hensey and Gautier, 1998). Neither treatment exhibits strong effects on the relative number of TUNEL-positive cells (Figure 5A, panels 7 and 8). We conclude that the inhibition of XPak3 activity results in an expansion of the neural plate, which is a consequence of increased proliferative activity.
Fig. 5. XPak3 is involved in cell cycle regulation. (A) Pigmented embryos were injected into one blastomere of two- or four-cell stage embryos with either XPak3-myr RNA or XPak3-MO; these embryos were allowed to develop to the stages indicated. The arrowhead shows the injected side: (1) 50 pg of Xpak3-myr induce cell cycle inhibition before MBT (100%, n = 75); (2) 25 pg give a similar phenotype after MBT (100%, n = 83). (3) Microinjection of 2.5–5 pmol of XPak3-MO induces neural plate expansion (67%, n = 116); (4–6) immunostained embryos showing increased signal on the injected side (arrowhead) for phosphorylated histone H3 (54%, n = 54), BrdU (58%, n = 48) and PCNA (79%, n = 62). (7 and 8) TUNEL staining of embryos injected with 10 pg of XPak3-myr RNA or 2.5 pmol of XPak3-MO shows no significant effect on apoptosis; rather, apoptotic cell death appears to correlate with increased proliferation. (B) Embryos were injected into one blastomere of two-cell stage embryos with either XPak3-myr RNA alone or XPak3-myr RNA and XPak3-MO together. By stage 10–11, XPak3-myr-injected embryos stopped growing (100%, n = 107), while XPak3-myr/XPak3-MO-co-injected ones continued to develop (79%, n = 95). Embryos were fixed at the stages indicated. (C) Deletion mutants of XPak3 were generated and tested for their influence on cell cycle regulation and neuronal differentiation. The corresponding phenotypes are indicated.
Injection of intermediate doses (12.5 pg) of XPak3-myr mRNA was found to induce developmental arrest at gastrula/neurula stages. Strikingly, XPak3-MO, which is specific for the endogenous transcript but unable to block expression of the injected construct, was used successfully to rescue this effect (Figure 5B, panels 1 and 2), suggesting that the endogenous maternal and/or zygotic XPak3 mRNA acts synergistically with the injected, constitutively active construct in cell cycle arrest.
In order to investigate which of the functional domains in XPak3 are responsible for the cell cycle arrest activity described above, a series of XPak3 deletion mutants, all containing the myristylation signal at their N-terminus, was generated (Figure 5C). Overexpression of different deletion mutants maintaining either the PXXP/CRIB or the kinase/Gbg motifs, respectively, reveals that they are equally capable of inducing cell cycle arrest; however, without overt effect on neuronal differentiation. Interest ingly, an internal fragment (ED/Pix), which contains the Pix-binding site and the AID, was found to induce neural plate expansion, as well as a reduction of N-tubulin expression in 46% of the injected embryos. Furthermore, this same deletion mutant was found to impair formation of the anterior–posterior axis in ∼40% of the injected embryos (data not shown), perhaps indicating an inhibitory cross-reaction with XPak1, which is strongly expressed in the notochord (see above). Thus, this deletion analysis reveals that the PXXP/CRIB motifs of the regulatory domain or the kinase domain are sufficient to induce cell cycle arrest, whereas the ED/Pix fragment seems to exert a dominant-inhibitory effect. We conclude that cell cycle arrest is not sufficient to induce neuronal differentiation.
Discussion
Results reported in this communication define XPak3 as a functional link between the proneural gene network and cell cycle inhibition, which is a prerequisite for neuronal differentiation. We propose that X-Ngnr-1, a key transcription regulator of neurogenesis, indirectly activates XPak3, which promotes cell cycle withdrawal and thus allows neuronal differentiation to proceed. This hypothesis is supported by three major arguments: (i) the spatial and temporal patterns of XPak3 expression, as well as its ectopic activation upon misexpression of proneural regulators; (ii) the inhibitory effect of a constitutively active variant of XPak3 on cell cycle activity that correlates with premature neuronal differentiation; and (iii) hyperproliferation of neural tissue that correlates with an inhibition of neuronal differentiation, as observed upon reducing XPak3 activity in Xenopus embryos.
