Abstract
Human factor C1 (HCF-1) is needed for the expression of herpes simplex virus 1 (HSV-1) immediate-early genes in infected mammalian cells. Here, we provide evidence that HCF-1 is required for spliceosome assembly and splicing in mammalian nuclear extracts. HCF-1 interacts with complexes containing splicing snRNPs in uninfected mammalian cells and is a stable component of the spliceosome complex. We show that a missense mutation in HCF-1 in the BHK21 hamster cell line tsBN67, at the non-permissive temperature, inhibits the protein’s interaction with U1 and U5 splicing snRNPs, causes inefficient spliceosome assembly and inhibits splicing. Transient expression of wild-type HCF-1 in tsBN67 cells restores splicing at the non-permissive temperature. The inhibition of splicing in tsBN67 cells correlates with the temperature-sensitive cell cycle arrest phenotype, suggesting that HCF-1-dependent splicing events may be required for cell cycle progression.
Keywords: factor C1/HCF-1/HSV/pre-mRNA splicing/spliceosome
Introduction
Factor C1, or HCF-1, is a host cell nuclear protein that is made up of a family of polypeptides that are processed from a 230 kDa precursor by site-specific proteolysis. The HCF-1 polypeptides resulting from site-specific proteolyses form a non-covalent heterogenous complex made up of N- and C-terminal fragments of the protein ranging in size from 68 to 180 kDa (Wilson et al., 1993, 1995; Kristie et al., 1995). The structure of the HCF-1 protein is characterized by the presence of two highly charged domains located at the N- and C-termini of the protein. The protein also has a basic domain that is ∼450 amino acids downstream from the N-terminus and an acidic domain located upstream from the highly charged C-terminal domain (Wilson et al., 1993). HCF-1 contains six kelch-repeat β-propeller domains characteristic of proteins in the kelch superfamily. These proteins are structurally and functionally diverse, making the deduction of function from structure impractical (see the review by Adams et al., 2000).
Pre-mRNA splicing is an essential post-transcriptional step in the pathway of gene expression in eukaryotes. Splicing occurs in the eukaryotic cell nucleus through a process that involves the removal of non-coding sequences (introns) from the pre-mRNA and the joining of adjacent or alternate coding sequences (exons) to produce mature mRNA. The splicing reaction takes place via a two-step transesterification mechanism involving sequential nucleophilic attack on phosphodiester bonds at the splice junctions and concomitant formation of spliced mRNA and an excised intron (for a review, see Staley and Guthrie, 1998). The mature mRNA is then exported to the cytoplasm for use in protein synthesis. Splicing is catalysed by a large ribonucleoprotein complex made up of >50 proteins called the spliceosome. The spliceosome complex contains four small nuclear ribonucleoprotein (snRNP) particles (U1, U2, U5 and U4/U6), each of which contains the corresponding snRNAs and a set of specific and common proteins (reviewed by Krämer, 1995, 1996; Will and Lührmann, 1997). The spliceosome complex also contains multiple non-snRNP-associated proteins that are essential for complex assembly and splicing catalysis (Will and Lührmann, 1997; Staley and Guthrie, 1998). Spliceosome assembly involves the sequential assembly of the snRNP particles and other proteins onto the pre-mRNA substrate prior to catalysis (reviewed in Reed and Palandjian, 1997).
Previous work on the function of HCF-1 has concentrated mainly on its role in herpes simplex virus (HSV) infection. In virus-infected cells, the expression of the HSV α/immediate-early (α/IE) genes is regulated by a multiprotein complex containing HCF-1. HCF-1 interacts with the viral transcription regulator VP16 to form a stable heterodimeric complex. The association of VP16 with HCF-1 assists its interaction with the cellular transcription factor Oct-1 on HSV DNA containing HSV IE promoters to form a transcription activator complex called the VP16-induced complex (VIC) (reviewed in O’Hare, 1993; Herr, 1998). Several other studies have shown that HCF-1 will associate with transcription factors including LZIP (Freiman and Herr, 1997), Sp1 (Gunther et al., 2000), CREB/ATF, luman and Zhangfei (Lu et al., 1997, 1998; Lu and Misra, 2000), suggesting a possible role for the protein in gene transcription. A recent study showing that HCF-1 associates with protein phosphatase 1 (Ajuh et al., 2000) implies that the cellular function of this protein may involve phosphorylative regulation. A role for HCF-1 in the cell division cycle has also been suggested following observations that a single proline to serine missense mutation in the VP16 interaction domain of the protein causes cell cycle arrest in the hamster cell line tsBN67 (Goto et al., 1997). It has also been shown recently that an association between HCF-1 and chromatin is disrupted upon temperature-induced cell cycle arrest. This could imply that an interaction between HCF-1 and chromatin is required for cell cycle progression (Wysocka et al., 2001). However, it is not clear whether the relationship between HCF-1 and cell cycle progression is direct, or instead is an indirect effect of HCF-1 affecting the expression or function of bona fide cell cycle regulatory factors.
In spite of all the associations and observations described above, the molecular function of HCF-1 in eukaryotic cells is not yet clear. In this report, we present data that indicate that HCF-1 is needed for pre-mRNA splicing. These results provide the first direct evidence for a cellular function for HCF-1.
Results
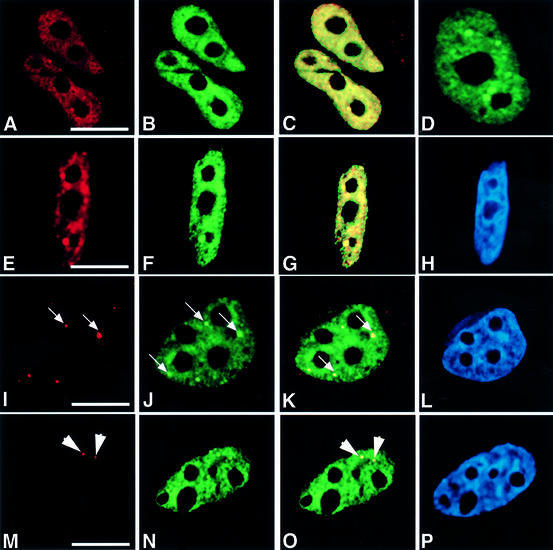
Factor C1 co-localizes with nuclear structures that contain pre-mRNA splicing factors
We have shown previously that complexes containing nuclear HCF-1 have an associated protein phosphatase 1 activity and that the C-terminal region of HCF-1 will interact directly in vitro with protein phosphatase 1 (Ajuh et al., 2000). Therefore, in order to analyse the cellular function of HCF-1, we decided to investigate its nuclear localization in various human cell lines. Localization studies were carried out using both transient expression of yellow fluorescent protein (YFP)-tagged HCF-1 and anti-HCF-1 antibodies (Ajuh et al., 2000; and references therein). In both cases, the protein showed a diffused speckled nuclear distribution, with accumulation in distinct nuclear bodies (Figure 1A and B). Co-localization of the endogenous HCF-1 (antibody staining) and YFP–HCF-1 indicates that the recombinant protein expressed from the pEYFP-HCF-1 plasmid localizes to the same nuclear structures as the endogenous protein (Figure 1C; data not shown). In order to ensure that the observed structures were not an artefact of the cell fixation method, the localization of exogenously expressed YFP-tagged HCF-1 was analysed in live cells (Figure 1D; data not shown). These data indicate that the HCF-1 protein in HeLa cells shows a similar diffuse and speckled distribution with accumulation in distinct nuclear bodies.
Fig. 1. HCF-1 co-localizes with nuclear structures that contain pre-mRNA splicing factors. HeLa cells were transfected with pEYFP-HCF, and indirect immunofluorescence experiments were performed on the transfected cells using various antibodies. The arrows point to gems, while the arrowheads point to Cajal bodies. (A) Light microscopic image of an indirect immunofluorescence experiment on HeLa cells using the polyclonal HC2 antibody. (B) A microscopic image obtained from the direct fluorescence of YFP–HCF-1 in the same cells as in (A). (C) Superimposed optical sections from (A) and (B). Co-localization of green and red results in the yellow colour. Note the presence of some green fluorescence that does not co-localize with the antibody staining. This may be due to the stronger fluorescence signal produced by the YFP–HCF-1 fusion protein. Secondly, YFP–HCF-1/cellular HCF-1 may exist in some complexes that, although visible by direct YFP fluorescence, are inaccessible to the anti-HCF-1 peptide antibodies used for cell staining. (D) Light microscopic image of a live HeLa cell expressing YFP–HCF-1 (green). (E, I and M) Microscopic images of indirect immunofluorescence experiments on HeLa cells using anti-Sm antibody (Y12), anti-SMN mAb (MANSMA1) and anti-p80 coilin mAb (5P10), respectively. (F, J and N) Microscopic images obtained from the direct fluorescence of YFP–HCF-1 in the same cells as in (E), (I) and (M), respectively. (G, K and O) Superimposed optical sections from (E) and (F), (I) and (J), and (M) and (N), respectively. (H, L and P) Microscopic images obtained from DAPI staining of the HeLa cells shown in (E), (I) and (M), respectively. Bar = 5 µm.
