Abstract
Taurine (2-aminoethanesulphonic acid), a naturally occurring, sulfur-containing amino acid, is found at high concentrations in mammalian plasma and tissues. Although taurine is involved in a variety of processes in humans, it has never been found as a component of a protein or a nucleic acid, and its precise biochemical functions are not fully understood. Here, we report the identification of two novel taurine-containing modified uridines (5-taurinomethyluridine and 5-taurinomethyl-2-thiouridine) in human and bovine mitochondrial tRNAs. Our work further revealed that these nucleosides are synthesized by the direct incorporation of taurine supplied to the medium. This is the first reported evidence that taurine is a constituent of biological macromolecules, unveiling the prospect of obtaining new insights into the functions and subcellular localization of this abundant amino acid. Since modification of these taurine-containing uridines has been found to be lacking in mutant mitochondrial tRNAs for Leu(UUR) and Lys from pathogenic cells of the mitochondrial encephalomyopathies MELAS and MERRF, respectively, our findings will considerably deepen our understanding of the molecular pathogenesis of mitochondrial encephalomyopathic diseases.
Keywords: mitochondria/mitochondrial disease/ post-transcriptional modification/taurine/tRNA
Introduction
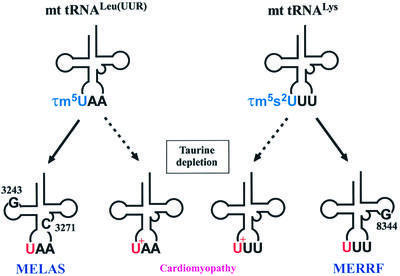
A characteristic structural feature of tRNAs is the presence of post-transcriptionally modified nucleosides at the anticodon first position (the ‘wobble’ position), which participate in codon–anticodon pairing (Curran, 1998). In mammalian mitochondrial (mt) tRNAs, uridine at the anticodon wobble position of the tRNALeu(UUR) and tRNALys undergoes such post-transcriptional modification, which is responsible for precise codon recognition. However, we recently discovered that the normal uridine modification does not occur in mt tRNALeu(UUR) with either an A3243G or U3271C mutation, or in mt tRNALys with an A8344G mutation obtained from human pathogenic cells of two mitochondrial encephalomyopathic diseases, MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes; Yasukawa et al., 2000a) and MERRF (myoclonus epilepsy associated with ragged-red fibers; Yasukawa et al., 2000b), respectively. Since it seemed likely that an unmodified uridine would cause either a misreading (on the basis of the mitochondrial wobble rule; Watanabe and Osawa, 1995) or a decoding deficiency, we examined the translational abilities of these mutant tRNAs using an in vitro mitochondrial translation system developed in our laboratory (Takemoto et al., 1995; Hanada et al., 2001). The results showed that mt tRNALys with the MERRF A8344G mutation was incapable of translating cognate codons due to a complete loss of codon–anticodon pairing on the ribosome (Yasukawa et al., 2001), strongly implying that in addition to the pathogenic point mutation itself, deficient decoding arising from the modification defect is significantly involved in this mitochondrial dysfunction. This is the first known case of a human disease apparently caused by the loss of a post-transcriptional modification.
To understand the molecular mechanisms of MELAS and MERRF pathogenesis, which will hopefully lead to the development of appropriate therapeutic measures for these mitochondrial diseases, the chemical structures of the modified uridines in mammalian mt tRNAs for Leu(UUR) and Lys need to be ascertained. In this study, we describe the identification and determination of novel taurine-containing uridine derivatives from bovine and human mt tRNAs. Although taurine has never been found as a component of a protein or a nucleic acid, the unique chemical structures of the derivatives prompt us to speculate that dietary taurine is a direct substrate for them.
Taurine, one of the most abundant amino acids in mammalian plasma and tissues, is known to have pleiotropic effects including modulation of calcium fluxes, maintenance of photoreceptor cells, modulation of neuronal excitability, osmoregulation and cell proliferation (Huxtable, 1992). However, its precise biochemical functions are still not fully understood. We demonstrate here that the novel modified uridines in mt tRNAs are synthesized by direct incorporation of taurine supplied to the medium. This is the first evidence that taurine is a component of RNA, a finding that provides us with new clues to understanding its biological functions and subcellular localization, as well as the molecular pathogenesis of mitochondrial diseases.
Results
Chemical structures of novel taurine-containing uridines
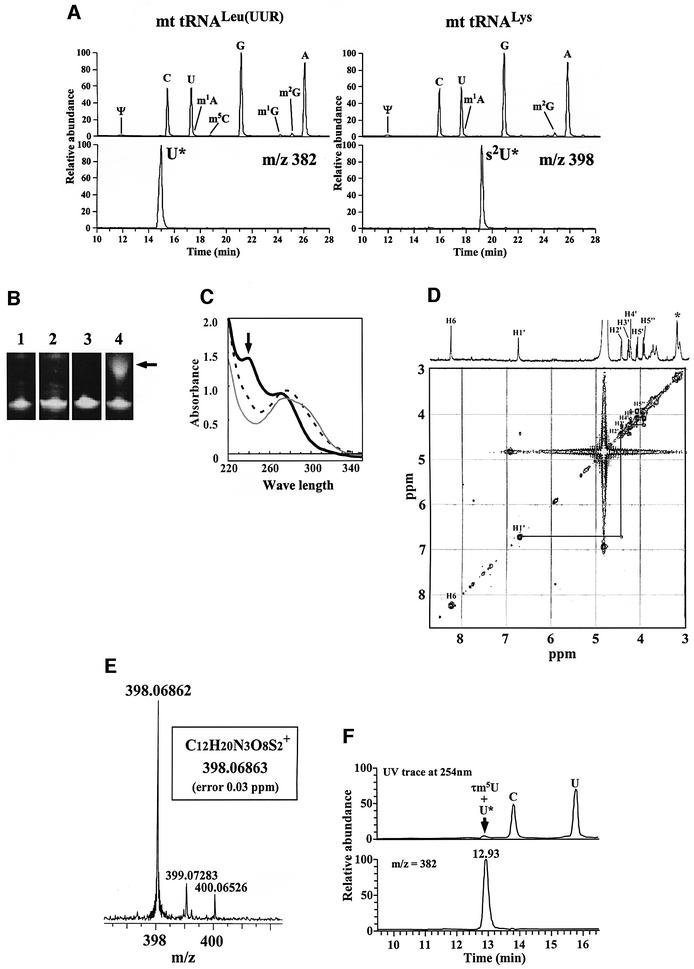
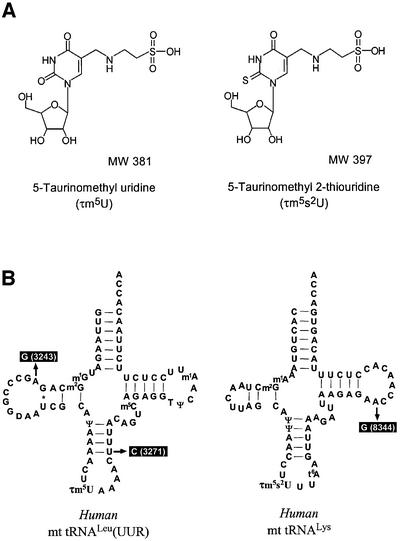
