Abstract
The evolution of alternate modes of development may occur through genetic changes in metamorphic timing. This hypothesis was examined by crossing salamanders that express alternate developmental modes: metamorphosis vs. paedomorphosis. Three strains were used in the crossing design: Ambystoma tigrinum tigrinum (Att; metamorph), wild-caught A. mexicanum (Am; paedomorph), and laboratory Am (paedomorph). Att/Am hybrids were created for each Am strain and then backcrossed to their respective Am line. Previous studies have shown that a dominant allele from Att (metAtt) and a recessive allele from lab Am (metlab) results in metamorphosis in Att/Am hybrids, and metAtt/metlab and metlab/metlab backcross genotypes are strongly associated with metamorphosis and paedomorphosis, respectively. We typed a molecular marker (contig325) linked to met and found that metAtt/metlab and metAtt/metwild were associated with metamorphosis in 99% of the cases examined. However, the frequency of paedomorphosis was 4.5 times higher for metlab/metlab than for metwild/metwild. We also found that metAtt/metwild and metwild/metwild genotypes discriminated distributions of early and late metamorphosing individuals. Two forms of phenotypic variation are contributed by met: continuous variation of metamorphic age and expression of discrete, alternate morphs. We suggest that the evolution of paedomorphosis is associated with genetic changes that delay metamorphic timing in biphasic life cycles.
ALTERNATE modes of development are often observed within and between closely related species. A number of classic examples have been studied, including flight and flightless forms of insects, feeding and nonfeeding echinoderm larvae, and metamorphic and nonmetamorphic salamanders (Roff 1986; Matsuda 1987; Raff 1996; West-Eberhard 2004). These examples indicate a taxonomically widespread potential for the evolution of discrete, morphological characteristics that allow for novel and alternative life histories. In each of these cases, the phenotypic transition is explainable using “heterochronic” terms that describe how development evolves through changes in the timing in which characters are expressed during ontogeny (Gould 1977). For example, the evolution of salamander paedomorphosis presumably required a genetic change that blocked the initiation of metamorphosis, and this resulted in larval-form adults. Although such description is useful for explaining evolutionary shifts in development, it provides little insight about the structure of genetic architectures that underlie heterochronic, evolutionary transitions.
Like the majority of frogs and toads, many salamanders undergo an obligate metamorphosis that allows for the exploitation of both aquatic and terrestrial habitats during ontogeny. However, some salamander species express an alternate developmental mode in which they forego metamorphosis and remain in the aquatic habitat throughout their lifetimes (Figure 1). Nonmetamorphic forms are termed paedomorphic because they maintain juvenile features of the ancestral condition as they mature reproductively into large, larval forms (Gould 1977). The exemplar of salamander paedomorphosis is the Mexican axolotl (Ambystoma mexicanum). Ambystoma mexicanum (Am) belongs to a group of several closely related species collectively known as the tiger salamander species complex (Shaffer and McKnight 1996). Salamanders of this complex occupy a variety of North American breeding habitats ranging from temporary vernal pools to large permanent lakes. Among these habitats, populations are highly variable for metamorphic timing and expression of paedomorphosis. Some populations express metamorphosis (e.g., A. tigrinum tigrinum, Att) or paedomorphosis like Am, while in other populations both phenotypes are observed at varying frequencies. Presumably, the expression of paedomorphosis is an opportunistic strategy that allows individuals to more successfully colonize relatively permanent aquatic niches (Wilbur and Collins 1973; Sprules 1974). Paedomorphic tiger salamanders are found in newly created habitats like cattle watering troughs and wastewater treatment ponds (Rose and Armentrout 1975; Collins 1981), as well as in stable, large lake systems (Shaffer 1984).
Figure 1.—
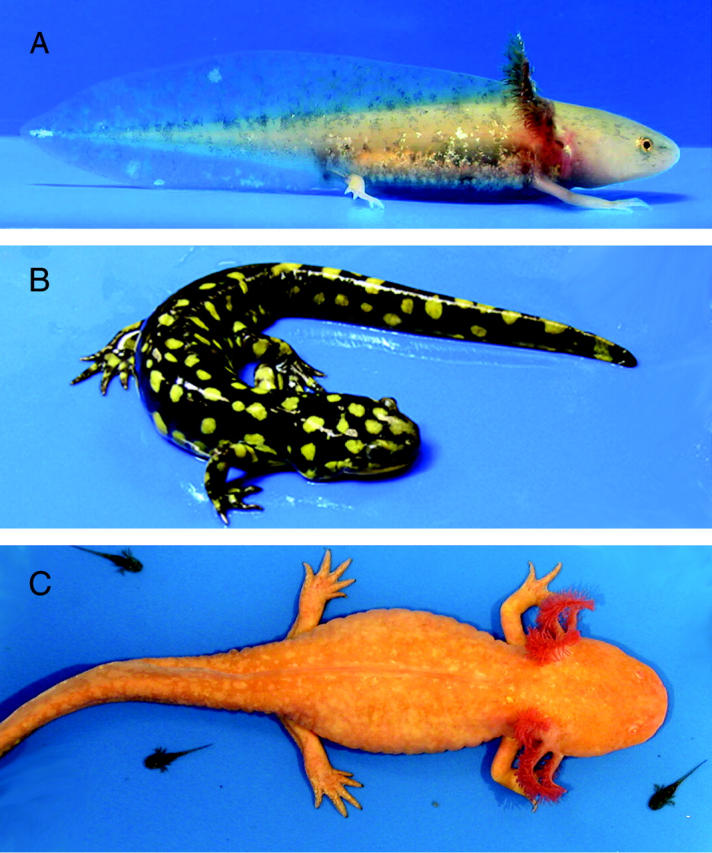
Larval and adult phases of Ambystoma. (A) Larval A. mexicanum. (B) Adult A. t. tigrinum (metamorphic). (C) Adult A. mexicanum (paedomorphic).