Most pre-mRNA splicing factors localize in nuclear structures called speckles and/or nuclear bodies (Cajal bodies and gems) in mammalian cells (reviewed in Lamond and Earnshaw, 1998; Mistelli and Spector, 1998; Gall, 2000). Because the HCF-1 nuclear structures observed above are similar to those described previously for pre-mRNA splicing factors, HeLa cells transfected with pEYFP-HCF-1 (Figure 1F, J and N) were counter-stained with either the snRNP-specific monoclonal antibody (mAb) Y12 (Figure 1E), the anti-SMN antibody (MANSMA1) (Figure 1I) or anti-p80 coilin mAb (5P10) (Figure 1M). SMN (survival of motor neurons) protein and p80 coilin are markers for the nuclear bodies called gems and Cajal bodies, respectively. Overlays of images obtained from the antibody-stained and the YFP–HCF-1-transfected cells show that HCF-1 co-localizes with splicing snRNPs, gems and Cajal bodies in HeLa cell nuclei (Figure 1G, K and O). Similar results were obtained when the co-localization experiments were performed using anti-HCF-1 antibodies to detect endogenous HCF-1 in untransfected cells (data not shown). Experiments performed using other cell lines, including primary lens epithelium cells (PLEBs), the breast cancer cell line MCF7 and neuroblastoma (IMR-32) cells (ATCC), also showed a nuclear localization pattern for HCF-1 that consists of diffuse and speckled nuclear staining, as well as accumulation in Cajal bodies (data not shown). However, no co-localization was observed between HCF-1 and PML bodies (data not shown). PML bodies are nuclear domains that are disrupted specifically in patients suffering from human acute promyelocytic leukaemia and do not contain pre-mRNA processing factors (reviewed in Lamond and Earnshaw, 1998). The presence of HCF-1 in speckles and Cajal bodies or gems, which contain RNA processing factors, suggests that the protein complex may have a role in RNA processing.
HCF-1 interacts with nuclear complexes that contain splicing snRNPs and SMN
We next investigated the possible interaction in vitro between HCF-1 and complexes containing splicing snRNPs in HeLa nuclear extract. Anti-HCF-1 antibodies were used to immunoprecipitate HCF-1-containing protein complexes from HeLa nuclear extract. The proteins pulled down were separated by SDS–PAGE, transferred onto nitrocellulose and probed with anti-HCF-1 antibodies (Figure 2A) and the anti-Sm protein antibody Y12 (Figure 2B). These results show that anti-HCF-1 antibodies will co-immunoprecipitate Sm proteins from HeLa nuclear extracts (Figure 2B, lanes 3–5), whereas control pre-immune IgG does not (Figure 2B, lane 2). We also observed the reciprocal result that antibodies to Sm proteins (Y12) as well as antibodies to the 2,2,7-trimethyl guanosine caps of snRNAs present in splicing snRNP complexes [anti-2,2,7-trimethyl-guanosine (Calbiochem)] will both immunoprecipitate HCF-1 from HeLa nuclear extract (data not shown).
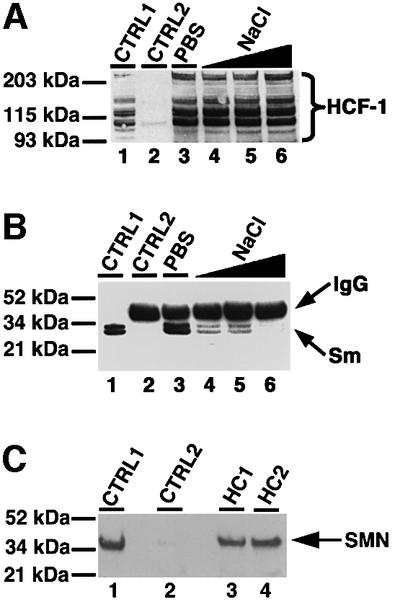
Fig. 2. HCF-1 interacts with complexes that contain splicing snRNPs and SMN. (A) Immunoprecipitation using anti-HCF-1 antibody (HC2). Lane 1 (marked CTRL1) is the positive control and contained HeLa nuclear extract. CTRL2 (lane 2) is a control immunoprecipitation using pre-immune IgG. Lanes 3–6 contained immunoprecipitates using HC2 antibodies, except that the immunoprecipitate in lane 3 was washed with PBS containing 0.01% Triton X-100 whereas the immunoprecipitates in lanes 4–6 were washed with PBS/Triton X-100 as above but with increasing concentrations of NaCl (i.e. 200, 300 and 400 mM), respectively. The beads containing immunoprecipitated proteins were loaded onto a 4–12% SDS–polyacrylamide gradient gel (Novex) and probed by western blotting using anti-HCF-1 antibodies. HCF-1 bands on the blot were revealed by ECL (Amersham-Pharmacia) and are shown by the bracket on the right of (A). (B) The experiment and the lane markings of the panel are identical to (A) except that the immunoblot was probed with the anti-Sm antibody Y12 instead of anti-HCF-1 antibodies. (C) A 10 µg aliquot of HC1 (lane 3) or HC2 (lane 4) was used to immunoprecipitate HCF-1 from ∼0.2 mg of HeLa nuclear extract. CTRL1 is a positive control lane containing ∼25 µg of HeLa nuclear extract. CTRL2 (lane 2) is a negative control immunoprecipitation reaction using 10 µg of pre-immune IgG. The immunoblot was probed with anti-SMN mAbs (MANSMA1), and the SMN bands revealed by ECL.
Because SMN interacts with Sm proteins common to the four splicing snRNP particles and also plays a role in RNA processing (Liu et al., 1997; Friesen and Dreyfuss, 2000; Mourelatos et al., 2001; Selenko et al., 2001; and references therein), we next investigated its possible association with the HCF-1 complex. HCF-1 immunoprecipitates were blotted to membranes as described above and then probed with anti-SMN antibodies (Figure 2C). The results show that anti-HCF-1 antibodies will co-immunoprecipitate SMN from HeLa nuclear extract (Figure 2C, lanes 3 and 4) whereas pre-immune IgG does not (Figure 2C, lane 2). Taken together, the above data suggest an interaction between HCF-1 and complexes containing splicing snRNPs. These results are consistent with the accumulation of HCF-1 in speckles, Cajal bodies and gems.
HCF-1 is a component of the spliceosome complex
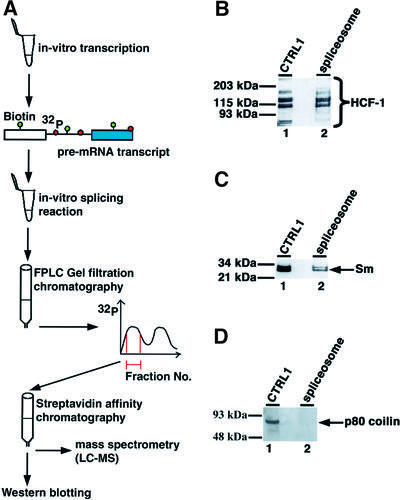
Because HCF-1 interacts with complexes that contain splicing snRNPs, we next investigated whether it is a component of the spliceosome complex assembled in vitro. The spliceosome complex was purified (Figure 3A) from in vitro splicing reactions performed using HeLa nuclear extracts (Reed et al., 1988; Reed, 1990; Neubauer et al., 1998). The purified spliceosomal proteins were separated on a denaturing SDS–polyacrylamide gel before immunoblotting and probing with anti-HCF-1 antibodies. The results obtained from this experiment show that HCF-1 is a stable component of the purified human spliceosome complex (Figure 3B, lane 2). As a positive control, the purified spliceosome complexes were also probed with antibodies to known spliceosomal proteins, e.g. the Sm proteins (Figure 3C, lane 2, data not shown). The purified spliceosome complexes were also probed with antibodies to other non-spliceosomal nuclear proteins, e.g. p80 coilin (an abundant nucleoplasmic and nuclear body protein component) that is not a spliceosomal protein, as a negative control (Figure 2D). This confirmed that coilin did not co-purify with the spliceosome complex. These results indicate that HCF-1 stably associates with the human spliceosome complex and may function in the pre-mRNA splicing pathway.
Fig. 3. HCF-1 is a component of the spliceosome complex in HeLa nuclear extract. Spliceosomes purified as described in Materials and methods were separated on a 10 or 4–12% SDS–polyacrylamide gradient gel. (A) Cartoon showing the assembly and purification scheme used to purify the human spliceosome complex. The pre-mRNA transcript was labelled with both biotin-UTP and [32P]GTP. (B) The spliceosomal proteins were blotted onto nitrocellulose filters and probed with anti-HCF-1 antibodies. Lane 1 is a control lane containing ∼25–30 µg of nuclear extract. Lane 2 contained the purified spliceosome complex. (C and D) The contents of the lanes are identical to those in (A) except that the blots were probed with the anti-Sm antibody (Y12) and anti-p80 coilin mAb (5P10), respectively.