We succeeded in isolating mitochondrial disease-related mt tRNAs for Leu(UUR) and Lys from bovine liver by means of an improved solid-phase DNA probe technique (Wakita et al., 1994; see Materials and methods). Subsequent liquid chromatography/mass spectrometry (LC/MS) analysis revealed the two previously unknown nucleosides to be a uridine derivative (U*; molecular mass, 381 Da) in mt tRNALeu(UUR) and its 2-thio derivative (s2U*; 397 Da) in mt tRNALys, respectively (Figure 1A). The presence of the 2-thio derivative in mt tRNALys was demonstrated in two ways: (i) specific retardation of mt tRNALys migration on phenyl-mercuric gel electrophoresis (Igloi, 1988) due to the thiocarbonyl group (Figure 1B), and (ii) a prominent peak at 240 nm in the UV spectrum of the purified nucleoside under an alkaline condition (Figure 1C), which is known to be a characteristic feature of 2-thiouridine derivatives (Watanabe et al., 1974). NMR analysis of the nucleoside revealed no H6 proton cross peak in the 1H-COSY spectrum, indicating the presence of a substituent at position 5 in the uracil base. No modification was found in the ribose portion as all ribose protons and expected cross peaks were assigned (Figure 1D). The molecular weight of s2U* was determined with a high degree of precision by using a Fourier-transform ion cyclotron resonance (FT-ICR) mass spectrometer with a 7-tesla magnet (Figure 1E). Its atomic composition was ascertained to be C12H19N3O8S2, also with excellent accuracy (0.03 p.p.m.). These findings indicate that the main modification occurs in the uracil base, the most plausible structure in both cases being a taurinomethyl possessing a sulfonic acid group derived from taurine (Figure 2A). The two nucleosides were thus named 5-taurinomethyluridine (τm5U) and 5-taurinomethyl-2-thiouridine (τm5s2U). The former was chemically synthesized by the Mannich reaction (Jones et al., 1982) as described in Materials and methods. The novel nucleoside was determined to be τm5U by comparison with the synthetic product: LC/MS revealed that U* from mt tRNALeu(UUR) was co-eluted with synthetic τm5U at the same retention time (Figure 1F). In addition, their CID (collision-induced dissociation) fragment patterns and NMR spectra were clearly identical (data not shown).
Fig. 1. (A) Total nucleoside analysis by LC/MS using ESI/iontrap mass spectrometry of purified bovine mt tRNAs for Leu(UUR) (left panels) and Lys (right panels), respectively. The upper chromatograms show UV traces at 254 nm for both tRNAs; modified nucleosides that were identified are indicated by the symbols recommended in the RNA Modification Database (see Figure 2 legend). The lower mass chromatograms show the mass ranges m/z 381.5–382.5 and 397.5–398.5 for U* and s2U*, respectively. (B) Phenyl-mercuric gel electrophoresis of purified mt tRNAs to detect specific retardation due to the thiocarbonyl group. Lanes 1 and 3, 10% PAGE (with 7 M urea) analyses for purified mt tRNALeu (UUR) and mt tRNALys, respectively. Lanes 2 and 4, 10% PAGE analyses with (N-acryloylamino)phenyl-mercuric chloride for purified mt tRNALeu (UUR) and mt tRNALys, respectively. The arrow indicates the retarded band in mt tRNALys. Some tRNA bands (not all) are known to up-shift on this gel (Igloi, 1988). (C) UV spectra of purified s2U* under acidic (0.1 N HCl, gray line), neutral (10 mM HEPES–KOH pH 7.0, dotted line), and alkaline (0.1 N NaOH, bold line) conditions. The arrow indicates a prominent peak at 240 nm under the alkaline condition. (D) 1H-COSY spectrum of purified s2U* in D2O. The x and y-axes represent chemical shift p.p.m. values. All the ribose protons (H1′, H2′, H3′, H4′, H5′ and H5′′) and their cross peaks are assigned. In the case of the uracil base, no H5 proton and no H6 proton cross peak are observed. Methylene protons derived from the 5-substituent in the uracil base are asterisked. (E) High-resolution mass spectrum of purified s2U* by ESI/FT-ICR mass spectrometry (Bruker Daltonics Bio APEX II 70e). The deduced atomic composition (proton adduct form) is indicated in the box. (F) Confirmation of the novel nucleoside (U*) by co-injection with synthetic τm5U using ESI/iontrap mass spectrometry. The purified U* and synthetic τm5U were analyzed by LC/MS using an ESI/iontrap mass spectrometer. The upper panel is the UV trace for nucleoside analysis of mt tRNA and synthetic τm5U. The lower panel is the mass chromatogram for U* from mt tRNA and synthetic τm5U (m/z 382). The purified U* and the synthetic τm5U were co-eluted at the same retention time.
Fig. 2. (A) Chemical structures of τm5U and τm5s2U with their molecular weights. (B) Cloverleaf structures of human mt tRNALeu(UUR) and mt tRNALys with modified nucleosides: τm5U, τm5s2U, 1-methyladenosine (m1A), 1-methylguanosine (m1G), 2-methylguanosine (m2G), 5-methylcytidine (m5C), pseudouridine (Ψ), 5-methyluridine (T) and N6-threoninocarbonyladenosine (t6A). Symbols and names of known modified nucleosides comply with the RNA Modification Database (McCloskey and Crain, 1998; http://medstat.med.utah.edu/RNAmods/). The pathogenic point mutations of MELAS (3243 and 3271) and MERRF (8344) are indicated.