XPak3 acts as cell cycle regulator in the context of primary neurogenesis
Paks define a family of protein kinases with numerous functions ranging from the control of the MAP kinase cascade to cytoskeletal rearrangements, which also provide a link to the cell cycle activities ascribed to these enzymes (Bagrodia and Cerione, 1999). Similar to what we report for XPak3 with respect to its function in the context of cell cycle regulation, the two other, closely related but differentially expressed Pak variants in Xenopus, XPak1 and XPak2, previously had been reported to be involved in cell cycle control, negatively regulating the G2/M phase transition during the process of oocyte maturation. Interestingly, and similar to the results that we have obtained upon misexpression of an activated form of XPak3, microinjection of a truncated, catalytically active form of Pak1 was reported to induce cleavage arrest in early Xenopus embryos (Rooney et al., 1996). Further more, it has been demonstrated that a constitutively active deletion mutant derived from XPak1 blocks progesterone-induced oocyte maturation (Faure et al., 1997, 1999). A negative regulatory function in meiotic cell cycle progression has also been reported for XPak2. Phosphorylation of XPak2, which may be directly linked to maturation-promoting factor (MPF) activity, was reported to result in its inactivation, allowing for maturation to proceed to completion (Cau et al., 2000). At present, we do not know which partner molecules would be relevant for the cell cycle regulatory activities that we describe for XPak3 in the context of neuronal differentiation, but the more common functional link of different Pak variants to cell cycle control indicates that these different Pak proteins may be elements of similarly organized functional networks. Interestingly, it has been found that the Drosophila Pak is required for photoreceptor axon guidance (Hing et al., 1999). We find that in the development stages following primary neurogenesis, XPak3 is strongly expressed throughout the central nervous system and in the developing eye; in the late stages, XPak3 may serve additional functions, perhaps including axonal guidance. In Xenopus, this question remains to be addressed experimentally.
Cell cycle regulatory activities in the context of different aspects of neural development in Xenopus have been ascribed to p27XIC1, a Cdk inhibitor (Ohnuma et al., 1999). During earlier phases of development, regulation of p27XIC1 expression may depend on XBF-1, a winged-helix transcription factor that is expressed specifically in the anterior neural plate. Similar to the effects that we have observed upon inactivation of XPak3, high concentrations of XBF-1 result in suppression of neuronal differentiation and expansion of the undifferentiated neuroectoderm, as well as in a direct suppression of p27XIC1 gene transcription (Bourguignon et al., 1998; Hardcastle and Papalopulu, 2000). However, the pattern of expression for these genes does not match that characteristic for N-tubulin, i.e. primary neurogenesis.
Ectopic activation of Delta/Notch signalling in Xenopus embryos also produces a phenotype that is reminiscent of that obtained upon XPak3 loss of function as described in this study, namely inhibition of neuronal differentiation and expansion of the neural plate (Coffman et al., 1993). This situation correlates with our observation of the strong inhibitory effect that ectopic Notch has on the expression of XPak3 in Xenopus embryos. Interestingly, Notch activity has been directly linked to a positive regulation of proliferation; Ronchini and Capobianco (2001) have reported on a direct transcriptional activation of cyclin D1 upon activation of Notch signalling. It is also interesting to note that XBF-1/p27XIC1-mediated proliferation control in the neural plate does not operate via increased Delta/Notch signalling (Bourguignon et al., 1998), while it may positively modulate premature cell cycle exit in the retina (Ohnuma et al., 2002). Thus, at least two mechanistically different pathways appear to exist that ensure cell cycle withdrawal as a prerequisite for separate aspects of neuronal differentiation.