We examined the genetic contribution of a major-effect QTL (met) that is strongly associated with the discrete expression of metamorphosis vs. paedomorphosis in interspecific crosses using Att and a laboratory strain of Am (Voss and Shaffer 1997). Previous studies have shown that a dominant allele from Att (metAtt) and a recessive allele from lab Am (metlab) results in metamorphosis in Att/Am hybrids and that metAtt/metlab and metlab/metlab backcross genotypes are strongly associated with metamorphosis and paedomorphosis, respectively (Voss 1995; Voss and Shaffer 1997, 2000) (Table 1). Here we describe a newly identified and highly informative expressed sequence tag marker for met called contig325. This marker is informative in the majority of Ambystoma species (data not shown) and thus represents an important new candidate for studies of developmental timing variation in natural populations. We also describe a large, newly created backcross population using wild-caught Am individuals called WILD2. WILD2 and the smaller WILD1 backcrosses (Voss and Shaffer 2000) may differ from lab Am (LAB) backcrosses as a result of differences in the effects of met alleles and/or genetic background effects. To test this idea, we examined contig325 within the context of all available backcross populations (LAB, WILD1, and WILD2) to infer genetic changes that have modified the paedomorphic response of the natural Am population during domestication of the laboratory strain at Indiana University (The Axolotl Colony). Because WILD2 is the largest Att/Am backcross resource ever obtained (N = 457), we were also able to accurately and reliably assess the genetic contribution of met to a second form of phenotypic variation: continuous variation in metamorphic timing. Our results show that met contributes genetically to both discrete and continuous forms of metamorphic timing variation. This result suggests a linkage between the evolutionary maintenance of biphasic life cycles and the evolution of alternate developmental modes.
TABLE 1.
Nomenclature for backcrosses, expectedmet genotypes, expectedcontig325 genotypes, and expected morph phenotypes
| Cross ID | Backcross hybrid × Am | met genotype | contig325 genotype | Morph type |
|---|---|---|---|---|
| LAB | (Att/Am-lab) × Am-lab | metAtt/metlab | 325Att/325Am | Met |
| metlab/metlab | 325Am/325Am | Paed | ||
| WILD1 | (Att/Am-wild1) × Am-wild1 | metAtt/metwild1 | 325Att/325Am | Met |
| metwild1/metwild1 | 325Am/325Am | Paed | ||
| WILD1 | (Att/Am-wild2) × Am-wild2 | metAtt/metwild2 | 325Att/325Am | Met |
| metwild2/metwild2 | 325Am/325Am | Paed |
Dominant metAtt alleles derive from the same Att strain. Recessive metlab and metwild alleles derive from different Am strains. Contig325 is a species-specific marker locus linked to met. Morph types are based upon a single gene model. Att, A. tigrinum tigrinum; Am, A. mexicanum. Met, metamorph; Paed, paedomorph.
MATERIALS AND METHODS
Genetic crosses:
We compared phenotypic and genotypic segregation ratios that were obtained from three different backcross resources. Three strains were used to make the backcrosses: wild-caught Att, wild-caught Am, and laboratory Am (Figure 1; Table 1). The LAB and WILD1 backcrosses have been described (Voss 1995; Voss and Shaffer 2000). For LAB, Am were obtained from the Indiana University Axolotl Colony strain; for WILD1, Am were collected from their natural habitat at Lake Xochimilco, Mexico D.F., Mexico. In these and the WILD2 crosses described below, Att were obtained from the same source population in Tennessee (Charles Sullivan).