We confirmed that these taurine-containing uridines are actually located at the anticodon wobble position of each tRNA by Donis-Keller’s RNA sequencing method (Donis-Keller, 1980), the post-labeling method (Kuchino et al, 1987) and mass spectrometric analysis. RNase T1-digested fragments containing anticodon wobble position, CAΨAAAACU[τm5U]AAACΨUUUAUACCm5CAGp (mol. wt 8416) and CACΨAACCU[τm5s2U]UUt6AAGp (mol. wt 5059) were specifically detected from bovine mt tRNALeu(UUR) and mt tRNALys, respectively (data not shown). These two taurine-containing uridines were also found in human mt tRNAs for Leu(UUR) and Lys (Figure 2B).
Direct incorporation of taurine into human mitochondrial tRNA
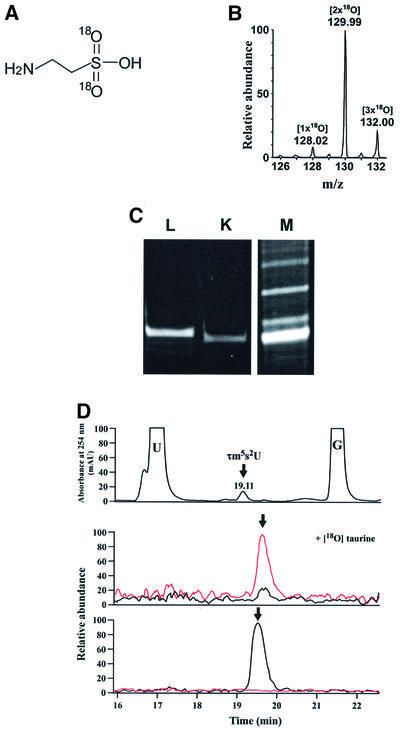
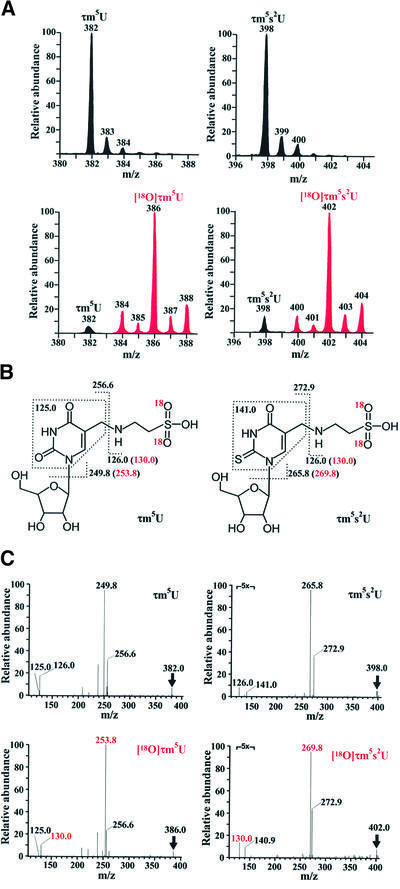
Taurine, an abundant amino acid in mammalian plasma, is widely used as an ingredient of nutritional supplements. However, though taurine is clearly pleiotropic in its effects on the human body (Huxtable, 1992), its exact functions in biochemical terms are still to be elucidated. Since there has been no report so far of taurine being a component of any protein or nucleic acid, we attempted to determine whether it is a direct constituent of the two taurine-containing modified uridines that we identified in the mt tRNAs. For this purpose, we synthesized a stable isotopic taurine with [18O]oxygens in the sulfonic acid group (Figure 3A); 80% of the synthetic taurine had two [18O]oxygens, while the remaining 7% and 13% had one and three, respectively (Figure 3B). HeLa cells were grown for 48 h in a medium containing the [18O]taurines thus synthesized. The cells were then harvested and the mt tRNAs for Leu(UUR) and Lys were isolated (Figure 3C). When purified mt tRNALys was subjected to nucleoside analysis, τm5s2U incorporating [18O]taurine was clearly identified in the LC/MS mass chromatogram (Figure 3D); τm5s2U with a 4 Da increase in its mass (m/z 402) was detected at the same retention time as natural τm5s2U (m/z 398). Likewise, in mt tRNALeu(UUR), τm5U incorporating [18O]taurine (m/z 386) was detected at the same retention time as natural τm5U (m/z 382) (see Figure 4A). As shown in the LC/MS mass spectra obtained at the elution points of τm5U and τm5s2U (Figure 4A), 2–6 Da increases were observed in the molecular masses of τm5U and τm5s2U, respectively, obtained from purified mt tRNAs for Leu(UUR) and Lys, and the ratio of synthetic isotopic taurines with different numbers of [18O]oxygen (Figure 3B) was exactly preserved both in τm5U and τm5s2U, demonstrating that the sulfonic acid groups of τm5(s2)U in mt tRNAs came directly from the taurine supplied in the medium. The CID spectra of τm5U and τm5s2U possessing two [18O]oxygens confirmed that fragment ions containing the sulfonic acid group specifically exhibited a 4 Da increase in mass due to the presence of the two [18O]oxygens (Figure 4B and C). These results provided direct evidence that taurine in the medium was incorporated into mitochondria without decomposition to become a component of the taurine-containing uridines in the mt tRNAs.
Fig. 3. (A) Stable isotopic taurine with two [18O]oxygens. (B) Mass spectrum for synthetic [18O]taurine. Judging from the peak intensities, 80% of the synthetic taurine had two [18O]oxygens (m/z 130), while the remaining 7 and 13% had one (m/z 128) and three (m/z 132), respectively. (C) Polyacrylamide gel electrophoretic analysis of purified human Leu(UUR) (L) and Lys (K) mt tRNAs from HeLa cells cultured with [18O]taurine. Each tRNA was visualized by ethidium bromide staining. The crude RNA fraction (M) is shown as a marker. (D) Nucleoside analysis of mt tRNALys by LC/MS to detect τm5s2U incorporating [18O]taurine. Top panel: UV trace at 254 nm with τm5s2U indicated. Middle and lower panels: mass chromatograms of τm5s2U from HeLa cells cultured in the presence (middle) and absence (lower) of [18O]taurine in the mass ranges m/z 397.5–398.5 (black line) for natural τm5s2U and m/z 401.5–402.5 (red line) for [18O]taurine- containing τm5s2U. The longer retention time of τm5s2U in the mass chromatograms compared with that in the UV trace (19.11 min) arose from a time lag between the UV detector and mass spectrometer in the LC/MS system.