Functional domains in XPak3
Interactions between the N- and C-terminal protein domains of Pak type kinases have been implicated in the negative regulation of their biological activities. Mechanistically, it has been demonstrated recently that Pak1 forms trans-inhibited homodimers in vivo that dissociate upon GTPase binding (Parrini et al., 2002). Recruitment of the Paks to the cell membrane by Nck or other adaptors in response to specific signals also promotes activation (Bokoch et al., 1996; Lu et al., 1997; Hing et al., 1999; Lu and Mayer, 1999). Different signalling pathways can alleviate the autoregulatory inhibition of Paks (Knaus and Bokoch, 1998; Bagrodia and Cerione, 1999). It will be interesting to define the signalling system(s) that regulates Pak3 activity during Xenopus neurogenesis. However, at present, we have no experimental information on this issue. We have observed that misexpression of wild-type XPak3 does not create a phenotypic effect, while adding a membrane anchor uncovers the activities of XPak3 with respect to cell cycle regulation and neuronal differentiation. The same functions can be exerted by a number of different XPak3 fragments. In the case of N-terminal truncations that maintain the kinase domain, it seems likely that these fragments are constitutively active because they lack the ability to form inactive homodimers. For C-terminal truncations that lack the kinase domain, a different mechanism of activation must apply. Such fragments might interact with the endogenous XPak3, thereby disrupting the trans-inhibited homodimers and allowing for the formation of active heterodimers that contain one copy of the endogenous wild-type protein and one copy of the respective N-terminal fragment. How ever, none of these deletion constructs was sufficient to phenocopy the premature neuronal differentiation. Interestingly, an internal fragment appears to act in a dominant-negative fashion, as it exerts the same effects as the morpholino antisense oligonucleotide. It may titrate out regulators of Pak kinase activity that are members of the PIX family (Manser et al., 1998), or it may bind and inhibit the kinase activity as described earlier (Frost et al., 1998). The same fragment also impairs anterior–posterior axis formation, suggesting that XPak1, which is expressed exclusively in the notochord during gastrulation, may be inhibited via the same mechanism.
XPak3 expression is a downstream target within the proneural gene network
We have shown that XPak3 is expressed in scattered cells in areas of the neural plate where primary neurons are being born, as marked by the expression of N-tubulin. Temporally, XPak3 expression in these areas anticipates that of N-tubulin and follows that of X-Ngnr-1. Embryo microinjection experiments suggest that the same factors which can control N-tubulin expression can regulate XPak3 expression. Both genes are activated by X-Ngnr-1 or its downstream targets X-NeuroD and Xebf3. However, compared with X-Ngnr-1 or X-NeuroD, Xebf3 only weakly induces XPak3 in a few scattered cells, suggesting that additional factors may also be required for a more robust activation of XPak3. Both XPak3- and N-tubulin-encoding genes also appear to be negatively regulated by lateral inhibition; animal cap experiments reveal that XPak3 expression is mediated via a regulatory transcription control region that is not directly responsive to lateral inhibition. Taken together, our data place XPak3 downstream of X-Ngnr-1/X-NeuroD/Xebf3 and upstream of N-tubulin within the proneural gene network.
Results obtained upon ectopic activation of XPak3, as well as upon inhibition of XPak3 activity in Xenopus embryos, provide further evidence for a function of this protein in the context of primary neurogenesis. Mis expression of a constitutively active variant of XPak3 enhances and accelerates neurogenesis, whereas suppression of XPak3 reduces neurogenesis, as reflected by the effects on the expression of N-tubulin as a neuronal differentiation marker. Interestingly, Pak3 function has been linked to disturbed neural function in humans; it is mutated in a particular form of X-linked mental retardation, caused by a mutation which results in premature translation termination, thus disrupting kinase function and preventing Gbg motif formation (Allen et al., 1998), or caused by a point mutation (Arg67 for cysteine) that inhibits G-protein-binding activity (Bienvenu et al., 2000). The phenotypic effects observed as a consequence of these different mutations may be the result of disturbed neuronal differentiation.
Materials and methods
Isolation of XPak3 cDNA
Large-scale whole-mount in situ hybridization was performed for screening a tadpole head λZAP express phage cDNA library, in principle as described by Hollemann et al. (1999). Briefly, single recombinant phages were eluted in 96-well microplates. Fluorescein-labelled antisense RNA probes were transcribed from templates obtained by PCR amplification of cDNA inserts from these single phages. Four sets of flat-bottomed 24-well devices for simultaneous whole-mount in situ hybridization were used per round of screening. cDNA clones with a suggestive expression pattern were sequenced and matched with the DDBJ/EMBL/GenBank sequence information. The XPak3 cDNA clone was selected on the basis of its expression similarity to N-tubulin. The DDBJ/EMBL/GenBank accession No. for XPak3 is AF485330.