The WILD2 backcrosses were created to obtain the largest-ever segregating population for genetic analysis of Ambystoma (Smith 2002). WILD2 was created using Am individuals collected from Lake Xochimilco to make F1 hybrids and first generation descendants of wild-caught Am to make backcrosses. The F1 hybrids were generated from a single cross and backcross offspring were generated using three male Am and four female Att/Am hybrids. A total of nine backcross families compose WILD2 (Table 2). Artificial insemination was used in all crosses (Armstrong and Duhon 1989; Voss 1995).
TABLE 2.
WILD2 backcrosses showing parentage, morph segregation, and mean age at metamorphosis forcontig325 genotypes
| Parents
|
Offspring
|
Mean age at metamorphosis
|
|||||
|---|---|---|---|---|---|---|---|
| Cross ID | F1 | P2 | Met | Paed | % Paed | 325Att/325Am | 325Am/325Am |
| 1 | F1.1 | P2.1 | 69 | 5 | 0.07 | 175.4 | 217.4 |
| 2 | F1.2 | P2.1 | 50 | 7 | 0.12 | 171.3 | 199.1 |
| 3 | F1.3 | P2.1 | 38 | 3 | 0.07 | 183.2 | 206.7 |
| 4 | F1.4 | P2.1 | 65 | 11 | 0.15 | 177.1 | 204.8 |
| 5 | F1.1 | P2.2 | 56 | 6 | 0.10 | 173.2 | 210.1 |
| 6 | F1.2 | P2.2 | 80 | 3 | 0.04 | 164.5 | 203.8 |
| 7 | F1.3 | P2.2 | 44 | 1 | 0.02 | 165.1 | 214.6 |
| 8 | F1.4 | P2.2 | 35 | 4 | 0.10 | 159.9 | 200.2 |
| 9 | F1.2 | P2.3 | 16 | 4 | 0.20 | 156.7 | 209.2 |
| Total | 453 | 44 | 0.09 | 170.9 | 207.1 | ||
Rearing conditions:
The rearing methods for offspring from LAB and WILD1 were described previously (Voss 1995; Voss and Shaffer 2000). Here we describe rearing conditions for WILD2 offspring. At 21 days postfertilization, larvae were released from their eggs and placed individually in 5-oz paper cups of artificial pond water. Throughout the course of this experiment all individuals were maintained in a single room within which the temperature fluctuated from 19°–22°. Individuals were reared in separate containers and rotated within the room after water changes to reduce effects of spatial temperature variation. Larvae were fed freshly hatched Artemia twice daily for their first 30 days posthatching. After day 20, posthatching diets were supplemented with small (<1 cm) California black worms (Lumbriculus). During this time, individuals were provided with fresh water and cups after every third feeding. On day 30, larvae were transferred to 16-oz plastic bowls, after which they were fed California black worms exclusively and water was changed every third day. Finally, at 80 days posthatching all individuals were transferred to 4-liter plastic containers and were otherwise maintained under the same regimen as the previous 50 days, until completion of metamorphosis or the end of the experiment (day 350). The majority of backcross offspring were euthanized, as described above, upon completion of metamorphosis or at day 350. At this time, individuals were dissected and tissue samples (liver and/or blood) were collected for DNA isolation. A few individuals were not euthanized and are currently being maintained for use in future studies. For these individuals, tissue samples were collected as tail clips.
Phenotypic scores:
Individuals were scored as metamorphs upon complete resorption of all external gills (gills <1.0 mm in length). Age at metamorphosis was recorded as the number of days from fertilization to completion of metamorphosis. For WILD2, the experiment was terminated on day 350, at which point no individuals had completed metamorphosis within the previous 3 weeks. All remaining individuals showed no sign of having initiated metamorphosis (no apparent regression of the tail fin or external gills) and were scored as paedomorphs.
Genotyping:
A total of 98, 112, and 457 individuals from LAB, WILD1, and WILD2, respectively, were genotyped for contig325, a molecular marker that was isolated as a result of ongoing EST and genetic linkage mapping projects that generate genome resources for Ambystoma research (http://salamander.uky.edu). This marker was isolated from an Am tail regeneration blastema cDNA library (Putta et al. 2004). Additional coding sequence for this EST was obtained by 5′-RACE and assembled with existing EST sequences. The resulting 985-bp DNA sequence shows strong similarity to a human nerve growth factor receptor precursor (sequence data not shown; NP_002498, bit score = 164; BLASTX).
A 221-bp DNA fragment corresponding to contig325 was amplified from all individuals under standard PCR conditions (150 ng DNA, 50 ng each primer, 1.2 mm MgCl2, 0.3 units Taq polymerase, 1× PCR buffer, 200 mm each of dATP, dCTP, dGTP, and dTTP; thermal cycling at 94° for 4 min; 33 cycles of 94° for 45 sec, 60° for 45 sec, 72° for 30 sec; and 72° for 7 min). DNA was isolated from all individuals using a previously described phenol extraction method (Voss 1993). Primer sequences for amplifying contig325 are forward, 5′-GTGAAGTCAGTGATGAAAGTCCATGT-3′, and reverse, 5′-CTAGGATACCAGTGGGAGAGTGTAAT-3′. Genotypes were assayed by restriction digestion of PCR products with a diagnostic AluI restriction enzyme (New England Biolabs, Beverly, MA) and agarose gel electrophoresis.