Intact HCF-1 is required for in vitro pre-mRNA splicing
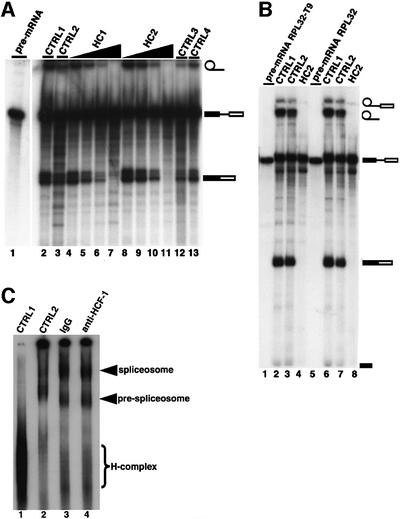
We next investigated a possible role for the protein in the pre-mRNA splicing mechanism. Attempts to immunodeplete HCF-1 quantitatively from HeLa nuclear extract in order to determine its role in the splicing mechanism were unsuccessful. We were only able to remove ∼70% of the HCF-1 polypeptides from HeLa nuclear extracts using our anti-HCF-1 antibodies. The reasons for this incomplete level of immunodepletion are unclear. It is possible, however, that HCF-1 is involved in multiple interactions with other nuclear proteins that obscure the epitopes recognized by the anti-HCF-1 antibodies, thus reducing the total amount of HCF-1 in the extracts available for antibody binding at any one time. At the levels of HCF-1 depletion obtained, we did not observe a significant reduction in overall splicing efficiency in HeLa nuclear extracts (data not shown). It is known, however, that similar levels of depletion of most other well-characterized splicing factors also do not result in the inhibition of splicing. Thus, experiments were performed where instead of immunodepleting HCF-1 from HeLa nuclear extracts, anti-HCF-1 peptide antibodies (Kristie et al., 1995; Ajuh et al., 2000) were added directly to pre-mRNA splicing reactions to determine whether the presence of antibody molecules bound to HCF-1 in nuclear extract will block splicing. As shown in Figure 4, increasing amounts of two different anti-HCF-1 peptide antibodies (affinity purified using their respective cognate peptides) were added to splicing reactions performed using pre-mRNA transcribed in vitro (Lamond et al., 1987). Addition of anti-HCF-1 antibodies, i.e. either HC1 (Figure 4, lanes 4–7) or HC2 (Figure 4, lanes 8–11), inhibits in vitro pre-mRNA splicing, whereas splicing remained active when pre-immune IgG was added to the reactions [i.e. an identical amount of pre-immune IgG to the highest amount of anti-HCF-1 antibodies used (Figure 4, lane 3)]. When the anti-HCF-1 antibodies were pre-incubated with a 10-fold molar excess of their cognate peptides, prior to addition to the splicing reactions, their inhibitory effect was blocked (Figure 4, lanes 12 and 13). This indicates that the splicing inhibition is specific and most probably caused by the binding of the anti-HCF-1 antibodies to the HCF-1 protein complex in the nuclear extracts.
Fig. 4. Inhibition of pre-mRNA splicing by anti-HCF-1 antibodies. (A) HeLa nuclear extracts (∼50 µg) were pre-incubated with 0.15–1.2 pmol anti-HCF-1 antibodies before addition to the splicing reaction. Lane 2 (CTRL1) is a control splicing reaction to which no antibodies were added. Lane 3 (CTRL2) is a control splicing assay containing 1.2 pmol of rabbit pre-immune IgG. Lanes 4–7 indicate splicing reactions that contained 0.15, 0.30, 0.6 and 1.2 pmol of the HC1 antibody, respectively. Lanes 8–11 represent splicing assays that contained 0.15, 0.30, 0.6 and 1.2 pmol of the HC2 antibody, respectively. Lanes 12 and 13 (CTRL3 and CTRL4) are control reactions that contained 1.2 pmol each of HC1 and HC2, respectively, except that here each anti-HCF-1 antibody had been pre-incubated with its cognate peptide before addition to the splicing reaction. The symbols on the right represent the input pre-mRNA and splicing products. (B) Inhibition of splicing of the RPL32 pre-mRNAs by anti-HCF-1 antibodies. Pre-mRNAs transcribed in vitro from the pGEM-RPL32 and pGEM-RPL32:T9 plasmids (Vilardell and Warner, 1997; and references therein) were used in splicing reactions containing 1.2 pmol of the HC2 antibody or pre- immune IgG. RPL32 pre-mRNA was used in the reactions in lanes 1–4, while RPL32:T9 pre-mRNA was used in splicing reactions in lanes 5–8. Lanes 3 and 7 are control reactions that contained pre- immune IgG. HC2 antibody was added to the reactions in lanes 4 and 8. No antibodies were added to samples in lanes 2 and 6. (C) Spliceosome assembly is unaffected by anti-HCF-1 antibodies in splicing reactions. Pre-mRNA splicing reactions were performed in vitro for 45–60 min using HeLa nuclear extract that had been pre-incubated for 15–20 min at 30°C with 1.2 pmol of the HC2 antibody or pre-immune IgG. The splicing complexes were separated on a native agarose–polyacrylamide composite gel. Lane 1 is a control reaction performed on ice. No antibodies were added to the reaction in lane 2. The reactions in lanes 3 and 4 contained pre-immune IgG and HC2 antibodies, respectively.
We next investigated whether the inhibitory effect of anti-HCF-1 antibodies on pre-mRNA splicing is template specific. Pre-mRNA was prepared by in vitro transcription from pGEM-RPL32 or pGEM-RPL32:T9 pre-mRNA plasmid templates (Vilardell and Warner, 1997; and references therein) and the hnRNP A1-533 series plasmids (Chabot et al., 1997). These plasmid vectors contain either wild-type or site-specific mutants of the genes for ribosomal protein L32 and hnRNP A1, respectively. When anti-HCF-1 antibodies were added to splicing reactions containing the pre-mRNAs derived from the RPL32 pre-mRNAs (Figure 4B, lanes 4 and 8) or the hnRNP A1-533 series plasmids (data not shown), splicing was inhibited. Addition of identical amounts of pre-immune IgG (Figure 4B, lanes 3 and 7) did not inhibit splicing. These results indicate that the requirement for HCF-1 in pre-mRNA splicing affects multiple templates, at least in vitro.
The above observations that anti-HCF-1 antibodies will inhibit pre-mRNA splicing led us to investigate whether anti-HCF-1 antibodies inhibit splicing by disrupting the spliceosome assembly pathway. Thus in vitro splicing experiments were performed and spliceosome complexes resolved using an agarose–polyacrylamide composite gel. One set of reactions contained inhibitory amounts of anti-HCF-1 antibodies (Figure 4C, lane 4), while another set contained an identical amount of pre-immune IgG (Figure 4C, lane 3). The results obtained from these experiments indicate that spliceosome assembly is not blocked by the presence of anti-HCF-1 antibodies in the reaction. We also found that the anti-HCF antibodies were incorporated into the stalled spliceosome complex along with HCF-1 (data not shown). Taken together, these results suggest a possible role for HCF-1 in either splicing catalysis or a late stage of spliceosome assembly.
A point mutation in HCF-1 causes inefficient spliceosome assembly and inhibition of catalysis
The BHK21 hamster cell line tsBN67 contains a missense proline to serine mutation at position 134 in HCF-1 that causes cell cycle arrest at G0/G1 when the cells are maintained at 39.5°C, the non-permissive temperature (Goto et al., 1997). A feature of tsBN67 cells is that they stop proliferating after a lag of a few cell divisions at the non-permissive temperature. Figure 5A shows the rate of growth of BHK21 cells and tsBN67 cells at 39.5°C. The results are consistent with previous observations for the growth of the cell lines at 39.5°C (Goto et al., 1997). We therefore investigated whether the mutation in HCF-1 would also affect splicing activity at the non-permissive temperature. BHK21 cells and tsBN67 cells were both transferred to the non-permissive temperature of 39.5°C and nuclear extracts prepared from the cells at different time points. The extracts were then used for in vitro splicing experiments (Figure 5B). The results indicate that when nuclear extracts were prepared from ts cells just after the lag period (∼10 h or more), these extracts are not able to splice pre-mRNA (Figure 5B, lanes 7–10), whereas extracts from control BHK21 cells were still able to splice pre-mRNA irrespective of how long they were maintained at 39.5°C (Figure 5B, lane 3). When tsBN67 cells were maintained at the permissive temperature, nuclear extracts prepared from these cells efficiently spliced pre-mRNA (Figure 5B, lane 4). These results indicate that the mutation in HCF-1 may be responsible for the splicing deficiency observed in the tsBN67 extracts (Figure 5B, lanes 7–10) and are consistent with an essential role for HCF-1 in pre-mRNA splicing.
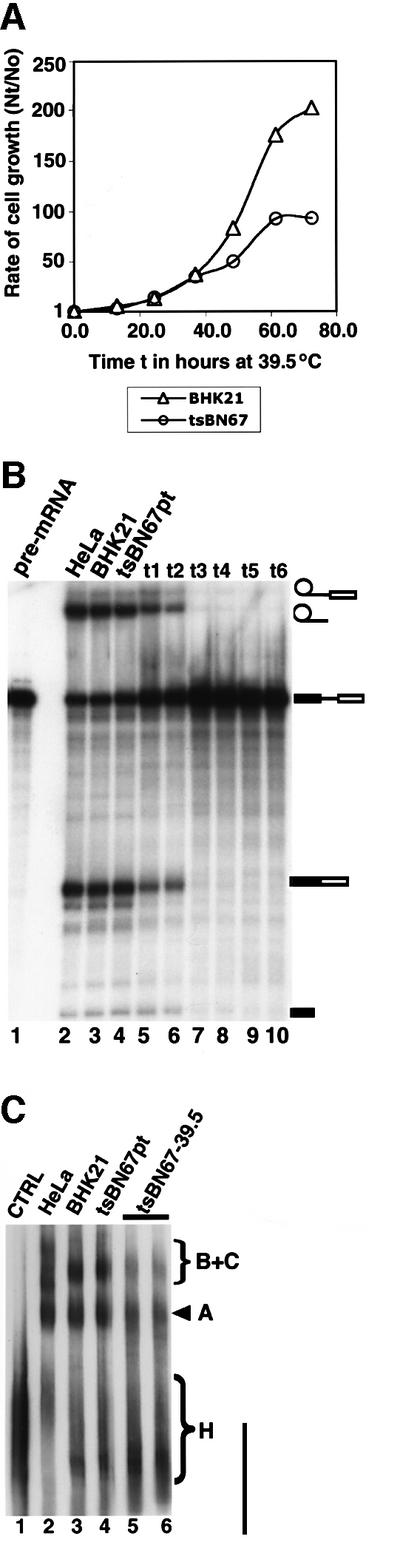
Fig. 5. Nuclear extracts from the hamster BHK21 and tsBN67 cell line containing a missense mutation in HCF-1 are incompetent in splicing. (A) Analysis of the growth rates of BHK21 and tsBN67 cells at 39.5°C. A total of 2 × 104 tsBN67 and BHK21 cells were seeded into 10 cm dishes and grown at 37°C. After 48 h, the temperature was shifted to 39.5°C. The cells were harvested at 12 h intervals by trypsin ization and counted using a haemocytometer. The cell counts obtained at each time point (Nt) were divided by the cell count (N0) at the time the cells were shifted to the non-permissive temperature, and plotted against the time in hours. (B) Nuclear extracts were prepared from the BHK21 and tsBN67 cells maintained at 39.5°C at 18–24 h intervals (t1–t6; lanes 5–10). About 45–50 µg of nuclear extract from each time point were used in each splicing experiment. Lane 1 contained the input pre-mRNA. Lane 2 is a positive control of a splicing reaction using HeLa nuclear extract. The lane marked BHK21 (lane 3) contained nuclear extract from BHK21 cells maintained indefinitely at 39.5°C. Lane 4 contained nuclear extract from tsBN67 cells maintained at the permissive temperature. (C) Nuclear extracts from tsBN67 cells are inefficient in spliceosome assembly. Pre-mRNA splicing reactions were performed in vitro for 45–60 min. The splicing complexes were then separated on a native agarose–polyacrylamide composite gel. Lane 1 is a control reaction using BHK21 nuclear extract and performed on ice. Lane 2 contained a splicing reaction using HeLa nuclear extract. Lanes 3 and 4 contained BHK21 and tsBN67 nuclear extracts prepared as in lanes 3 and 4 of (B) above. Lanes 5 and 6 contained tsBN67 nuclear extracts from time points t3 and t4.