Fig. 4. (A) Mass spectra of τm5U from mt tRNALeu(UUR) (left panels) and τm5s2U from mt tRNALys (right panels) derived from HeLa cells cultured in the presence (lower panels) and absence (top panels) of [18O]taurine. Black and red peaks indicate natural and [18O]taurine- containing τm5(s2)U, respectively. (B) Chemical structures of τm5U and τm5s2U with indication of fragment ions generated by collision-induced dissociation (CID). The red m/z values in parentheses indicate fragment ions containing two [18O]oxygens. (C) CID spectra of τm5U (left panels) and τm5s2U (right panels) with (lower panels) or without (top panels) [18O]taurine. The parent ion in each spectrum is indicated by an arrow. Figures in red in the lower spectra are the m/z values of fragment ions with two [18O]oxygens, which correspond to the values shown in (B).
Judging from the relative abundances in the spectra of the natural (black) and isotopic (red) taurine-containing uridines (Figure 4A, lower panels), τm5U from mt tRNALeu(UUR) and τm5s2U from mt tRNALys consisted of 96 and 91% [18O]taurine, respectively. Although a small quantity of taurine is known to be synthesized from cysteine de novo in human cells, our experiment clearly proved that taurine supplied to the medium was efficiently incorporated into the mt tRNAs in spite of the presence of medium cysteine, suggesting that plasma taurine is a major constituent of the taurine-containing uridines, even in the human body.
Taurine uptake by mitochondria
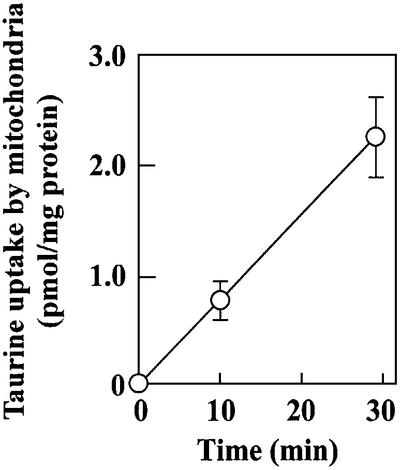
Our findings revealed a new pathway for the transport of cytoplasmic taurine to mitochondria, implying the existence of a putative mitochondrial taurine transporter responsible for cytoplasmic taurine uptake by mitochondria, where it is used to synthesize τm5(s2)U. Although there has been no report of taurine transport into mitochondria, when we examined mitochondrial taurine uptake in vitro using the standard method for taurine uptake by intact cells (Uchida et al., 1992; Qian et al., 2000), we found that taurine was time-dependently incorporated into isolated bovine mitochondria (Figure 5). This suggests active transport of cytoplasmic taurine into mitochondria, because it is known that lipophobic taurine does not penetrate the lipid bilayer by diffusion (Huxtable, 1992).
Fig. 5. Taurine uptake by isolated bovine mitochondria. The values were normalized by the mitochondrial protein weight.
Decoding activity of mitochondrial tRNA with taurine-containing wobble uridine
In previous studies, we found that taurine modification was lacking at the wobble uridine of mutant tRNAs from pathogenic cells obtained from patients with MELAS and MERRF (Yasukawa et al., 2000a,b, 2001). To estimate the effect of the C5 taurine modification at the wobble position of mt tRNALys on the decoding activity and to clarify whether it does contribute significantly to the defective tRNA function observed in mitochondrial diseases, we carried out a ribosome-binding experiment with modified wild-type tRNA missing the s2U modification but still containing the C5 taurine modification. A small amount of tRNA having τm5U without the s2U modification was found to occur naturally in purified bovine mt tRNALys as a modification intermediate (data not shown), which we succeeded in purifying from wild-type mt tRNALys with τm5s2U by phenyl-mercuric gel electrophoresis (Figure 6A). We used this in the ribosome-binding experiment, which was carried out with the AAA codon-programmed small subunit ribosome as described previously (Yasukawa et al., 2001). The results showed that while the absence of the s2U modification considerably reduced AAA codon binding, mt tRNALys with τm5U still retained a high level of efficiency (63% activity of τm5s2U) compared with its transcript (Figure 6B). We thus concluded that in addition to the s2U modification, the C5 taurine modification confers codon-binding efficiency.
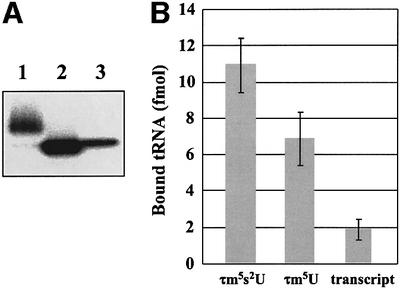
Fig. 6. (A) Phenyl-mercuric gel electrophoresis of mt tRNAsLys with and without the 2-thio group. Lane 1, bovine mt tRNALys with τm5s2U (wild type); lane 2, bovine mt tRNALys with τm5U (modification intermediate); lane 3, transcript of bovine mt tRNALys. 5′-32P-labeled tRNAs (0.125 pmol) were loaded onto the gel and visualized by an image analyzer (Fuji Photo Film). The different intensity of each band is due to the labeling efficiency. (B) Binding affinity of bovine mt tRNAsLys with and without the 2-thio group for the AAA codon- programmed ribosomal small subunit. The average values for three independent experiments are plotted; bars indicate the SD.
Discussion
Our findings are the first reported instance of any modified nucleoside having a sulfonic acid group in a side chain, and it will be of great interest to clarify the decoding properties of these nucleosides in relation to the strong electrostacity of the sulfonic acid group. Since we have also found these two taurine-containing uridines in ascidian mt tRNA counterparts (Kondow et al., 1999; A.Kondow, T.Suzuki and K.Watanabe, unpublished observations), it is likely they are common to vertebrate and protochordate mitochondria. Meanwhile, modified uridine at the wobble position of yeast mt tRNA was reported as 5-carboxymethyaminomethyuridine (cmnm5U; Osawa, 1995). In addition, we found that Caenorhabditis elegans mt tRNAs have cmnm5U at the wobble position (M.Sakurai, T.Ohtsuki, T.Suzuki and K.Watanabe, in preparation). Since glycine is attached through a methylene group at position 5 in the uracil base of cmnm5U instead of taurine, cmnm5U and τm5U both have negatively charged side chains. Further, by analogy with the findings of the present study, glycine is presumably a direct substrate for cmnm5U synthesis. Thus, there appears to be a relationship between the mitochondrial decoding property and the evolutionarily conserved chemical character of modified uridines in mt tRNAs.