Embryos, whole-mount in situ assays and histology
Wild-type and albino X.laevis embryos were obtained by hormone-induced egg laying and in vitro fertilization using standard methods as described by Harland (1991). Whole-mount in situ hybridization was performed in principle as described by Harland (1991), with modifications as reported in Hollemann et al. (1999). Whole-mount PCNA staining was performed essentially as described by Paganelli et al. (2001). Whole-mount BrdU staining was performed essentially as decribed by Hardcastle and Papalopulu (2000). Whole-mount phospho-histone H3 staining was performed essentially as described by Saka and Smith (2001). Whole-mount TUNEL was carried out as described by Hensey and Gautier (1998), with the exception that the vitelline membrane was removed by proteinase K treatment, at a concentration of 0.5 µl/ml for 15 min at room temperature. For sectioning, stained and post-fixed embryos were gelatin embedded and vibratome sectioned at 30 µm thickness. Images from embryos and mounted sections were visualized using Normarski optics and recorded with a CCD camera (Sony).
Microinjection and animal cap explants
The entire XPak3-coding region was subcloned into the pCS2+ vector (Turner and Weintraub, 1994). The myr-tagged version was engineered by subcloning the open reading frame (ORF) into the pCS2+ vector and ligating an oligonucleotide (5′-AATTGCCATGGGTTCATCGAAGTCCAAGCCAAAGGACCCATCGCAACGACGACGG-3′) coding for a myristylation signal (MGSSKSKPKDPSQRRR) into the EcoRI site of the resulting construct. XPak3 deletion constructs were generated by PCR, and by restriction digestion with EcoRI in the case of the myristylated N-terminal fragment. After linearization with NotI, the DNA template was transcribed in vitro with SP6 RNA polymerase using the mMESSAGE mMACHINE kit (Ambion). Variable amounts of XPak3 or XPak3-myr capped-RNA, either alone or together with 50 pg of LacZ RNA (Chitnis et al., 1995), were injected in a volume of ∼5 nl into one or two blastomeres of two-cell stage Xenopus embryos. At various stages, as indicated in the text, the injected embryos were either fixed for histological analysis, or fixed, stained with X-gal and analysed by whole-mount in situ hybridization. The antisense morpholino oligo nucleotide XPak3-MO (5′-CAGCACTCCTCACAGCCTCCTGGC-3′) (Gene Tools LLC) was diluted to 1 mM in RNase-free water, aliquoted and stored at –20°C. For each injection, an equivalent of 2.5–5 pmol in 5 nl was administered per blastomere of two- or four-cell stage embryos.
For the animal cap assays, embryos were injected at the two-cell stage into the animal pole of both blastomeres with the following capped RNAs: 100 pg of X-Ngnr-1 (Ma et al., 1996) and 300 pg of X-Notch ICD (Chitnis et al., 1995). Animal caps were dissected from stage 9 embryos, cultivated in 0.5× MBS and harvested until control siblings had reached the appropriate stage.
RT–PCR
The Qiagen RNeasy mini kit was used for RNA isolation from animal caps. All RNA preparations were treated with DNase I (Qiagen) and checked with 32 cycles of histone H4-specific PCR (Niehrs et al., 1994) for DNA contamination. RT–PCR was carried out using the Gene Amp RNA PCR kit from Perkin-Elmer. The following primers, annealing temperatures and cycle numbers were utilized: histone H4, forward (F) 5′-CGGGATAACATTCAGGGTATCACT-3′ and reverse (R) 5′-ATCCATGGCGGTAACTGTCTTCCT-3′, 58°C, 24 cycles (Niehrs et al., 1994); N-tubulin, F 5′-ACACGGCATTGATCCTACAG-3′ and R 5′-AGCTCCTTCGGTGTAATGAC-3′, 58°C, 32 cycles (Oschwald et al., 1991); XPak1, F 5′-GTGTCTGAAGCAGCAGAGGTCCCT-3′ and R 5′-GTCTACGCTGCTTTCTGCTCCATC-3′, 60°C, 30 cycles (Faure et al., 1997); XPak2, F 5′-GTCTGTGCCCTGCACCTAATGCAA-3′ and R 5′-CTTTGCAGCACTGTCTGAATCTCC-3′, 60°C, 31 cycles (Cau et al. 2000); XPak3, F 5′-TGAATGCAAAGACTGCATCTGAGC-3′ and R 5′-GCTCGTGCTTGAGTTTGAGTTTTC-3′, 60°C, 33 cycles. PCR products were separated on 2% agarose gels.
Analysis using the hormone-inducible form of XNgnr-1 (hGR Ngn) was performed as described by Perron et al. (1999).