We next investigated whether the inability of extracts from tsBN67 cells at 39.5°C to splice pre-mRNA was due to inefficient assembly of the spliceosome complex. Splicing reactions were performed using BHK21 and tsBN67 cell extracts as above, and the spliceosome complexes separated on a native gel (Figure 5C). The results from this experiment show that spliceosome assembly (especially the formation of the B and C com plexes) either is inefficient, or the resulting complexes are unstable in nuclear extracts from tsBN67 cells grown at 39.5°C (Figure 5C, lanes 5 and 6). Conversely, extracts from BHK21 cells maintained at 39.5°C and from tsBN67 cells at the permissive temperature were very competent in spliceosome assembly (Figure 5C, lanes 3 and 4, respectively). Taken together, these results suggest an important role for HCF-1 in the efficient assembly of the spliceosomal B and C complexes.
Nuclear extracts from BHK21 and tsBN67 cells induced to arrest at the G0/G1 stage of the cell cycle can splice pre-mRNA
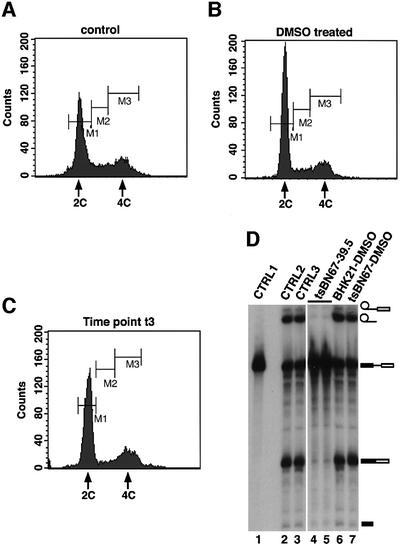
To confirm that the inability to splice pre-mRNA observed in nuclear extracts prepared from tsBN67 cells cultured at the non-permissive temperature was not caused by a non-specific splicing defect of cells arrested at the G0/G1 stage of the cell cycle, BHK21 and tsBN67 cells were treated at the permissive temperature with dimethyl sulfoxide (DMSO). DMSO has been shown to be able to induce G0/G1 arrest in many mammalian cell types (Takase et al., 1992; Boquest et al., 1999; Kudo et al., 2002). We therefore monitored cell cycle arrest of tsBN67 and BHK21 cells caused by DMSO treatment as well as tsBN67 cells incubated at 39.5°C up to time point t3 by fluorescence-activated cell sorting (FACS) analysis (Figure 6A–C). Time point t3 corresponds to the first time point at which splicing inhibition was observed in the nuclear extracts from tsBN67 cells shifted to 39.5°C. The FACS analysis data (Figure 6B; data not shown) indicate that DMSO treatment causes G0/G1 cell cycle arrest in BHK21 and tsBN67 cells to the same extent as has been shown previously for an asynchronous population of tsBN67 cells maintained at 39.5°C (Goto et al., 1997). In our experiments, we observed that on average, >71% of cells were arrested at G0/G1 upon DMSO treatment. We also found that at t3, ∼66% of tsBN67 cells were arrested at G0/G1 (Figure 6C; data not shown) and ∼15–20% less in untreated cells (Figure 6A and data not shown). These observations are consistent with previous analyses of these cell lines (Goto et al., 1997).
Fig. 6. Nuclear extracts from BHK21 and tsBN67 cells arrested at G0/G1 using DMSO can splice adeno-pre-mRNA efficiently. BHK21 and tsBN67 cells were maintained for 48 h at 37°C in culture medium containing 1% (v/v) DMSO. No DMSO was added to control cells. (A–C) Cell cycle analysis of tsBN67 cells. The labels M1, M2 and M3 represent cells at the G0/G1, S and G2/M phases of the cell cycle, respectively. (A) DNA content FACS analysis of untreated cells. (B) DNA content FACS analysis of DMSO-treated cells. (C) DNA content FACS analysis of tsBN67 cells incubated at 39.5°C up to time point t3. (D) Nuclear extracts were prepared from treated and untreated cells for use in splicing experiments. Lane 1 contained 30% of the input pre-mRNA transcript. Lanes 2 and 3 contained control nuclear extracts from untreated BHK21 and tsBN67 cells, respectively. Lanes 4 and 5 contained a splicing reaction using nuclear extract from tsBN67 cells maintained at the non-permissive temperature (time point t3), whereas lanes 6 and 7 contained nuclear extracts from DMSO-treated BHK21 and tsBN67 cells, respectively.
Nuclear extracts were prepared from DMSO-treated and control untreated BHK21 and tsBN67 cells maintained at the permissive or non-permissive temperature. The extracts were used subsequently for in vitro splicing experiments. Results from these experiments indicate that nuclear extracts obtained from the DMSO-treated cells were as efficient in pre-mRNA splicing as extracts prepared from untreated cells (Figure 6D, compare lanes 2 and 3 with lanes 6 and 7). These data demonstrate that the inability of nuclear extracts from tsBN67 cells maintained at 39.5°C to splice pre-mRNA is most likely to be due to the HCF-1 mutation and not the result of a non-specific splicing defect in cells stopped at G0/G1.
Wild-type HCF-1 will restore splicing to tsBN67 nuclear extracts
tsBN67 cells were transfected with either a pEYFP-HCF fusion plasmid containing wild-type human HCF-1 or the plasmid pEYFP-C1 that contains YFP alone. The cells were then maintained at 39.5°C up to the end of the lag proliferation period (i.e. t3 of Figure 5B) before preparation of nuclear extracts. Figure 7A shows that extracts from the tsBN67 cells transfected with pEYFP-HCF-1 could splice pre-mRNA (lanes 7 and 8), whereas extracts from cells transfected with pEYFP-C1 were unable to splice the pre-mRNA (lanes 9 and 10). However, the splicing efficiency of cells transfected with the pEYFP-HCF plasmid was not as high as observed for either BHK21 or control tsBN67 extracts (lanes 3 and 4, respectively), presumably due to incomplete transfection efficiencies (40–50% of cells expressed pEYFP-HCF-1 as monitored by fluorescence microscopy; data not shown) and possibly also due to the presence of mutant HCF-1 in these extracts. To check that the wild-type HCF-1 is expressed in transfected cells, the nuclear extracts were probed by western blotting using anti-HCF-1 (Figure 7B) and anti-YFP (Figure 7C) antibodies. (Note that the anti-HCF-1 antibodies used here recognize the C-terminal polypeptides of HCF-1 whereas anti-YFP antibodies recognize the N-terminal polypeptides of HCF-1 because the construct pEYFP-HCF contains YFP as an N-terminal fusion of HCF-1.) The results show that YFP and wild-type HCF-1 fused to YFP are expressed in the transfected tsBN67 cells (Figure 7C, lanes 2 and 3, doublet bands, respectively). The doublet of bands (YFP–HCF-1n, Figure 7C, lane 3) representing the N-terminal fragments of processed HCF-1 polypeptides are identical to those previously observed (Wysocka et al., 2001). These data indicate that YFP–HCF-1 can restore the ability to splice pre-mRNA to tsBN67 nuclear extracts, whereas YFP alone cannot.