Linkage analysis:
Linkage and QTL mapping studies were performed using the software package MapMakerQTXb19 (http://www.mapmanager.org/mmQTX.html; Meer et al. 2004). Linkage distance and arrangement among contig325 and previously described amplified fragment length polymorphisms (AFLP) (Voss and Shaffer 1997) was estimated using the Kosambi mapping function at a linkage threshold of P = 0.001. The maximum-likelihood position of the met QTL was estimated using the interval mapping function. Significance thresholds for interval mapping were obtained through 10,000 permutations of trait values among backcross progeny. Associations between contig325 genotypes and phenotypic variation were measured using the marker regression function.
RESULTS
Identification of a highly informative EST marker for met:
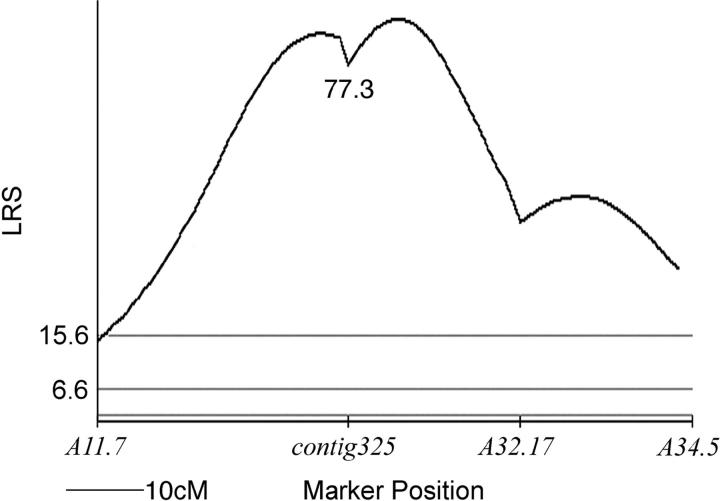
The met QTL was originally identified in LAB using AFLPs and interval mapping (Voss and Shaffer 1997). In LAB, metAtt/metlab and metlab/metlab segregate as highly penetrant genotypes for metamorphosis and paedomorphosis, respectively; in fact, cosegregation of associated AFLPs and morph phenotypes was statistically consistent with simple Mendelian inheritance. However, given the anonymous nature of AFLPs and the nonspecific way in which these markers are generated, we identified a more informative and user-friendly marker for met: an expressed sequence tag that we refer to as contig325. To show correspondence of contig325 to met, we genotyped individuals from LAB. We observed a higher association between contig325 and met than was observed with the most closely linked AFLP marker (estimated proportion recombinants: AFLP32.17 = 0.15, N = 70; contig325 = 0.07, N = 91). Genotypes at contig325 explain 71% of discrete variation for segregation of metamorphosis vs. paedomorphosis in LAB. Interval mapping shows that contig325 is located near the maximum inflexion point of the previously determined AFLP LOD profile for met (Figure 2).
Figure 2.—
Likelihood-ratio statistic (LRS) plot for association of paedomorphosis with genetic factors in the met QTL region. The LRS for the contig325 marker is shown. Horizontal shaded lines represent LRS thresholds for suggestive (1.3), significant (6.6), and highly significant associations (15.6) as estimated using Map Manager QTXb19.
Survival of offspring in the newly created WILD2 mapping panel:
Overall, a high proportion (91%) of WILD2 backcross offspring survived from hatching to completion of metamorphosis or failed to metamorphose by day 350. There were no significant differences in survival probability among crosses. In total, the nine crosses generated 497 backcross offspring that survived through completion of metamorphosis or to the end of the experiment as paedomorphs.
Segregation of discrete developmental modes and contig325 in WILD2 and WILD1:
We observed the segregation of metamorphs and paedomorphs in all nine WILD2 crosses (Table 2). Segregation ratios were not significantly heterogeneous (G = 13.32, d.f. = 8, P = 0.10) among crosses; therefore segregation ratios were pooled for hypothesis testing. The majority of offspring generated (453 of 497) metamorphosed before day 350. In total, only 44 (9%) of the offspring exhibited paedomorphosis and ratios were significantly different from the simple Mendelian expectation of 1:1 (G = 392, d.f. = 1, P = 4 × 10−87, N = 497). Significantly lower-than-expected numbers of paedomorphs (19%) were also observed in WILD1 (Voss and Shaffer 2000). Thus, results from WILD1 and WILD2 indicate that the proportion of paedomorphs is significantly lower in backcrosses using wild-caught Am, relative to laboratory Am.