In previous studies (Yasukawa et al., 2000a,b), we found a lack of taurine modification at the wobble uridine of mt tRNALeu(UUR) and tRNALys from pathogenic cells of MELAS and MERRF, respectively. We also demonstrated that the modification deficiency of mutant tRNALys with the MERRF 8344 mutation causes defective translation of both AAA and AAG codons due to weak codon–anticodon interaction on the ribosomal small subunit (Yasukawa et al., 2001). Since the 2-thio group of wobble uridine is known to confer stable codon–anticodon interaction (Ashraf et al., 1999), loss of the 2-thio group of mt tRNALys with the MERRF 8344 mutation is assumed to be one of the main reasons for the weak binding to cognate codons. In the case of MELAS, mt tRNALeu(UUR) has been shown to have τm5U at the wobble position, indicating that a decoding disorder arising from lack of the C5 taurine modification is also a causative factor of mitochondrial diseases. Yarian et al. (2002) reported that the C5 modification (mnm5U34) in the anticodon stem loop derived from Escherichia coli tRNA clearly stabilizes codon–anticodon interaction at both the ribosomal A and P sites. The fact that misreading of asparagine codons by tRNALys was greatly reduced in trmU and trmE mutants, respectively, containing hypomodified mnm5U34 and s2U34 instead of the fully modified mnm5s2U34 (Hagervall et al., 1998), indicates that both the 2-thio group and C5 modification confer efficient codon recognition. In the present work, we observed that absence of the s2U modification considerably reduced AAA codon binding, but mt tRNALys with τm5U retained relatively high efficiency compared with its transcript (Figure 6B). Also, we previously reported that the binding efficiency of MERRF tRNA lacking both the 2-thio and C5 taurine modifications was reduced 10-fold (Yasukawa et al., 2001). The binding efficiency of the MERRF tRNA was at the same level as that of its human transcript (our unpublished observations). Thus, our findings demonstrate the apparent contribution of the C5 taurine modification in efficient codon recognition.
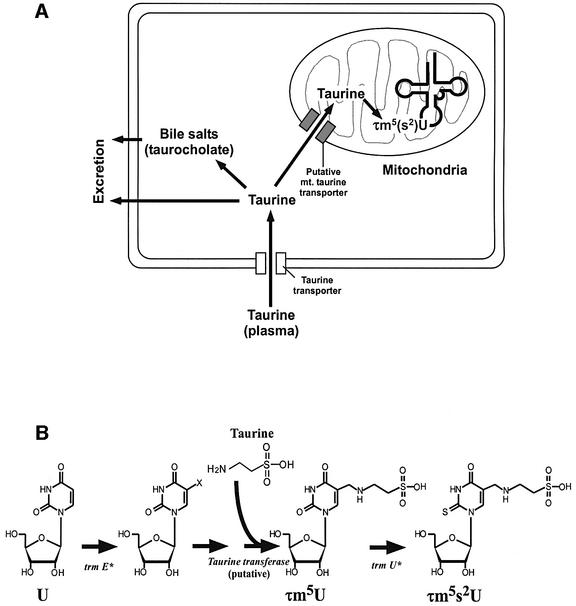
In human body fluids, including plasma, taurine concentrations range from 10 to 100 µM, while intracellular concentrations can rise to several hundred times as great (Huxtable, 1992). The taurine concentration gradient across the cell membrane is maintained by a high-affinity taurine transporter (Uchida et al., 1992; Figure 7A). Most cytoplasmic taurine is known to be excreted as such, or in bile salts that are taurine-conjugated metabolites of cholesterol such as taurocholate (Huxtable, 1992). Our results strongly suggest a new pathway for the transport of cytoplasmic taurine into mitochondria (Figure 7A). Time-dependent taurine uptake by isolated bovine mitochondria suggests active transport of taurine across the mitochondrial membrane. In practice, since cytosolic taurine is known to accumulate at concentrations up to 40 mM (Huxtable, 1992), mitochondrial taurine uptake is considered to be more efficient in cells. In this regard, taurine has been shown to be localized in most subcellular compartments, including mitochondria (Lobo et al., 2000).
Fig. 7. (A) Schematic depiction of catabolic flow of intracellular taurine. Plasma taurine is taken up through the taurine transporter. Cytoplasmic taurine is excreted as such or in the form of bile salts like taurocholate. Cytoplasmic taurine is imported into mitochondria through a putative mitochondrial taurine transporter to be used as a constituent for τm5(s2)U synthesis in mt tRNAs. (B) Possible biosynthetic pathway of τm5(s2)U. Mitochondrial trmE (trmE*) is involved in the initial step of τm5(s2)U synthesis. After some unidentified steps, mitochondrial taurine is directly incorporated into mt tRNAs to build τm5U. Mitochondrial trmU (trmU*) is responsible for 2-thio group synthesis of τm5s2U.
The biosynthesis of τm5(s2)U is considered to involve several modification enzymes, including a putative taurine transferase (Figure 7B). The first step of τm5(s2)U biosynthesis is likely to be similar to an xm5U-type modification with a methylene group at the root of position 5, such as mnm5s2U (5-methylaminomethyl 2-thio uridine) in bacterial tRNAs or cmnm5U in yeast mt tRNAs (Björk, 1995). In E.coli, the trmE gene is known to be responsible for the initial step of this type of modification (Elseviers et al., 1984). It can be assumed that the pathogenic point mutations of MELAS or MERRF abolish the tRNA recognition by the mitochondrial trmE homolog (trmE*) (Figure 7B). Thus, each of the point mutations (3243, 3271 or 8344) is considered to be selected as a negative determinant in the initial step of taurinomethyl synthesis.
It is of interest that taurine appears to be an essential nutrient for cats and possibly for primates, including humans (Hayes et al., 1975; Geggel et al., 1985; Hayes, 1985). As noted earlier, taurine is involved in a variety of important processes, including synthesis of bile salts, modulation of calcium fluxes, maintenance of photoreceptor cells, modulation of neuronal excitability, antioxidation, osmoregulation and cell proliferation (Huxtable, 1992). Although taurine-containing small peptides such as glutamyl-taurine (Marnela et al., 1985) have been isolated from brain, taurine has not thus far been found as a component of a protein or a nucleic acid, and its precise biochemical functions are still obscure. In the case of humans, infants and young children biosynthesize very little taurine, so dietary taurine is essential for normal human development (Sturman, 1993). The cat and the fox have no biosynthetic pathway for this amino acid, and, in both of these animals, a deficiency of dietary taurine has been shown to cause cardiomyopathy (Pion et al., 1987; Moise et al., 1991), which significantly is a major manifestation of human mitochondrial encephalomyopathies (Wallace, 2000). Although taurine plays crucial roles in myocardial functions such as calcium flux modulation and cardiac contractility, our evidence strongly suggests that incomplete modification of τm5(s2)U in mt tRNAs due to a low plasma taurine level is likely to be one of the main causative factors of cardiomyopathy in cats, in a similar way to mitochondrial encephalomyopathies in humans (Figure 8). Further study of taurine import into mitochondria and the biosynthetic pathway(s) of τm5(s2)U will shed new light on the biochemical role(s) of taurine.