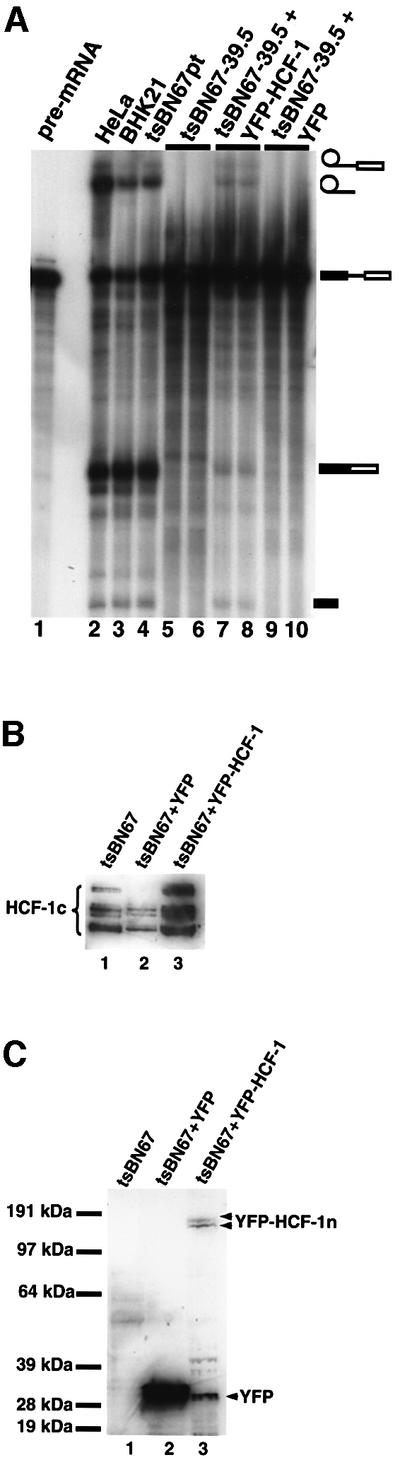
Fig. 7. Transfection of tsBN67 cells maintained at the non-permissive temperature restores splicing to the tsBN67 extracts. (A) tsBN67 cells were transfected with wild-type HCF-1 in the plasmid pEYFP-HCF and the transfection efficiency monitored by fluorescence microscopy (data not shown). Transfected cells were maintained at 39.5°C up to the time point t3 above. Nuclear extracts were then prepared from the cells and used for in vitro splicing assays. Lane 1 contained input pre-mRNA. Lane 2 contained a splicing reaction using HeLa nuclear extract. The lane marked BHK21 (lane 3) contained nuclear extract from BHK21 cells maintained at 39.5°C. Lane 4 contained nuclear extract from tsBN67 cells maintained at the permissive temperature. Lanes 5 and 6 represent duplicate splicing reactions using tsBN67 nuclear extracts (from cells maintained at 39.5°C) prepared at t3. The lanes marked 7 and 8 contained splicing reactions using nuclear extracts from tsBN67 cells transfected with wild-type HCF-1 plasmid, maintained at 39.5°C, and extracts prepared at time t3 (10–12 h after lag period). Lanes 9 and 10 contained control reactions using extracts from tsBN76 cells treated identically to the cells used for the reactions in lanes 7 and 8 except that the tsBN67 cells used here were transfected with a plasmid that contained only YFP. (B and C) Nuclear extracts from transfected and untransfected cells were probed with (B) anti-HCF-1 and (C) anti-YFP antibodies (BD Biosciences, UK). The samples in (B) and (C) are identical. The lanes marked 1 contained nuclear extracts from untransfected cells while the lanes marked 2 and 3 contained tsBN67 cells transfected with pEYFP-C1 and pEYFP-HCF, respectively. Note that ∼3- to 4-fold less nuclear extract was loaded for the lanes marked 2 in order to compensate for the high signal from the YFP. Note also that only the doublet of bands representing the N-terminal HCF-1 fragments was detected in (C, lane 3) because the YFP protein is fused to the N-terminus of HCF-1 in the pEYFP-HCF construct, whereas the antibodies HC1 and HC2 recognize the C-terminal fragments of the processed HCF-1 polypeptides.
The mutation in HCF-1 impairs its association with U1 and U5 splicing snRNPs
Because anti-HCF-1 antibodies co-immunoprecipitate Sm proteins, SMN and all the five splicing snRNAs from HeLa nuclear extracts (Figure 8A, lane 3; data not shown), we next investigated whether the mutation in HCF-1 will affect the interaction between the protein and splicing snRNPs. Immunoprecipitations were performed using anti-HCF-1 antibodies and nuclear extracts from control tsBN67 cells as well as BHK21 and tsBN67 cells main tained at 39.5°C. Co-immunoprecipitated RNAs were analysed by northern blotting using [32P]GTP-labelled snRNA probes. The results show that the interactions between U1 or U5 snRNPs and HCF-1 are blocked or reduced in extracts from tsBN67 cells that have been grown at 39.5°C (Figure 8A, lane 6), whereas these interactions were unaffected in extracts from control BHK21 cells grown at 39.5°C or tsBN67 cells maintained at the permissive temperature (Figure 8A, lanes 4 and 5, respectively). These results indicate that the inefficient assembly of spliceosomes and splicing inhibition observed in tsBN67 extracts may be caused by disruption of the interactions between U1 or U5 snRNPs and HCF-1 in the extracts containing mutant HCF-1.
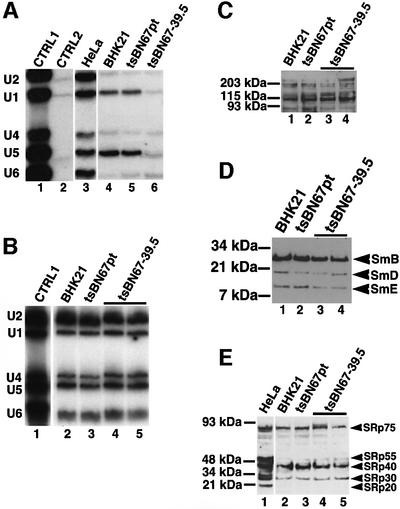
Fig. 8. The association between HCF-1 and U1 or U5 splicing snRNPs is inhibited in tsBN67 nuclear extracts. (A) Anti-HCF-1 antibody immunoprecipitates using ∼10 µg of antibody and ∼0.25 mg of nuclear extract were probed with 32P-labelled snRNA probes. Lane 1 is a positive control containing snRNAs in HeLa nuclear extract. Lane 2 contained immunoprecipitates using pre-immune IgG, whereas lanes 3–6 contained anti-HCF-1 antibody immunoprecipitates from HeLa cells, BHK21 cells (maintained at 39.5°C), tsBN67 cells maintained at the permissive temperature and tsBN67 cells maintained at 39.5°C, respectively. (B) Total nuclear RNA was extracted from ∼100 µg of nuclear extracts and the snRNAs in the extracts separated on an 8 M urea– polyacrylamide gel and transferred onto a nylon membrane by northern blotting. The snRNA bands were visualized by probing the membrane with [32P]GTP-labelled snRNA probes and autoradiography. Lanes 1–3 contained snRNAs in nuclear extracts from HeLa cells, BHK21 cells maintained at 39.5°C and tsBN67 cells maintained at the permissive temperature, respectively. Lanes 4 and 5 contained duplicate samples of RNA prepared from non-splicing-competent nuclear extracts of tsBN67 cells maintained at 39.5°C. (C–E) About 30 µg of nuclear extracts from BHK21 cells maintained at 39.5°C (lane 1), tsBN67 cells maintained at the permissive temperature (lane 2) and tsBN67 cells maintained at 39.5°C (lanes 3 and 4, i.e. duplicate), respectively, were probed with anti-HCF-1 antibodies (C), anti-Sm antibodies (D) and anti-SR protein antibodies (E).
In order to be certain that maintaining tsBN67 cells at 39.5°C did not lead to reduced levels of U1 and U5 snRNA expression, total RNA was extracted from identical amounts of nuclear extracts prepared from BHK21 cells and tsBN67 cells maintained at the permissive and non-permissive temperatures. The snRNAs present in the extracts were analysed by northern blotting using [32P]GTP-labelled snRNA probes (Figure 8B). The results from this experiment show that the levels of all the snRNAs are similar in nuclear extracts from cells maintained at the permissive or non-permissive temperatures (Figure 8B, compare lanes 2 and 3 with lanes 4 and 5). Taken together, these results suggest a functionally important interaction between HCF-1 and the U1 and U5 snRNPs in efficient spliceosome assembly.
To check whether extracts from tsBN67 cells had lower levels of some essential splicing factors when grown at 39.5°C, identical amounts of nuclear extracts from BHK21 and tsBN67 cells maintained at the permissive and non-permissive temperatures were analysed by western blotting using anti-HCF-1, Y12 (splicing snRNP-specific antibody) and anti-SR protein antibodies (Figure 8C–E), respectively. SR proteins are essential pre-mRNA splicing factors that are needed for the activation of constitutive splicing and the regulation of alternative splicing in mammals (reviewed by Manley and Tacke, 1996; Valcarcel and Green, 1996). These results show that the levels of the essential splicing factors analysed are similar in all the extracts. Thus, while we cannot exclude that some splicing factor that was not tested here has a reduced expression level, it is most likely that the inability of extracts from tsBN67 cells to splice pre-mRNA is caused by disruptions of essential interactions between mutant HCF-1 and other splicing factors (e.g. U1 and U5 snRNPs) rather than diminished expression levels of some specific splicing factors in these extracts.
Discussion
To date, the only well-characterized molecular function of HCF-1 has been related to its role in promoting the expression of the HSV IE genes upon infection of cells by the virus. The cellular and/or biochemical function of HCF-1 in uninfected cells is not yet clear. Because HCF-1 interacts with some cellular transcription factors, it has been suggested previously that this protein may have a role in transcriptional regulation of certain genes (Freiman and Herr, 1997; Lu et al., 1997, 1998; Gunther et al., 2000; Lu and Misra, 2000). However, which specific cellular genes may have their transcription regulated by HCF-1 has not been established. Here we provide evidence of a novel function for HCF-1 as a cellular pre-mRNA splicing factor. HCF-1 associates with complexes containing pre-mRNA splicing factors in vivo and in nuclear extract. The protein is a stable component of the mammalian spliceosome complex. The results also demonstrate that HCF-1 may be required for efficient spliceosome assembly and catalysis to occur in mammalian nuclear extracts. This work therefore constitutes a first characterization of a molecular function for HCF-1 in a biochemical pathway.