To determine if met contributed to the segregation of discrete developmental modes in WILD2, we genotyped all individuals for contig325 (325) (Table 3). Inheritance of 325Att/325Am, and thus presumably of metAtt/metwild2 (Table 1), yielded the expected metamorphic phenotype in >99% of the cases. The 325Am/325Am genotype (presumably marking metwild2/metwild2 ) was not as penetrant for the paedomorphic phenotype as only 17% of individuals in this genotypic class were paedomorphs. However, inheritance of metwild2/metwild2 is apparently necessary for expression of paedomorphosis as only one paedomorph inherited a metAtt/metwild2 genotype. To investigate linkage results between WILD2 and WILD1, which had previously been examined using only the informative AFLP makers (Voss and Shaffer 2000), we genotyped individuals from WILD1 for contig325. The pattern of segregation for contig325 in WILD1 was the same as that observed for WILD2. All but one individual that inherited 325Att/325Am was metamorphic and 325Am/325Am yielded incomplete penetrance for paedomorphosis (only 16 of 51 325Am/325Am genotypes were paedomorphic). Observation of the same pattern of segregation between WILD1 and WILD2 suggests no sex linkage or maternal effect on the segregation of genotypes and phenotypes because the crossing designs were reversed to create WILD1 and WILD2 backcrosses (i.e., F1 hybrids were male in creating WILD1 but female in creating WILD2). Overall, our results show that 325Att/325Am is strongly associated with the metamorphic phenotype; this association did not vary across LAB, WILD1, or WILD2. However, the proportion of 325Am/325Am genotypes that were associated with paedomorphosis was 4.5 times higher in LAB than in WILD1 and WILD2. This indicates a genetic difference in the basis of paedomorphosis between the natural and domestic strains of Am.
TABLE 3.
Segregation ofcontig325 genotypes
| Cross type | Morph | 325Att/325Am | 325Am/325Am |
|---|---|---|---|
| LAB | Met | 52 | 5 |
| Paed | 2 | 39 | |
| WILD1 | Met | 60 | 35 |
| Paed | 1 | 16 | |
| WILD2 | Met | 219 | 196 |
| Paed | 1 | 41 |
Continuous variation for age at metamorphosis and association with met:
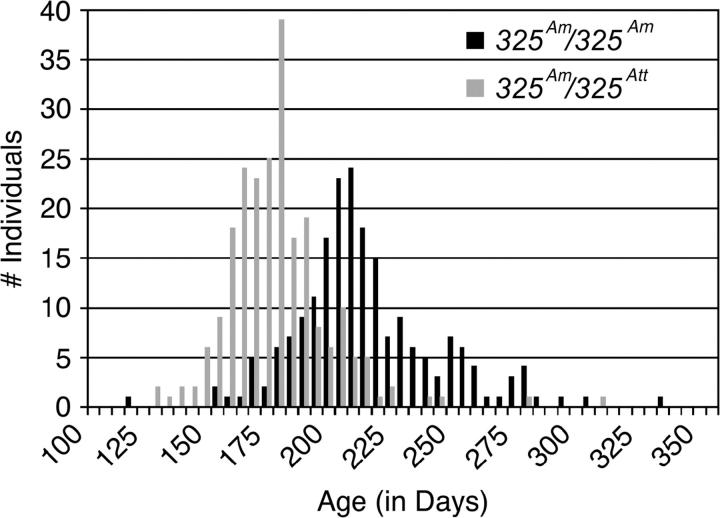
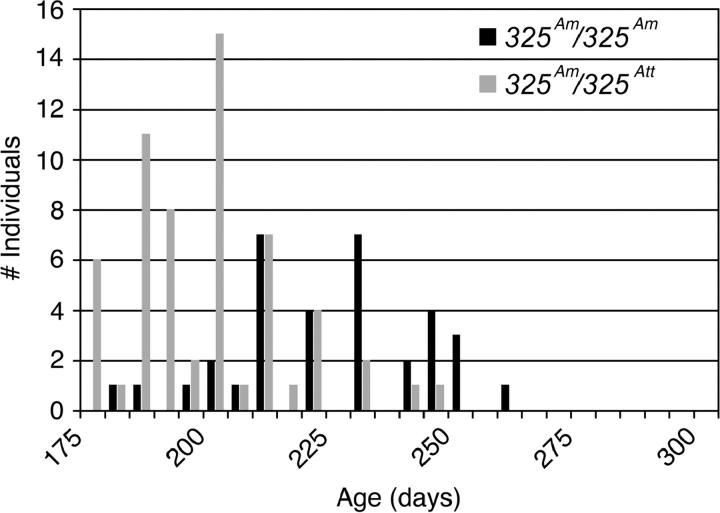
The large number of metamorphic individuals in both genotypic classes from the WILD2 panel provided the opportunity to test for association between contig325 and a second form of metamorphic timing variation: age at metamorphosis. We examined timing of metamorphosis only for those individuals that did undergo a metamorphosis. Age at metamorphosis varied continuously from 115 to 300 days in WILD2. Plotting metamorphic ages separately for 325Att/325Am and 325Am/325Am revealed two overlapping yet distinct distributions (Figure 3; Table 2). The means of these two distributions, 171 days and 207 days, differ significantly (t = 14.48, d.f. = 413, P < 0.0001), with 325Am/325Am individuals metamorphosing on average 36 days later than 325Att/325Am individuals. A similar difference in age at metamorphosis between genotypic classes was also observed within the WILD1 panel (t = 6.99, d.f. = 93, P < 0.0001) with 325Am/325Am individuals metamorphosing on average 25 days later than 325Att/325Am individuals (Figure 4). Our replicated result indicates that met, which is strongly associated with discrete variation for metamorphosis/paedomorphosis, is also strongly associated with continuous variation for metamorphic age.