Acknowledgments
Acknowledgements
We are grateful to Tina Berneking for expert technical assistance. We also thank Kristine Henningfeld for revising this manuscript, G.G.Consalez for providing the Xebf3 construct, and other colleagues from the Xenopus community for providing the different plasmids utilized in this study. This work was supported by funds from the Deutsche Forschungsgemeinschaft to T.P.
References
- Allen C.M., Gleeson,J.G., Bagrodia,S., Partington,M.W., MacMillan,J.C., Cerione,R.A., Mulley,J.C. and Walsh,C.A. (1998) Pak3 mutation in nonsyndromic X-linked mental retardation. Nat. Genet., 20, 25–30. [DOI] [PubMed] [Google Scholar]
- Bagrodia S. and Cerione,R.A. (1999) Pak to the future. Trends Cell Biol., 9, 350–355. [DOI] [PubMed] [Google Scholar]
- Bagrodia S., Taylor,S.J., Creasy,C.L., Chernoff,J. and Cerione,R.A. (1995) Identification of a mouse p21Cdc42/Rac activated kinase. J. Biol. Chem., 270, 22731–22737. [DOI] [PubMed] [Google Scholar]
- Bellefroid E.J., Bourguignon,C., Holleman,T., Ma,Q., Anderson,D.J., Kintner,C. and Pieler,T. (1996) X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell, 87, 1191–1202. [DOI] [PubMed] [Google Scholar]
- Bienvenu T. et al. (2000) Missense mutation in Pak3, R67C, causes X-linked nonspecific mental retardation. Am. J. Med. Genet., 93, 294–298. [DOI] [PubMed] [Google Scholar]
- Bokoch G.M., Wang,Y., Bohl,B.P., Sells,M.A., Quilliam,L.A. and Knaus,U.G. (1996) Interaction of the Nck adapter protein with p21-activated kinase (PAK1). J. Biol. Chem., 271, 25746–25749. [DOI] [PubMed] [Google Scholar]
- Bourguignon C., Li,J. and Papalopulu,N. (1998) XBF-1, a winged helix transcriprion factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development, 125, 4889–4900. [DOI] [PubMed] [Google Scholar]
- Cau J., Faure,S., Vigneron,S., Labbé,J.C., Delsert,C. and Morin,N. (2000) Regulation of Xenopus p21-activated kinase (X-PAK2) by Cdc42 and maturation-promoting factor controls Xenopus oocyte maturation. J. Biol. Chem., 275, 2367–2375. [DOI] [PubMed] [Google Scholar]
- Chitnis A. and Kintner,C. (1996) Sensitivity of proneural genes to lateral inhibition affects the pattern of primary neurons in Xenopus embryos. Development, 122, 2295–2301. [DOI] [PubMed] [Google Scholar]
- Chitnis A., Henrique,D., Lewis,J., Ish-Horowicz,D. and Kintner,C. (1995) Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature, 375, 761–766. [DOI] [PubMed] [Google Scholar]
- Coffman C., Harris,W. and Kintner,C. (1990) Xotch, the Xenopus homolog of Drosophila Notch. Science, 249, 1438–1441. [DOI] [PubMed] [Google Scholar]
- Coffman C.R., Skoglund,P., Harris,W.A. and Kintner,C.R. (1993) Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell, 73, 659–671. [DOI] [PubMed] [Google Scholar]
- Dan I., Watanabe,N.M. and Kusumi,A. (2001) The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol., 11, 220–230. [DOI] [PubMed] [Google Scholar]
- Daniels R.H. and Bokoch,G.M. (1999) p21-activated protein kinase: a crucial component of morphological signaling? Trends Biochem. Sci., 24, 350–355. [DOI] [PubMed] [Google Scholar]
- Daniels R.H., Zenke,F.T. and Bokoch,G.M. (1999) αPix stimulates p21-activated kinase activity through exchange factor-dependent and -independent mechanisms. J. Biol. Chem., 274, 6047–6050. [DOI] [PubMed] [Google Scholar]
- Faure S., Vigneron,S., Dorée,M. and Morin,N. (1997) A member of the Ste20/PAK family of protein kinases is involved in both arrest of Xenopus oocytes at G2/prophase of the first meiotic cell cycle and in prevention of apoptosis. EMBO J., 16, 5550–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Vigneron,S., Galas,S., Brassac,T., Delsert,C. and Morin,N. (1999) Control of G2/M transition in Xenopus by a member of the p21-activated kinase (PAK) family: a link between protein kinase A and PAK signaling pathways? J. Biol. Chem., 274, 3573–3579. [DOI] [PubMed] [Google Scholar]