Fig. 8. Schematic depiction of modification defects in mitochondrial tRNAs for Leu(UUR) and Lys caused by point mutations related to the mitochondrial encephalomyopathies MELAS (nucleotide positions 3243 or 3271) and MERRF (8344), and possible relationship between taurine dietary deficiency and speculated modification impairment as a cause of cardiomyopathy in the cat. U+ stands for a putative intermediate of τm5(s2)U.
Materials and methods
Purification of individual mitochondrial tRNAs from bovine liver
The crude RNA fraction was extracted from bovine liver according to the literature (Nishimura, 1971). The tRNA fraction (3.4 g) was obtained by anion-exchange column chromatography using DEAE–Sepharose Fast Flow (8 × 73 cm; Amersham Biosciences) with a linear gradient of NaCl and MgCl2, 25 l of elution buffer A (20 mM Tris–HCl pH 7.5, 200 mM NaCl and 8 mM MgCl2) and elution buffer B (20 mM Tris–HCl pH 7.5, 450 mM NaCl and 16 mM MgCl2) with gravitational flow, and a flow rate of ∼6.5 ml/min. The fractions containing mt tRNAs for Leu(UUR) and Lys were determined by dot hybridization (Yokogawa et al., 1989) using the following synthetic DNA probes: bL1 for bovine tRNALeu(UUR), 5′-TGTTAAGGAGAGGATTTGAATCTCTGGATA-3′ and bK1 for bovine tRNALys, 5′-GTTAGTGCTATATAGCTTCTTAGTG-3′.
The pooled fractions were combined, precipitated with ethanol and dissolved in a binding buffer (1.2 M NaCl, 30 mM HEPES–KOH pH 7.5 and 15 mM EDTA). To isolate individual mt tRNAs with the highest efficiency, we devised and successfully improved an original solid-phase DNA probe method (Wakita et al., 1994), which we have named ‘chaplet’ column chromatography. A biotinylated DNA probe, as described above, complementary to each tRNA was immobilized on avidin Sepharose. Two columns, one for each individual mitochondrial tRNA, were connected in tandem and the DEAE tRNA fraction was circulated through the chaplet column by a peristaltic pump at a temperature of 65°C to entrap the target mt tRNAs. After washing out non-specific tRNA with a wash buffer (0.6 M NaCl, 15 mM HEPES–KOH pH 7.5 and 7.5 mM EDTA), each individual tRNA was eluted separately from its respective column with a low-salt buffer (20 mM NaCl, 0.5 mM HEPES–KOH pH 7.5 and 0.25 mM EDTA) at 65°C. The individual tRNAs were purified homogeneously. A detailed description of this method will be reported in a specialized journal.
Purification of individual mitochondrial tRNAs from HeLa cells
The crude RNA fraction was extracted from HeLa cells according to the literature (Nishimura, 1971; Chomczynski and Sacchi, 1987). The tRNA fraction was obtained by anion-exchange column chromatography using DEAE–Sepharose Fast Flow. The subsequent procedures were the same as described above. Synthetic DNA probes with 3′ biotin complementary to each tRNA were hL1 for human mt tRNALeu(UUR), 5′-GCGATT ACCGGGCTCTGCCATCTTAAC-3′ and hK1 for human mt tRNALys, 5′-TCACTGTAAAGAGGTGTTGG-3′.
Liquid chromatography mass spectrometry
An LCQ ion trap (IT) mass spectrometer (ThermoFinnigan) equipped with an electrospray ionization (ESI) source and a MAGIC 2002 liquid chromatography system (Michrom BioResources) were used to analyze nucleosides from the mitochondrial tRNAs. Purified tRNAs (0.01– 0.05 A260 units) were digested into nucleosides at 37°C for 1 h in 10 µl of a reaction mixture containing 20 mM HEPES–KOH pH 7.5, 10 µg/ml nuclease P1 and 0.5 U/ml bacterial alkaline phosphatase. The hydrolysates were analyzed by LC/MS as follows. An ODS reversed-phase column with a 3 × 10 mm pre-column cartridge (Inertsil ODS-3, 2.1 × 250 mm; GL Sciences) was connected online to the electrospray interface. The conditions for the chromatography were determined as described by Pomerantz and McCloskey (1990). Collision-induced dissociation (CID) spectra were obtained by an LC/MS/MS experiment using a data-dependent scan. The MS/MS setting was as follows: isolation width, m/z 1.0; activation amplitude,15%; activation time, 30 msec.
NMR analysis
1H-COSY spectrum of purified s2U* in D2O was obtained using a Bruker Daltonics AMX-500 at 298 K.
FT-ICR mass spectrometry
Purified s2U* (5 pmol/µl) from mt tRNALys was subjected to ESI/FT-ICR mass spectrometry by infusion at a flow rate of 5 µl/min. The data were obtained using a Bruker Daltonics Bio APEX II 70e FT mass spectrometer with the kind help of Dr I.Kudaka (Bruker Daltonics).
Phenyl-mercuric gel electrophoresis
The presence of the 2-thio derivative in mt tRNA for Lys was verified by electrophoresis in acrylamide gel immobilized with a phenyl-mercuric compound developed by Igloi (1988). To analyze purified mt tRNAs, we used 10% acrylamide gel containing 7 M urea and 0.05 mg/ml (N-acryloylamino) phenyl-mercuric chloride, which was kindly provided by Mr N.Shigi of our laboratory.
Chemical synthesis of 5-taurinomethyluridine
2′,3′-O-isopropyrideneuridine (142 mg, 0.5 mmol), taurine (313 mg, 2.5 mmol), paraformaldehyde (75 mg, 2.5 mmol) and triethylamine (0.35 ml, 2.5 mmol) were suspended in water (5 ml). The mixture was then heated and stirred at 110°C for 24 h. The cooled products were evaporated under reduced pressure. The residue was dissolved in formic acid/water (98:2 v/v; 5 ml) and the solution was allowed to stand at room temperature for 1 h. It was then concentrated under reduced pressure and the residue was co-evaporated with ethanol. The product was purified by reversed-phase column chromatography (Sep-Pak tC18, Waters; 10 g). The appropriate fractions eluted from the column with water/acetonitrile (95:5 v/v) were combined and evaporated under reduced pressure. The residue was lyophilized from water to give τm5U (triethylammonium salt; 0.24 g, 25%).