Figure 3.—
Distribution of ages at metamorphosis for WILD2 plotted separately for contig325 genotypes.
Figure 4.—
Distribution of ages at metamorphosis for WILD1 plotted separately for contig325 genotypes.
DISCUSSION
Novel developmental modes may evolve as a result of genetic changes in developmental timing or heterochrony (Gould 1977; Roff 1986; Matsuda 1987; Ambros 1988; Raff 1996; West-Eberhard 2004). The paedomorphic developmental mode of the Mexican axolotl (Am) is a classic example of heterochrony. Paedomorphosis in Am presumably evolved as a result of a genetic change that blocked the initiation of metamorphosis in a biphasic ancestor, and this resulted in larval-form adults. In support of this idea, we found that within interspecific crosses using Am and metamorphic Att, the segregation of genotypes at a major-effect QTL (met) was associated with the expression of metamorphosis vs. paedomorphosis. This result supports the long-held idea that paedomorphosis in Am evolved via saltation (Goldschmidt 1940; Gould 1977, 1981; Tompkins 1978; Ambros 1988; McKinney and McNamara 1991; Voss 1995; Voss and Shaffer 1997; Futuyma 1998). However, we also identified differences in gene effect that have evolved rapidly between the laboratory and wild strains of Am (Voss and Shaffer 2000), and we found that met contributed to a second form of phenotypic variation: continuous variation in age at metamorphosis. This later result indicates that expression of paedomorphosis is associated with genetic changes that alter developmental timing (contra Raff and Wray 1989; Raff 1996). Below, we review the primary results and then explain how a genetic architecture that contributes to both continuous and discrete phenotypic variation supports a more gradual selection model for the evolution of paedomorphosis.
Genetic basis of discrete variation: expression of metamorphosis vs. paedomorphosis:
A conceptual framework for understanding how polygenes give rise to discrete phenotypic variation is the threshold model (Falconer 1989). Under this model, the expression of alternate phenotypes depends upon an individual's liability value relative to a threshold value, with liability values above and below the threshold yielding alternate phenotypes. We suggest that contig325 makes a major contribution to the liability or threshold underlying the expression of metamorphosis vs. paedomorphosis. Within LAB, both 325Att/325Am and 325Am/325Am were highly predictive of their expected phenotypes, indicating highly significant linkage to a single locus (met) (χ2 = 84.97, d.f. = 1, N = 98, P < 0.001; Table 3). Thus, in the LAB genetic background, the threshold for expressing metamorphosis vs. paedomorphosis is traversed by the segregation of alternate met genotypes at a single locus. Apparently, 325Att/325Am is not sensitive to genetic background because this genotype was also highly predictive of metamorphosis in WILD1 and WILD2. Thus, in both LAB and WILD genetic backgrounds, substitution of a single Am met allele with a dominant Att met allele rescued the metamorphic phenotype in essentially all cases.
In contrast to 325Att/325Am, the penetrance of 325Am/325Am for paedomorphosis varied between LAB and the WILD backcrosses. This suggests that metlab and metwild1,2 contribute differently to the underlying genetic architecture or that LAB and WILD genetic backgrounds influence the probability of paedomorphosis differently. Although we cannot differentiate between these two possibilities, the genetic basis of paedomorphosis clearly differs between the natural population and a recently derived laboratory strain of Am, thus indicating the potential for rapid evolution of genetic architecture. This supports the idea that the simple Mendelian basis of paedomorphosis in LAB evolved recently during the domestication of Am (Voss and Shaffer 2000; see also Malacinski 1978). Although paedomorphosis is expressed by both the wild strain and the laboratory strain, our results indicate that selection has canalized expression of paedomorphosis to a greater degree in the laboratory strain, as assayed by our interspecific crossing design. Thus, although paedomorphosis has been cited as a classic example of heterochrony by a major gene effect, our study shows that factors beyond a single major gene are important in discrete trait expression in Am.