The nuclear localization pattern for HCF-1 is consistent with its role as a splicing factor because it localizes in nuclear structures that previously have been shown to contain pre-mRNA splicing factors (reviewed in Lamond and Earnshaw, 1998; Mistelli and Spector, 1998; Gall, 2000). The diffuse and punctate nuclear localization of HCF-1 has been observed previously in HeLa cells (Kristie et al., 1995), although the authors did not confirm the identity of these nuclear structures by co-localization experiments. As recently demonstrated, not all punctate or speckled nuclear patterns co-localize with splicing factors (Fox et al., 2002). We observe that HCF-1 co-localizes with Cajal bodies and gems in hamster and several different human cancer and primary cell lines. These nuclear bodies contain proteins involved in pre-mRNA processing (Meister et al., 2000; and references therein) and may function in a nuclear RNP maturation pathway (Sleeman et al., 1999).
HCF-1 is conserved across species and is expressed in all mammalian cell types (Johnson et al., 1999), suggesting that the protein may have a general cellular function, e.g. in either gene regulation or expression in the cell nucleus. However, there are a few tissues in which HCF-1 expression is not detectable (Kristie et al., 1995; Kristie, 1997). This may be due to very low levels of expression of this protein in the above-mentioned tissues or, alternatively, to the replacement of HCF-1 in these cells with other factors of similar function. In sensory neurons, where HCF-1 is expressed, the protein is not detectable in the nucleus. It has been shown that in trigeminal ganglia stained with anti-HCF-1 antibodies, the protein is localized in the cytoplasm and relocalizes to the nucleus upon reactivation from latency of HSV in infected cells (Kristie et al., 1999). The above observations indicate that either the level of HCF-1 present in the nuclei of these tissues is very low, thus making detection difficult, or there may be some redundancy in the function of HCF-1, in pre-mRNA splicing, in these cell types. The function of HCF-1 in these cells could be carried out by other proteins, e.g. the recently described HCF-1-like proteins (Johnson et al., 1999; Zhou et al., 2001). Further studies are required to investigate whether these HCF-1-like proteins can perform a similar cellular function(s) to that of HCF-1.
Although HCF-1 was found to interact with complexes containing splicing snRNPs, it is unlikely that the protein is a bona fide and stable snRNP particle component because when the immunoprecipitation washes were performed under stringent conditions (>400 mM salt), the HCF-1–snRNP association was disrupted. Indeed, standard purification protocols for snRNPs use column wash buffers containing salt concentrations >400 mM (Bach et al., 1990). Thus, although HCF-1 interacts with complexes containing splicing snRNPs, this interaction is not as stable as those that exist between the known core components of splicing snRNP particles and may be mediated by other factors. The co-immunoprecipitation of SMN from nuclear extracts by anti-HCF-1 antibodies is consistent with this hypothesis. SMN interacts with Sm proteins, and several recent studies have suggested a role for the SMN protein in pre-mRNA processing (Pellizzoni et al., 1998; Meister et al., 2000; Mourelatos et al., 2001). Mutations in the SMN protein can lead to the common neurodegenerative disease condition called spinal muscular atrophy (SMA) (Young et al., 2001; reviewed by Paushkin et al., 2002).
We have previously analysed the major components of the human spliceosome using two-dimensional SDS–PAGE, mass spectrometry and expressed sequence tag (EST) database searches (Neubauer et al., 1998). However, using such two-dimensional gel systems, a lot of proteins with relatively high molecular weights (>100 kDa) are not resolved efficiently and thus remained unidentified. Also, less abundant proteins in the spliceosome complex may be missed during the analyses. Most of the HCF-1 polypeptides in HeLa nuclear extract as determined using anti-HCF-1 antibodies or HCF-1 purification (Wilson et al., 1993) have molecular weights >110 kDa. Thus these HCF-1 polypeptides in the spliceosome complex may have been missed in the above study due to the difficulties encountered in resolving high molecular weight proteins in the two-dimensional gel system. In this study, we used a one-dimensional SDS–PAGE system capable of resolving large proteins for separating the components of the spliceosome complex. Results from all these experiments consistently showed that HCF-1 is a specific and stable component of the human spliceosome complex. Also, in a recent parallel analysis of spliceosomal proteins by LC/MS–MS without prior SDS–PAGE separation of the components, the presence of HCF-1 was confirmed in the purified spliceosome complex (Rappsilber et al., 2002).
Mutations in several pre-mRNA processing factors have been linked to defects in the cell division cycle, e.g. CDC5L/hCDC5 (Ohi et al., 1998; Bernstein and Coughlin, 1998), Prp17, Syf1p, Syf2p and Syf3 (Russell et al., 2000), SAP130/SF3b130 (Habara et al., 2001), Prp4p kinase (Schwelnus et al., 2001), Schizosaccharomyces pombe PRP5, PRP6 (Potashkin et al. 1998), PRP8 (Lundgren et al., 1996) and U2AF65 (Beales et al., 2000; and references therein). Therefore, it would be interesting to investigate whether the proline to serine missense mutation at position 134 in HCF-1 of the BHK21 hamster cell line tsBN67 that causes cell cycle arrest at G0/G1 when the cells are maintained at the non-permissive temperature (Goto et al., 1997) would affect splicing. We found that interactions between HCF-1 and U1 and/or U5 snRNPs were disrupted or destabilized in extracts from tsBN67 cells, and this disruption may be the cause of the observed deficiency in the assembly of stable spliceosome complexes. Thus HCF-1 may function in the spliceosome assembly pathway by interacting with splicing snRNPs to stabilize their association with the maturing spliceosome complex. The observation that antibodies to HCF-1 will inhibit splicing catalysis but not prevent spliceosome assembly in nuclear extracts containing wild-type HCF-1 suggests a possible function for the protein in the first step of splicing, perhaps through interactions with other non-snRNP splicing factors.
In order to demonstrate that the inefficient splicing observed in nuclear extracts from tsBN67 cells maintained at the non-permissive temperature is not the result of some general non-specific splicing deficiency in G0/G1-arrested cells, we treated BHK21 and tsBN67 cells with DMSO, a chemical widely used to induce arrest in mammalian cells (Takase et al., 1992; Boquest et al., 1999; Kudo et al., 2002). We found that this treatment does not result in a loss of splicing activity in extracts from the treated cells. This observation means that the inefficient splicing observed in tsBN67 extracts prepared from cells shifted to the non-permissive temperature is most likely due to the mutation in HCF-1, a conclusion strongly supported by our finding that transient expression of YFP–HCF-1 can restore splicing activity in extracts from tsBN67 cells maintained at 39.5°C. Our results are also consistent with previous observations that the expression of some genes (that contain introns and need to be spliced) is up-regulated in mammalian cells that have been chemically induced to stop at the G0/G1 phase of the cell cycle. For example, treatment of murine F-MEL cells with DMSO up-regulates Bcl-XL gene expression (Hafid-Medheb et al., 1999). Tamoxifen induces G0/G1 arrest in pancreatic cancer cells with an associated up-regulation of the expression of p21WAF1 (Robinson et al., 1998). Curcumin has been shown to inhibit cell cycle progression of human ECV304 cells by up-regulating the expression of cyclin-dependent kinase inhibitor, p21WAF1/CIP1, p27KIP1 and p53 (Park et al., 2002). Exposure of the human U937 cell line to a combination of granulocyte– macrophage colony-stimulating factor (GM-CSF) and vitamin D3 results in an up-regulation of c-fos gene expression that is associated with a shift of cell population from the S phase to the G0/G1 phase (Kim et al., 1991). Our results and the above reports demonstrate that many cell types arrested at the G0/G1 phase of the cell cycle maintain the ability to splice pre-mRNA. Therefore, it is very unlikely that the splicing defect of nuclear extracts from tsBN67 cells cultured at 39.5°C is the result of a general non-specific pre-mRNA splicing deficiency in G0/G1-arrested cells.
HCF-1 has been best characterized as a facilitator of HSV infection and its establishment in host cells. In humans, after infection by the virus and replication at mucosal surfaces, HSV enters sensory nerve endings and is then transported to the neuronal cell bodies where a more restrictive replication cycle occurs, resulting in a latent infection of these neurons. Productive infection at the sites of latency can be reactivated by a variety of stimuli (reviewed by Quinn et al., 2000). HCF-1 has been shown to be required for the transcription of the HSV α/IE genes. Upon infection of susceptible cells by HSV, the first genes to be expressed are the α/IE genes (ICP0, ICP4, ICP22, ICP27 and ICP47), followed by the early (β) genes and then finally the late (γ) genes (reviewed by Weir, 2001). Only four of the >80 transcripts expressed during HSV infection contain introns. Interestingly, three of the four intron-containing transcripts are expressed from α/IE genes (reviewed by Weir, 2001). It is possible, therefore, that HCF-1 may also be required in virally infected cells for the efficient splicing of the above α/IE pre-mRNAs.
While our data indicate that HCF-1 acts as a splicing factor, this does not exclude it also having other cellular functions. Also, HCF-1 may not be required for the splicing of all pre-mRNAs in vivo. In uninfected cells, HCF-1 may be a component of a larger complex or a transcriptosome that plays a role in either the expression (i.e. both transcription and splicing) of specific cellular genes or the co-transcriptional splicing of pre-mRNAs. Furthermore, HCF-1 is a large protein and it is possible that it may be multifunctional in the cell, as demonstrated by its presence in the cytoplasm of trigeminal ganglia (Kristie et al., 1999). There is recent evidence pointing to coupling between transcription and pre-mRNA splicing mediated often by interactions between splicing factors and the C-terminal domain of RNA polymerase II (reviewed by Bentley, 2002). Thus, our data do not exclude a role for HCF-1 in transcription because the transcription factor WT1 has also been shown to function in splicing (reviewed by Englert, 1998). Although our results in this study indicate that HCF-1 is required for pre-mRNA splicing, further studies are needed to elucidate the possible role of HCF-1 in the splicing of HSV α/IE transcripts in vivo. These studies may provide new insight into the mechanisms of HSV infection and gene expression in host cells.