- Frost J.A., Khokhlatchev,A., Stippec,S., White,M.A. and Cobb,M.H. (1998) Differential effects of PAK1-activating mutation reveal activity-dependent and -independent effects on cytoskeletal regulation. J. Biol. Chem., 273, 28191–28198. [DOI] [PubMed] [Google Scholar]
- Hardcastle Z. and Papalopulu,N. (2000) Distinct effects of XBF-1 in regulating the cell cycle inhibitor p27(XIC1) and imparting a neural fate. Development, 127, 1303–1314. [DOI] [PubMed] [Google Scholar]
- Harland R. (1991) In situ hybridization: an improved whole mount method for Xenopus embryos. Methods Cell. Biol., 36, 685–695. [DOI] [PubMed] [Google Scholar]
- Harland R. (2000) Neural induction. Curr. Opin. Genet. Dev., 10, 357–362. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A. and Melton,D. (1997) Vertebrate neural induction. Annu. Rev. Neurosci., 20, 43–60. [DOI] [PubMed] [Google Scholar]
- Hensey C. and Gautier,J. (1998) Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev. Biol., 203, 36–48. [DOI] [PubMed] [Google Scholar]
- Hing H., Xiao,J., Harden,N., Lim,L. and Zipursky,S.L. (1999) Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell, 97, 853–863. [DOI] [PubMed] [Google Scholar]
- Hollemann T., Panitz,F. and Pieler,T. (1999) In situ hybridization techniques with Xenopus embryos. In Richter,J.D. (ed.), A Comparative Methods Approach to the Study of Oocytes and Embryos. Oxford University Press, New York, NY, pp. 279–290.
- Islam N., Poitras,L. and Moss,T. (2000) The cytoskeletal effector xPAK1 is expressed during both ear and lateral line development in Xenopus. Int. J. Dev. Biol., 44, 245–248. [PubMed] [Google Scholar]
- Knaus U.G. and Bokoch,G.M. (1998) The p21Rac/Cdc42-activated kinases (PAKs). Int. J. Biochem. Cell Biol., 30, 857–862. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N., Wettstein,D. and Kintner,C. (1999) Activation of Xenopus genes required for lateral inhibition and neuronal differentiation during primary neurogenesis. Mol. Cell. Neurosci., 14, 327–339. [DOI] [PubMed] [Google Scholar]
- Lamar E., Deblandre,G., Wettstein,D., Gawantka,V., Pollet,N., Niehrs,C. and Kintner,C. (2001) Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev., 15, 1885–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Hollenberg,S.M., Snider,L., Turner,D.L., Lipnick,N. and Weintraub,H. (1995) Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix–loop–helix protein. Science, 268, 836–844. [DOI] [PubMed] [Google Scholar]
- Leeuw T., Wu,C., Schrag,J.D., Whiteway,M., Thomas,D.Y. and Leberer,E. (1998) Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature, 391, 191–195. [DOI] [PubMed] [Google Scholar]
- Lei M., Lu,W., Meng,W., Parrini,M.C., Eck,M.J., Mayer,B.J. and Harrison,S.C. (2000) Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell, 102, 387–397. [DOI] [PubMed] [Google Scholar]
- Lu W. and Mayer,B.J. (1999) Mechanism of activation of Pak1 kinase by membrane localization. Oncogene, 18, 797–806. [DOI] [PubMed] [Google Scholar]
- Lu W., Katz,S., Gupta,R. and Mayer,B.J. (1997) Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr. Biol., 7, 85–94. [DOI] [PubMed] [Google Scholar]
- Ma Q., Kintner,C. and Anderson,D.J. (1996) Identification of neurogenin, a vertebrate neuronal differentiation gene. Cell, 87, 43–52. [DOI] [PubMed] [Google Scholar]
- Manser E., Leung,T., Salihuddin,H., Zhao,Z.S. and Lim,L. (1994) A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature, 367, 40–46. [DOI] [PubMed] [Google Scholar]