Genetic basis of continuous variation: variation in metamorphic age:
Because the WILD backcrosses yielded a large number of metamorphosing offspring reared under identical conditions, we were able to estimate the contribution of met to variation in metamorphic age. We found that metamorphic age varied significantly between 325Am/325Am and 325Att/325Am genotypic classes. This indicates that metwild/metwild delays timing of metamorphosis relative to metAtt/metwild. Because metwild/metwild was associated with paedomorphosis in WILD1,2 (all but two paedomorphs were metwild/metwild), our results show that both delayed metamorphosis and expression of paedomorphosis are associated with this genotype; we note that these associations were observed in the same genetic background. Conversely, an earlier metamorphosis was associated with the alternate metAtt/metwild genotype, again within the same WILD genetic backgrounds. This indicates that met alleles deriving from paedomorphic Am delay metamorphosis while met alleles from the metamorphic Att decrease the time to metamorphosis. We suggest that metamorphic age is a continuous variable that is closely associated with the underlying liability or threshold that determines the expression of alternate developmental modes. It is possible that met influences metamorphic timing via changes in the timing of the sensitive period for hormonal initiation of metamorphosis, as has been suggested for dung beetles (Onthophagus taurus) that express alternate male morphs (Moczek and Nijhout 2002). A comparative mapping project is underway to identify likely candidate genes in the vicinity of contig325 (http://salamander.uky.edu).
Evolutionary maintenance of the biphasic life cycle and evolution of paedomorphosis:
Our results suggest that two distinct evolutionary processes—(1) adaptation of biphasic life cycles through selection of metamorphic timing (Voss et al. 2003) and (2) evolution of novel paedomorphic developmental modes that isolate lineages and promote speciation (Shaffer 1984)—are apparently linked by a common genetic architecture. Selection for met alleles that increase or decrease age at metamorphosis is expected to allow the evolution of a continuum of metamorphic timing phenotypes. Because met did not account for all of the variation in metamorphic timing in WILD2, it is likely that other loci make a contribution to continuous variation (Voss et al. 2003). The average difference in metamorphic age that we observed between met genotypic classes was 36 days. This amount of variation may significantly affect larval survivorship in natural populations that use unpredictable, ephemeral ponds (Wilbur and Collins 1973). In more predictable ephemeral ponds, selection is expected to favor alleles that delay metamorphic timing because larvae that attain larger body sizes have increased survival probabilities after metamorphosis (Semlitsch et al. 1988). In our study, inheritance of the same met genotype was associated with delayed metamorphosis and expression of paedomorphosis. Because both of these life history strategies would be favored in a stable aquatic habitat, we propose that the evolution of paedomorphosis in Am occurred gradually via selection for delayed metamorphic timing. Overall, our results provide a framework for understanding how metamorphic timing and paedomorphic phenotypes can evolve to be fixed or variable within and between species and thus how microevolutionary processes lead to macroevolutionary patterns.
Acknowledgments
We thank the Axolotl Colony for animal resources and historical information and two anonymous reviewers for their comments. The work was supported by the U.S. National Science Foundation EPSCoR's (Experimental Program to Stimulate Competitive Research) support of a functional genomics initiative at the University of Kentucky and grants to S.R.V. by the U.S. National Science Foundation (IBN-0242833; IBN-0080112) and the National Institutes of Health (5 R24 RR16344-03).
We dedicate our work to the memory of Virginia Graue, without whose efforts in conserving natural axolotls we would have not gained the evolutionary insights reported in this article.
References
- Ambros, V., 1988 Genetic basis for heterochronic variation, pp. 269–286 in Heterochrony in Evolution, edited by M. L. McKinney. Plenum Press, New York.
- Armstrong, J. B., and S. T. Duhon, 1989 Induced spawnings, artificial spawnings, and other genetic manipulations, pp. 228–236 in Developmental Biology of the Axolotl, edited by J. B. Armstrong and G. M. Malacinski. Oxford University Press, New York.
- Collins, J. P., 1981. Distribution, habitats and life history variation in the tiger salamander, Ambystoma tigrinum, in east-central and southeast Arizona. Copeia 1981: 666–675. [Google Scholar]
- Falconer, D. S., 1989 Quantitative Genetics. Longman Scientific & Technical, Essex, UK.
- Futuyma, D. J., 1998 Evolutionary Biology. Sinauer Associates, Sunderland, MA.
- Goldschmidt, R. B., 1940 The Material Basis of Evolution. Yale University Press, New Haven, CT.
- Gould, S. J., 1977 Ontogeny and Phylogeny. Belkap Press, Cambridge, MA.
- Gould, S. J., 1981 Change in developmental timing as a mechanism of macroevolution, pp. 333–346 in Evolution and Development, edited by J. T. Bonner. Springer-Verlag, Berlin.