Chemical synthesis of [18O]taurine
The synthesis scheme was determined as described previously in the literature (Horning, 1955). Cystamine–2HCl (300 µmol) was dissolved in 20 mmol [18O]H2O (95.1 atom% of 18O; Isotec). Br2 (1.8 mmol) was slowly dropped into the solution, which was then stirred for 60 min to oxidize the cystamine. The product was dried in vacuo and dissolved in 500 µl normal water ([16O]H2O). The [18O]oxygen contents in the synthetic taurine were determined by mass spectrometric analysis (Figure 2B); synthetic taurine with one, two or three [18O]oxygens comprised ∼7, 80 and 13% of the total, respectively. The differences would have arisen from dissolved oxygen in the reaction mixture and H216O impurity in [18O]H2O.
Pulse labeling with [18O]taurine
HeLa cells (3 × 109 cells) grown until semi-confluence in a rich medium (DMEM/F12; Gibco) with 10% FBS were cultured for 48 h in the presence of the synthetic [18O]taurine in a medium containing DMEM (Gibco), 15 mM HEPES–KOH, 0.2 mM [18O]taurine, 100 U/ml penicillin, 0.1 mg/ml streptomycin (Gibco) and 10% dialyzed FBS (Gibco) (intrinsic taurine removal). Total RNA was extracted from the cells using Isogen (Nippon Gene). mt tRNAs were subsequently isolated as described above.
Taurine uptake by isolated bovine mitochondria
The conditions were determined as described in the literature (Uchida et al., 1992; Qian et al., 2000) with modifications. Mitochondria were prepared from fresh bovine liver according to the standard methods (Schwartzbach et al., 1996). Isolated mitochondria (3 mg, protein weight) were incubated at 37°C in 10 mM HEPES–KOH pH 7.5, 150 mM KCl, 5 mM MgCl2, 0.44 M mannitol, 0.2 mM DTT and 10 µM [14C]taurine (New England Nuclear). After washing three times with the buffer without taurine, mitochondria were dissolved in 1.5% Triton X-100 to measure the taurine incorporated by liquid scintillation counting.
Ribosome binding
Unmodified mt tRNALys was transcribed by T7 RNA polymerase (Milligan and Uhlenbeck, 1989) from template DNA synthesized by Klenow enzyme using two oligo DNAs: 5′-CCGGGTAATACGACTCACTATACACTAAGAAGCTATATAGCACTAACCTTTTAAGTTAGAGA-3′ and 5′-TGGTCACCAAGGAGAGTATATGGCTCTC AATCTCTAACTTAAAAGGTTAGTGCTATA-3′. Purified bovine mt tRNALys and its transcript were dephosphorylated by bacterial alkaline phosphatase (Takara Bio) and labeled at the 5′ end with 32P according to the literature (Midgley and Murray, 1985). 5′-32P-labeled mt tRNALys with τm5U was separated from that with τm5s2U by phenyl-mercuric gel electrophoresis as described above. The labeled tRNAs were eluted from the gel and quantified by measuring the absorbance at 260 nm. Ribosomal P-site binding was performed as described previously (Ashraf et al., 1999; Yasukawa et al., 2001) with a slight modification. Binding of labeled tRNAs to the cognate codon on the ribosomal 30S submit was carried out at 0°C for 1 h in 5 µl of a mixture consisting of 50 mM Tris–HCl pH 7.5, 30 mM MgCl2, 60 mM KCl, 1 mM DTT, 2 mM spermine, 0.2 µM E.coli 30S subunit, 0.1 µM 5′-32P-labeled tRNA and 0.1 µg/µl poly A (Sigma). The mixture was passed through a nitrocellulose filter (0.45 µm, Advantech) to quantify the bound tRNA as described previously (Ashraf et al., 1999).
Acknowledgments
Acknowledgements
We are grateful to Drs Ryan J.Huxtable (University of Arizona) and Yusei Miyamoto (University of Tokyo) for critical reading of the manuscript and many helpful suggestions. We thank Drs T.Ohtsuki, T.Hanada, T.Yasukawa, Mr N.Shigi and Mr Y.Kirino for their technical advice. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (Japan) (to T.S. and K.W.), a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to T.S.) and a grant from the Human Frontier Science Program (grant RG0349) (to T.S.).
References
- Ashraf S.S., Sochacka,E., Cain,R., Guenther,R., Malkiewicz,A. and Agris,P.F. (1999) Single atom modification (O→S) of tRNA confers ribosome binding. RNA, 5, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G. (1995) Biosynthesis and function of modified nucleosides. In Soll,D. and Rajbhandary,T.L. (eds) tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 165–205.
- Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Curran J.F. (1998) Modified nucleosides in translation. In Grosjean,H. and Benn,R. (eds), Modification and Editing of RNA. American Society for Microbiology, Washington, DC, pp. 493–516.
- Donis-Keller H. (1980) Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res., 8, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elseviers D., Petrullo,L.A. and Gallagher,P.J. (1984) Novel E.coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res., 12, 3521–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geggel H.S., Ament,M.E., Heckenlively,J.R., Martin,D.A. and Kopple,J.D. (1985) Nutritional requirement for taurine in patients receiving long-term parenteral nutrition. N. Engl. J. Med., 312, 142–146. [DOI] [PubMed] [Google Scholar]
- Hagervall T.G., Pomerantz,S.C. and McCloskey,J.A. (1998) Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol., 284, 33–42. [DOI] [PubMed] [Google Scholar]
- Hanada T., Suzuki,T., Yokogawa,T., Takemoto-Hori,C., Sprinzl,M. and Watanabe,K. (2001) Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells, 6, 1019–1030. [DOI] [PubMed] [Google Scholar]
- Hayes K.C. (1985) Taurine requirement in primates. Nutr. Rev., 43, 65–70. [DOI] [PubMed] [Google Scholar]
- Hayes K.C., Carey,R.E. and Schmidt,S.Y. (1975) Retinal degeneration associated with taurine deficiency in the cat. Science, 188, 949–951. [DOI] [PubMed] [Google Scholar]
- Horning E.C. (1955) Cysteic acid monohydrate. Organic Syntheses Collective. Vol. 3. John Wiley & Sons Inc., New York, NY, pp. 226–227.