Materials and methods
Plasmids
The plasmid constructs pBSAd1 (Konarska and Sharp, 1987), pBSAL4 (Lamond et al., 1987), hnRNP A1-533 series plasmids (Chabot et al., 1997), pGEM-RPL32 and pGEM-RPL32:T9 (Vilardell and Warner, 1997; and references therein) were used for preparing pre-mRNA templates for in vitro splicing. The plasmid construct pEYFP-HCF was made by digesting the plasmid pCGNHCF (Wilson et al., 1993) with the restriction endonucleases SnaBI and BamHI (Boerhinger). The full-length HCF-1 cDNA fragment was gel purified (Qiagen) and subcloned into SmaI–BamHI-digested pEYFP-C1 (Clontech) that contains the YFP gene.
Cell culture and transfection
Human and Syrian hamster (BHK21 and tsBN67) cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and 100 U/ml penicillin and streptomycin (Life Technologies Ltd). The BHK21 and tsBN67 (obtained from RIKEN Gene Bank, Tsukuba, Japan) cells were maintained as described previously (Goto et al., 1997; Wysocka et al., 2001). For immunofluorescence assays, cells were grown on coverslips and transfected using Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions.
Cell staining and immunofluorescence analyses
Cells were washed in phosphate-buffered saline (PBS) and fixed for 5 min in 3.7% (w/v) paraformaldehyde in PHEM buffer (60 mM PIPES pH 6.8, 27 mM HEPES pH 7.0, 4 mM MgSO4·7H2O and 13 mM EGTA) at room temperature. Permeabilization was performed with 1% Triton X-100 in PBS for 10 min at room temperature. Cells were incubated with primary antibodies diluted in PBS with 1% goat serum for 35 min to 1 h, washed three times for 10 min with PBS, incubated for 0.5–1 h with the appropriate secondary antibodies diluted in PBS with 1% goat serum and washed three times for 10 min with PBS. Antibodies used were Y12 mAb (anti-Sm) diluted 1:500 (Petterson et al., 1984), anti-p80 coilin mAb (5P10) diluted 1:10, MANSMA1 anti-SMN mAb (Young et al., 2000) diluted 1:10 and rabbit anti-HCF peptide antibodies HC1 or HC2 diluted 1:700. Tetramethylrhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse and Cy5-conjugated goat anti-rabbit secondary antibodies were also used (Jackson Immunochemicals). Microscopy and image analysis were carried out using a Zeiss Delta Vision Restoration microscope as described previously (Platani et al., 2000).
FACS analyses
Cells to be used for FACS analysis were harvested by trypsinization. The harvested cells were then washed with PBS, fixed with ice-cold 70% ethanol and left at 4°C for at least 3 h (sometimes up to several days). Fixed cells were collected by centrifugation. To determine cellular DNA content, the cell pellets were resuspended in PBS containing 100 µg/ml RNase A and stained with 25 µg/ml propidium iodide. After incubating the cells at 37°C for 30 min in the dark, the stained cell pellets were resuspended in PBS and the DNA content analysed by FACSort (Becton Dickinson).
Preparation of nuclear extracts from tissue culture cells
Tissue culture cells were grown in tissue culture dishes and harvested by scraping when they were ∼80–90% confluent. The cells were washed twice in PBS and the pellet resuspended in one packed cell volume of buffer NE1 [10 mM HEPES pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 1 M dithiothreitol (DTT)]. The suspension was incubated on ice for 15 min before being taken up and forced out five times through a 23 gauge syringe needle. The sample was then centrifuged in an Eppendorf microfuge for 20 s at full speed. The supernatant was discarded and the nuclear pellet resuspended in a two-thirds packed cell volume of buffer NE2 [20 mM HEPES pH 8.0, 1.5 mM MgCl2, 25% glycerol, 420 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] before incubation on ice with regular stirring for 30 min. Nuclear debris was separated by centrifugation, and the supernatant dialysed against nuclear extract buffer (20 mM HEPES pH 8.0, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF, 1 mM DTT).
Immunoprecipitation, SDS–PAGE and western blotting
Immunoprecipitations were performed from HeLa nuclear extracts, and SDS–PAGE and western blotting were performed as described previously (Ajuh et al., 2000). For western blotting, the following primary antibodies were used: 5P10 anti-p80 coilin mAb (dilution 1:100), anti-HCF peptide antibodies (HC1 and HC2) (Ajuh et al., 2000) (dilution 1:1000), Y12 mAb (anti-Sm) (Petterson et al., 1984) (dilution 1:2500), anti-SMN mAb (Transduction Laboratories), and mAb104, which recognizes the common epitope of SR proteins (Zahler et al., 1993). Protein bands were detected by using appropriate peroxidase-conjugated secondary antibodies (Pierce) and blots developed using the ECL kit (Amersham-Pharmacia) according to the manufacturer’s instructions. Samples to be used for northern blotting of snRNAs were treated as described previously (Rappsilber et al. 2001).
In vitro transcription and antisense probes
The pBSAd1 plasmid was digested with Sau3AI. The 533 plasmid constructs were digested with ScaI, pBSAL4 was digested with PvuII and the RPL32 constructs were digested with RsaI. All the digested plasmids were used for in vitro transcription under conditions similar to pBSAd1 (Konarska and Sharp, 1987) using the enzyme T3 RNA polymerase, except for the RPL32 constructs that were transcribed using T7 RNA polymerase (Promega). Preparation of antisense probes for snRNA and northern blot hybridization experiments was performed as described previously (Ryder et al., 1990).
Splicing assays and spliceosome purification
Splicing assays were performed using uniformly labelled, capped pre-mRNAs incubated with nuclear extract as described previously (Lamond et al., 1987). Nuclear extracts used in the splicing assays were obtained commercially from Computer Cell Culture Centre (Mons, Belgium). When the reactions were to be used for the analysis of splicing complexes, they were loaded onto a polyacrylamide–agarose composite gel and run for ∼5 h at 25 mA (Konarska and Sharp, 1987). The spliceosome complex was purified from the splicing reaction in a two-step process as described previously (Neubauer et al., 1998).
Acknowledgments
Acknowledgements
We thank Dr Josep Vilardell for the gift of the RPL32 plasmids, Dr Faiz Nasim for providing us with the hnRNP A1-533 series plasmids, Dr D.Gregory for the gift of the BHK21 cell line, Dr Winship Herr for providing us with the pCGNHCF plasmid, Dr G.E.Morris for the generous gift of the anti-SMN monoclonal antibody, MANSNA1, and all the members of the Lamond laboratory for their helpful advice. The Wellcome Trust supported this work. A.I.L is a Wellcome Trust Principal Research Fellow.
References
- Adams J., Kelso,R. and Cooley,L. (2000) The kelch superfamily of proteins: propellers of cell function. Trends Cell Biol., 10, 17–24. [DOI] [PubMed] [Google Scholar]
- Ajuh P.M., Browne,G.J., Hawkes,N.A., Cohen,P.T., Roberts,S.G. and Lamond,A.I. (2000) Association of a protein phosphatase 1 activity with the human factor C1 (HCF) complex. Nucleic Acids Res., 28, 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M., Krol,A. and Luhrmann,R. (1990) Structure-probing of U1 snRNPs gradually depleted of the U1-specific proteins A, C and 70k. Evidence that A interacts differentially with developmentally regulated mouse U1 snRNA variants. Nucleic Acids Res., 18, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales M., Flay,N., McKinney,R., Habara,Y., Ohshima,Y., Tani,T. and Potashkin,J. (2000) Mutations in the large subunit of U2AF disrupt pre-mRNA splicing, cell cycle progression and nuclear structure. Yeast, 16, 1001–1013. [DOI] [PubMed] [Google Scholar]
- Bentley D. (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol., 14, 336–342. [DOI] [PubMed] [Google Scholar]
- Bernstein H.S. and Coughlin,S.R. (1998) A mammalian homolog of fission yeast Cdc5 regulates G2 progression and mitotic entry. J. Biol. Chem., 273, 4666–4671. [DOI] [PubMed] [Google Scholar]
- Boquest A.C., Day,B.N. and Prather,R.S. (1999) Flow cytometric cell cycle analysis of cultured porcine fetal fibroblast cells. Biol. Reprod., 60, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Chabot B., Blanchette,M., LaPierre,I. and La Branche,H. (1997) An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol., 17, 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert C. (1998) WT1—more than a transcription factor? Trends Biochem. Sci., 23, 389–393. [DOI] [PubMed] [Google Scholar]
- Fox A.H., Lam,Y.W., Leung,A., Lyon,C., Andersen,J., Mann,M. and Lamond,A.I. (2002) Paraspeckles: a novel nuclear domain. Curr. Biol., 12, 13–25. [DOI] [PubMed] [Google Scholar]
- Freiman R.N. and Herr,W. (1997) Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev., 11, 3122–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen W.J. and Dreyfuss,G. (2000) Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN). J. Biol. Chem., 275, 26370–26375. [DOI] [PubMed] [Google Scholar]
- Gall J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell. Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Goto H., Motomura,S., Wilson,A.C., Freiman,R.N., Nakabeppu,Y., Fukushima,K., Fujishima,M., Herr,W. and Nishimoto,T. (1997) A single point mutation in HCF causes temperature sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev., 11, 726–737. [DOI] [PubMed] [Google Scholar]
- Gunther M., Laithier,M. and Brison,O. (2000) A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol. Cell. Biochem., 210, 131–142. [DOI] [PubMed] [Google Scholar]
- Habara Y., Urushiyama,S., Shibuya,T., Ohshima,Y. and Tani,T. (2001) Mutation in the prp12+ gene encoding a homologue of SAP130/SF3b130 causes differential inhibition of pre-mRNA splicing and arrest of cell-cycle progression in Schizosaccharomyces pombe. RNA, 7, 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafid-Medheb K., Poindessous-Jazat,V., Augery-Bourget,Y., Hanania,N. and Robert-Lezenes,J. (1999) Bcl-XL induction during terminal differentiation of Friend erythroleukaemia cells correlates with delay of apoptosis and loss of proliferative capacity but not with haemoglobinization. Cell Death Differ., 6, 166–174. [DOI] [PubMed] [Google Scholar]
- Herr W. (1998) The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol., 63, 599–607. [DOI] [PubMed] [Google Scholar]
- Johnson K.M., Mahajan,S.S. and Wilson,A.C. (1999) Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J. Virol., 73, 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.R., Abraham,N.G. and Lutton,J.D. (1991) Mechanisms of differentiation of U937 leukemic cells induced by GM-CSF and 1,25(OH)2 vitamin D3. Leukemia Res., 15, 409–418. [DOI] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A. (1987) Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell, 49, 763–774. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1995) The biochemistry of pre-mRNA splicing. In Lamond,A.I. (ed.), Pre-mRNA Processing. Springer-Verlag, Heidelberg, Germany, pp. 39–64.