- Manser E., Loo,T.H., Koh,C.G., Zhao,Z.S., Chen,X.Q., Tan,L., Tan,I., Leung,T. and Lim,L. (1998) PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell, 1, 183–192. [DOI] [PubMed] [Google Scholar]
- Niehrs C. and Pollet,N. (1999) Synexpression groups in eukaryotes. Nature, 402, 483–487. [DOI] [PubMed] [Google Scholar]
- Niehrs C., Steinbeisser,H. and De Robertis,E.M. (1994) Mesodermal patterning by a gradient of the vertebrate homeobox gene goosecoid. Science, 263, 817–820. [DOI] [PubMed] [Google Scholar]
- Ohnuma S., Philpott,A., Wang,K., Holt,C.E. and Harris,W.A. (1999) p27 Xic1, a Cdk inhibitor, promotes the determination of glial cells in Xenopus retina. Cell, 99, 499–510. [DOI] [PubMed] [Google Scholar]
- Ohnuma S., Hopper,S., Wang,K.C., Philpott,A. and Harris,W.A. (2002) Co-ordinating retinal histogenesis: early cell cycle exit enhances early cell fate determination in the Xenopus retina. Development, 129, 2435–2446. [DOI] [PubMed] [Google Scholar]
- Oschwald R., Richter,K. and Grunz,H. (1991) Localization of a nervous system-specific class II β-tubulin gene in Xenopus laevis embryos by whole mount in situ hybridization. Int. J. Dev. Biol., 35, 399–405. [PubMed] [Google Scholar]
- Paganelli A.R. et al. (2001) The Alzheimer-related gene presenilin-1 facilitates sonic hedgehog expression in Xenopus primary neurogenesis. Mech. Dev., 107, 119–131. [DOI] [PubMed] [Google Scholar]
- Parrini M.C., Lei,M., Harrison,S.C. and Mayer,B.J. (2002) Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell, 9, 73–84. [DOI] [PubMed] [Google Scholar]
- Perron M., Opdecamp,K., Butler,K., Harris,W.A. and Bellefroid,E. (1999) X-Ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc. Natl Acad. Sci. USA, 96, 14996–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzoli O., Bosetti,A., Croci,L., Consalez,G.G. and Vetter,M.L. (2001) Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev. Biol., 233, 495–512. [DOI] [PubMed] [Google Scholar]
- Ronchini C. and Capobianco,A.J. (2001) Induction of cyclin D1 transcription and CDK2 activity by Notch ic: implication for cell cycle disruption in transformation by Notch ic. Mol. Cell. Biol., 21, 5925–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney R.D., Tuazon,P.T., Meek,W.E., Carroll,E.J., Hagen,J.J., Gump,E.L., Monnig,C.A., Lugo,T. and Traugh,J.A. (1996) Cleavage arrest of early frog embryos by the G protein-activated protein kinase Pak1. J. Biol. Chem., 271, 21498–21504. [DOI] [PubMed] [Google Scholar]
- Saka Y. and Smith,J.C. (2001) Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev. Biol., 229, 307–318. [DOI] [PubMed] [Google Scholar]
- Sasai Y. and De Robertis,E.M. (1997) Ectodermal patterning in vertebrate embryos. Dev. Biol., 182, 5–20. [DOI] [PubMed] [Google Scholar]
- Sun Y., Nadal-Vincents,M., Misono,S., Lin,M.Z., Zubiga,A., Hua,X., Fan,G. and Greenberg,M.E. (2001) Neurogenin promotes neuro genesis and inhibits glial differentiation by independent mechanisms. Cell, 104, 365–376. [DOI] [PubMed] [Google Scholar]
- Tomita K., Moriyoshi,K., Nakanishi,S., Guillemot,F. and Kageyama,R. (2000) Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J., 19, 5460–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.L. and Weintraub,H. (1994) Expression of achaete-scute homolog in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev., 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- VanDoren M., Powell,P.A., Pasternak,D., Singson,A. and Posakony,J.W. (1992) Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaete. Genes Dev., 6, 2592–2605. [DOI] [PubMed] [Google Scholar]
- Wilson S.I. and Edlund,T. (2001) Neural induction: toward a unifying mechanism. Nat. Neurosci., 4, 1161–1168. [DOI] [PubMed] [Google Scholar]