- Huxtable R.J. (1992) Physiological actions of taurine. Physiol. Rev., 72, 101–163. [DOI] [PubMed] [Google Scholar]
- Igloi G.L. (1988) Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry, 27, 3842–3849. [DOI] [PubMed] [Google Scholar]
- Jones S.S., Reese,C.B. and Ubasawa,A. (1982) A convenient synthesis of 5-methyluridine from uridine. Synthesis, 4, 259. [Google Scholar]
- Kondow A., Suzuki,T., Yokobori,S., Ueda,T. and Watanabe,K. (1999) An extra tRNAGly(U*CU) found in ascidian mitochondria responsible for decoding non-universal codons AGA/AGG as glycine. Nucleic Acids Res., 27, 2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu,N. and Nishimura,S. (1987) Analysis of modified nucleosides and nucleotide sequence of tRNA. Methods Enzymol., 155, 379–396. [DOI] [PubMed] [Google Scholar]
- Lobo M.V., Alonso,F.J. and Martin del Rio,R. (2000) Immunocyto chemical localization of taurine in different muscle cell types of the dog and rat. Histochem. J., 32, 53–61. [DOI] [PubMed] [Google Scholar]
- Marnela K.M., Morris,H.R., Panico,M., Timonen,M. and Lahdesmaki,P. (1985) Glutamyl-taurine is the predominant synaptic taurine peptide. J. Neurochem., 44, 752–754. [DOI] [PubMed] [Google Scholar]
- McCloskey J.A. and Crain,P.F. (1998) The RNA modification database–1998. Nucleic Acids Res., 26, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgley C.A. and Murray,N.E. (1985) T4 polynucleotide kinase; cloning of the gene (pseT) and amplification of its product. EMBO J., 4, 2695–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- Moise N.S., Pacioretty,L.M., Kallfelz,F.A., Stipanuk,M.H., King,J.M. and Gilmour,R.F.,Jr (1991) Dietary taurine deficiency and dilated cardiomyopathy in the fox. Am. Heart J., 121, 541–547. [DOI] [PubMed] [Google Scholar]
- Nishimura S. (1971) In Cantoni,G.L. and Davies,D.R. (eds), Procedures in Nucleic Acid Research. Vol. 2. Harper & Row, New York, NY, pp. 542–564.
- Osawa S. (1995) Anticodon composition. Evolution of the Genetic Code. Oxford University Press, Oxford, UK, pp. 36–44.
- Pion P.D., Kittleson,M.D., Rogers,Q.R. and Morris,J.G. (1987) Myocardial failure in cats associated with low plasma taurine: a reversible cardiomyopathy. Science, 237, 764–768. [DOI] [PubMed] [Google Scholar]
- Pomerantz S.C. and McCloskey,J.A. (1990) Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol., 193, 796–824. [DOI] [PubMed] [Google Scholar]
- Qian X., Vinnakota,S., Edwards,C. and Sarkar,H.K. (2000) Molecular characterization of taurine transport in bovine aortic endothelial cells. Biochim. Biophys. Acta, 1509, 324–334. [DOI] [PubMed] [Google Scholar]
- Schwartzbach C.J., Farwell,M., Liao,H.X. and Spremulli,L.L. (1996) Bovine mitochondrial initiation and elongation factors. Methods Enzymol., 264, 248–261. [DOI] [PubMed] [Google Scholar]
- Sturman J.A. (1993) Taurine in development. Physiol. Rev., 73, 119–147. [DOI] [PubMed] [Google Scholar]
- Takemoto C., Koike,T., Yokogawa,T., Benkowski,L., Spremulli,L.L., Ueda,T.A., Nishikawa,K. and Watanabe,K. (1995) The ability of bovine mitochondrial transfer RNAMet to decode AUG and AUA codons. Biochimie, 77, 104–108. [DOI] [PubMed] [Google Scholar]
- Uchida S., Kwon,H.M., Yamauchi,A., Preston,A.S., Marumo,F. and Handler,J.S. (1992) Molecular cloning of the cDNA for an MDCK cell Na+- and Cl–-dependent taurine transporter that is regulated by hypertonicity. Proc. Natl Acad. Sci. USA, 89, 8230–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita K., Watanabe,Y., Yokogawa,T., Kumazawa,Y., Nakamura,S., Ueda,T., Watanabe,K. and Nishikawa,K. (1994) Higher-order structure of bovine mitochondrial tRNAPhe lacking the ‘conserved’ GG and TΨCG sequences as inferred by enzymatic and chemical probing. Nucleic Acids Res., 22, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. (2000) Mitochondrial defects in cardiomyopathy and neuromuscular disease. Am. Heart J., 139, S70–S85. [DOI] [PubMed] [Google Scholar]
- Watanabe K. and Osawa,S. (1995) tRNA sequences and variations in the genetic code. In Soll,D. and Rajbhandary,T.L. (eds) tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 225–250.
- Watanabe K., Oshima,T., Saneyoshi,M. and Nishimura,S. (1974) Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GTΨC in tRNA of an extreme thermophile. FEBS Lett., 43, 59–63. [DOI] [PubMed] [Google Scholar]
- Yarian C., Townsend,H., Czestkowski,W., Sochacka,E., Malkiewicz,A.J., Guenther,R., Miskiewicz,A. and Agris,P.F. (2002) Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem., 277, 16391–16395. [DOI] [PubMed] [Google Scholar]
- Yasukawa T., Suzuki,T., Suzuki,T., Ueda,T., Ohta,S. and Watanabe,K. (2000a) Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes. J. Biol. Chem., 275, 4251–4257. [DOI] [PubMed] [Google Scholar]
- Yasukawa T., Suzuki,T., Ishii,N., Ueda,T., Ohta,S. and Watanabe,K. (2000b) Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett., 467, 175–178. [DOI] [PubMed] [Google Scholar]
- Yasukawa T., Suzuki,T., Ishii,N., Ohta,S. and Watanabe,K. (2001) Wobble modification defect in tRNA disturbs codon–anticodon interaction in a mitochondrial disease. EMBO J., 20, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa T., Kumazawa,Y., Miura,K. and Watanabe,K. (1989) Purification and characterization of two serine isoacceptor tRNAs from bovine mitochondria by using a hybridization assay method. Nucleic Acids Res., 17, 2623–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]