- Krämer A. (1996) The structure and function of proteins in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 357–409. [DOI] [PubMed] [Google Scholar]
- Kristie T.M. (1997) The mouse homologue of the human transcription factor—factor C1 (host cell factor) conservation of forms and function. J. Biol. Chem., 272, 26749–26755. [DOI] [PubMed] [Google Scholar]
- Kristie T.M., Pomeranz,J.L., Twomey,T.C., Parent,S.A. and Sharp,P.A. (1995) The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem., 270, 4387–4394. [DOI] [PubMed] [Google Scholar]
- Kristie T.M., Vogel,J.L. and Sears,A.E. (1999) Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc. Natl Acad. Sci. USA, 96, 1229–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo H., Nakayama,T., Mano,Y., Suzuki,S., Sassa,S. and Sakamoto,S. (2002) Early progression from dimethyl sulfoxide-induced G(0)/G(1) arrest in L(1210) cells. Cell Biol. Int., 26, 211–215. [DOI] [PubMed] [Google Scholar]
- Lamond A.I. and Earnshaw,W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Konarska,M.M. and Sharp,P.A. (1987) A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev., 1, 532–543. [DOI] [PubMed] [Google Scholar]
- Liu Q., Fischer,U., Wang,F. and Dreyfuss,G. (1997) The spinal muscular atrophy disease gene product, SMN and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell, 90, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Lu R. and Misra,V. (2000) Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res., 28, 2446–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Yang,P., O’Hare,P. and Misra,V. (1997) Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol., 17, 5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Yang,P., Padmakumar,S. and Misra,V. (1998) The herpes virus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J. Virol., 72, 6291–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K., Allan,S., Urushiyama,S., Tani T., Ohshima,Y., Frendewey, D. and Beach,D. (1996) A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell., 7, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J.L. and Tacke,R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Meister G., Buhler,D., Laggerbauer,B., Zobawa,M., Lottspeich,F. and Fischer,U. (2000) Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet., 9, 1977–1986. [DOI] [PubMed] [Google Scholar]
- Mistelli T. and Spector,D.L. (1998) The cellular organization of gene expression. Curr. Opin. Cell Biol., 10, 323–331. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Abel,L., Yong,J., Kataoka,N. and Dreyfuss,G. (2001) SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J., 20, 5443–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- O’Hare P. (1993) The varion transactivator of herpes simplex virus. Semin. Virol., 4, 145–155. [Google Scholar]
- Ohi R., Feoktistova,A., McCann,S., Valentine,V., Look,A.T., Lipsick, J.S. and Gould,K.L. (1998) Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol., 18, 4097–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.J. et al. (2002) Curcumin inhibits cell cycle progression of immortalized human umbilical vein endothelial (ECV304) cells by up-regulating cyclin-dependent kinase inhibitor, p21WAF1/CIP1, p27KIP1 and p53. Int. J. Oncol., 21, 379–383. [PubMed] [Google Scholar]
- Paushkin S., Gubitz,A.K., Massenet,S. and Dreyfuss,G. (2002) The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol., 14, 305–312. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka,N., Charroux,B. and Dreyfuss,G. (1998) A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell, 95, 615–624. [DOI] [PubMed] [Google Scholar]
- Petterson I., Hinterberger,M., Mimori,T., Gottlieb,E. and Steitz,J.A. (1984) The structure of mammalian small nuclear ribonucleoproteins. Identification of multiple protein components reactive with anti-(U1) ribonucleoprotein and anti-Sm autoantibodies. J. Biol. Chem., 259, 5907–5914. [PubMed] [Google Scholar]
- Platani M., Goldberg,I., Swedlow,J.R. and Lamond,A.I. (2000) In vivo analysis of cajal body movement, separation and joining in live human cells. J. Cell Biol., 151, 1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin J., Kim,D., Fons,M., Humphrey,T. and Frendewey,D. (1998) Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet., 34, 153–163. [DOI] [PubMed] [Google Scholar]
- Quinn J.P., Dalziel,R.G. and Nash,A.A. (2000) Herpes virus latency in sensory ganglia—a comparison with endogenous neuronal gene expression. Prog. Neurobiol., 60, 167–179. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ajuh,P., Lamond,A.I. and Mann,M. (2001) SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J. Biol. Chem., 276, 31142–31150. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ryder,U., Lamond,A.I. and Mann,M. (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res., 12, 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. (1990) Protein composition mammalian spliceosomes assembled in vitro. Proc. Natl Acad. Sci. USA, 87, 8031–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. and Palandjian,L. (1997) Spliceosome assembly. In Krainer, A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 103–129.
- Reed R., Griffith,J. and Maniatis,T. (1988) Purification and visualisation of native spliceosomes. Cell, 53, 949–961. [DOI] [PubMed] [Google Scholar]
- Robinson E.K., Grau,A.M., Evans,D.B., Smid,C.M., Chiao,P.J., Abbruzzese,J.L. and Grimm,E.A. (1998) Cell cycle regulation of human pancreatic cancer by tamoxifen. Ann. Surg. Oncol., 5, 342–349. [DOI] [PubMed] [Google Scholar]
- Russell C.S., Ben-Yehuda,S., Dix,I., Kupiec,M. and Beggs,J.D. (2000) Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA, 6, 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder U., Sproat,B.S. and Lamond,A.I. (1990) Sequence-specific affinity selection of mammalian splicing complexes. Nucleic Acids Res., 18, 7373–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P., Sprangers,R., Stier,G., Buhler,D., Fischer,U. and Sattler,M. (2001) SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol., 8, 27–31. [DOI] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 15–26. [DOI] [PubMed] [Google Scholar]
- Schwelnus W., Richert,K., Opitz,F., Gross,T., Habara,Y., Tani,T. and Kaufer,N.F. (2001) Fission yeast Prp4p kinase regulates pre-mRNA splicing by phosphorylating a non-SR-splicing factor. EMBO rep., 2, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase K., Sawai,M., Yamamoto,K., Yata,J., Takasaki,Y., Teraoka,H. and Tsukada,K. (1992) Reversible G1 arrest induced by dimethyl sulfoxide in human lymphoid cell lines: kinetics of the arrest and expression of the cell cycle marker proliferating cell nuclear antigen in Raji cells. Cell Growth Differ., 8, 515–521. [PubMed] [Google Scholar]
- Valcarcel J. and Green,M.R. (1996) The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci., 21, 296–301. [PubMed] [Google Scholar]
- Vilardell J. and Warner,J.R. (1997) Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol., 17, 1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J.P. (2001) Regulation of herpes simplex virus gene expression. Gene, 271, 117–130. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) snRNP structure and function. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 130–173.
- Wilson A.C., LaMarco,K., Peterson,M.G. and Her,W. (1993) The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell, 74, 115–125. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Peterson,M.G. and Herr,W. (1995) The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev., 9, 2445–2458. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Reilly,P.T. and Herr,W. (2001) Loss of HCF-1–chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol., 21, 3820–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P.J., Le,T.T., thi Man,N., Burghes,A.H. and Morris,G.E. (2000) The relationship between SMN, the spinal muscular atrophy protein and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res., 256, 365–374. [DOI] [PubMed] [Google Scholar]
- Young P.J., Day,P.M., Zhou,J., Androphy,E.J., Morris,G.E. and Lorson,C.L. (2001) A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J. Biol. Chem., 277, 2852–2859. [DOI] [PubMed] [Google Scholar]
- Zahler A.M., Neugebauer,K.M., Lane,W.S. and Roth,M.B. (1993) Distinct functions of SR proteins in alternative pre-mRNA splicing. Science, 260, 219–222. [DOI] [PubMed] [Google Scholar]
- Zhou H.J., Wong,C.M., Chen,J.H., Qiang,B.Q., Yuan,J.G. and Jin,D.Y. (2001) Inhibition of LZIP-mediated transcription through direct interaction with a novel host cell factor-like protein. J. Biol. Chem., 276, 28933–28938. [DOI] [PubMed] [Google Scholar]