- Malacinski, G. M., 1978. The Mexican axolotl, Ambystoma mexicanum: its biology and developmental genetics, and its autonomous cell-lethal genes. Am. Zool. 18: 195–206. [Google Scholar]
- Matsuda, R., 1987 Animal Evolution in Changing Environments With Special Reference to Abnormal Metamorphosis. John Wiley & Sons, New York.
- McKinney, M. L., and K. J. McNamara, 1991 Heterochrony: The Evolution of Ontogeny. Plenum Press, New York.
- Meer, J. M., R. H. Cudmore, Jr. and K. F. Manly, 2004 MapManager QTX (http://www.mapmanager.org/mmQTX.html.)
- Moczek, A. P., and H. F. Nijhout, 2002. Developmental mechanisms of threshold evolution in a polyphenic beetle. Evol. Dev. 4: 252–264. [DOI] [PubMed] [Google Scholar]
- Putta, S., J. J. Smith, J. Walker, M. Rondet, D. W. Weisrock et al., 2004. From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genomics 5: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, R. A., 1996 The Shape of Life: Genes, Development, and the Evolution of Animal Form. University of Chicago Press, Chicago.
- Raff, R. A., and G. A. Wray, 1989. Heterochrony: developmental mechanisms and evolutionary results. J. Evol. Biol. 2: 409–434. [Google Scholar]
- Roff, D. A., 1986. The genetic basis of wing dimorphism in the sand cricket, Gryllus firmus and its relevance to the evolution of wing dimorphism in insects. Evolution 40: 1009–1020. [DOI] [PubMed] [Google Scholar]
- Rose, F. L., and D. Armentrout, 1975. Adaptive strategies of Ambystoma tigrinum Green inhabiting the Llano Estacado of West Texas. J. Anim. Ecol. 45: 713–739. [Google Scholar]
- Semlitsch, R. D., D. E. Scott and J. H. K. Pechmann, 1988. Time and size at metamorphosis related to adult fitness in Ambystoma talpoideum. Ecology 69: 184–192. [Google Scholar]
- Semlitsch, R. D., R. N. Harris and H. M. Wilbur, 1990. Paedomorphosis in Ambystoma talpoideum: maintenance of population variation and alternative life history pathways. Evolution 44: 1604–1613. [DOI] [PubMed] [Google Scholar]
- Shaffer, H. B., 1984. Evolution in a paedomorphic lineage. 1. An electrophoretic analysis of the Mexican ambystomatid salamanders. Evolution 38: 1194–1206. [DOI] [PubMed] [Google Scholar]
- Shaffer, H. B., and M. L. McKnight, 1996. The polytypic species revisited: genetic differentiation and molecular phylogenetics of the tiger salamander (Ambystoma tigrinum) (Amphibia: Caudata) complex. Evolution 50: 417–433. [DOI] [PubMed] [Google Scholar]
- Smith, J. J., 2002 Genetics in Ambystoma: metamorphic failure and sex determination. MS Thesis, Colorado State University, Fort Collins, CO.
- Sprules, W. G., 1974. The adaptive significance of paedogenesis in North American species of Ambystoma (Amphibia: Caudata): an hypothesis. Can. J. Zool. 52: 393–400. [Google Scholar]
- Tompkins, R., 1978. Genic control of axolotl metamorphosis. Am. Zool. 18: 313–319. [Google Scholar]
- Voss, S. R., 1993. Randomly amplified polymorphic DNA (RAPD) analysis of ambystomatid salamanders. Axolotl Newsl. 22: 28–32. [Google Scholar]
- Voss, S. R., 1995. Genetic basis of paedomorphosis in the axolotl, Ambystoma mexicanum: a test of the single-gene hypothesis. J. Hered. 86: 441–447. [Google Scholar]
- Voss, S. R., and H. B. Shaffer, 1997. Adaptive evolution via a major gene effect: paedomorphosis in the Mexican axolotl. Proc. Natl. Acad. Sci. USA 94: 14185–14189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss, S. R., and H. B. Shaffer, 2000. Evolutionary genetics of metamorphic failure using wild-caught vs. laboratory axolotls (Ambystoma mexicanum). Mol. Ecol. 12: 1217–1223. [DOI] [PubMed] [Google Scholar]
- Voss, S. R., K. L. Prudic, J. C. Oliver, H. B. Shaffer, 2003. Candidate gene analysis of metamorphic timing in ambystomatid salamanders. Mol. Ecol. 12: 1217–1223. [DOI] [PubMed] [Google Scholar]
- West-Eberhard, M. J., 2004 Developmental Plasticity and Evolution. Oxford University Press, New York.
- Wilbur, H. M., and J. P. Collins, 1973. Ecological aspects of amphibian metamorphosis. Science 182: 1305–1314. [DOI] [PubMed] [Google Scholar]