Abstract
MAD2 is a key component of the spindle checkpoint that delays the onset of anaphase until all the kinetochores are attached to the spindle. It binds to human p55CDC and prevents it from promoting destruction of an anaphase inhibitor, securin. Here we report the characterization of a novel MAD2-binding protein, CMT2. Upon the completion of spindle attachment, formation of the CMT2–MAD2 complex coincides with dissociation of the p55CDC–MAD2 complex. Overexpression of CMT2 in cells arrested by the spindle checkpoint causes premature destruction of securin and allows exit from mitosis without chromosome segregation. Depletion of CMT2 induces cell death following a transient delay in the onset of anaphase. These results indicate that CMT2 interacts with the spindle checkpoint and coordinates cell cycle events in late mitosis.
Keywords: CMT2/MAD2/mitosis/spindle checkpoint
Introduction
For accurate chromosome segregation, the spindle must be attached to all the kinetochores before sister chromatid separation, an action that occurs at anaphase. The spindle checkpoint is a surveillance mechanism that delays the onset of anaphase until completion of the spindle microtubules attachment to all the kinetochores (Nicklas, 1997; Skibbens and Hieter, 1998; Amon, 1999). Pioneering work in budding yeast allowed an increased understanding of the spindle checkpoint and its subsequent emergence as a molecular signaling cascade. A number of components of the checkpoint were identified, namely, Mad1, 2, 3, Bub1, 3 and Mps1 (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996). Studies in other eukaryotes indicated that most of the checkpoint components are conserved from yeast to human (Li and Benezra, 1996; Skibbens and Hieter, 1998; Amon, 1999), although additional proteins are required in human and fly (Abrieu et al., 2000; Basto et al., 2000; Chan et al., 2000).
Duplicated chromosomes remain together and form sister chromatids. The cohesin protein complex is responsible for holding sister chromatids (Guacci et al., 1997; Michaelis et al., 1997; Nasmyth, 2001). At anaphase, a subunit of the complex (budding yeast Scc1/Mcd1; fission yeast Rad21) is cleaved by a specific protease, separase (budding yeast Esp1; fission yeast Cut1) (Uhlmann et al., 1999, 2000; Tomonaga et al., 2000). For most of the cell cycle, separase is found in a complex with an anaphase inhibitor, securin (budding yeast Pds1, Yamamoto et al., 1996; fission yeast Cut2, Funabiki et al., 1996; human securin, Zou et al., 1999), and remains inactive. At the onset of anaphase, separase is released from the securin complex upon destruction of securin (Nasmyth et al., 2000; Nasmyth, 2001), which requires ubiqutin-dependent proteolysis. Anaphase-promoting complex (APC)/cyclosome (E3 ubiquitin ligase) together with its substrate-specific activator, human p55CDC/fission yeast Slp1/budding yeast Cdc20, attach ubiquitin to securin (Cohen-Fix et al., 1996; Visintin et al., 1997; Peters, 2002) and promote its proteolysis by the 26S proteasome.
Mad2 forms protein complexes with other components of the spindle checkpoint and plays a key role in the signaling cascade of the checkpoint. In interphase, it binds to Mad1 and is preferentially found on the nuclear periphery (Chen et al., 1999). This localization is dependent on Mad1. In a fission yeast strain lacking Mad1, Mad2 is no longer found on the nuclear periphery (Ikui et al., 2002). Upon the onset of mitosis, Mad2 translocates into the nucleus and is guided to unattached kinetochores by Mad1 (Chen et al., 1996, 1998, 1999). From early to mid-mitosis, Mad2 is found in a complex with its target, Cdc20/Slp1/p55CDC. Expression of Slp1/Cdc20 that is defective in binding to Mad2 abolishes the function of the checkpoint in a dominant manner in yeast (Hwang et al., 1998; Kim et al., 1998). Furthermore, addition of recombinant MAD2 purified from bacteria can inhibit APC activity in vitro (Fang et al., 1998). These results indicate that association by Mad2 prevents the target from promoting the proteolysis, and thus appears a central process in the signaling cascade of the spindle checkpoint. A recent study indicates that additional components form a large protein complex with MAD2, termed mitotic checkpoint complex (MCC; Sudakin et al., 2001). Although speculative, unattached kinetochores may provide sites for assembly of the complex of MAD2 and its target, which then disperses throughout the nucleus (Kallio et al., 1998; Waters et al., 1998; Howell et al., 2000). Supporting this, real-time image analysis indicates that MAD2 is a transient component of kinetochores at prometaphase with a half-life of ∼24–28 s (Howell et al., 2000). Furthermore, structural studies also indicate that MAD1 regulates the formation of MAD2–p55CDC complex at kinetochores, which serve as ‘folding factories’ (Sironi et al., 2001, 2002).
Upon the completion of the spindle attachment, MAD2 diminishes from the kinetochores. It has also been shown that MAD2 dissociates from p55CDC at or around the transition from metaphase to anaphase (Wassmann and Benezra, 1998). Considering that the complex between MAD2 and the target, CDC20, is stable and resistant to washing with 0.4 M KCl (Fang et al., 1998), one would speculate that an active mechanism is responsible for dissociation of MAD2 from the target at the transition from metaphase to anaphase.
Although yeast strains lacking Mad2 are viable, deletion of Mad2 in mouse causes cell lethality. Mad2-null mouse cells do not arrest in response to spindle damage, show widespread chromosome missegregation, and undergo apoptosis during initiation of gastulation (Dobles et al., 2000). Mouse embryo cells may be much more sensitive to aneuploidy, which would be caused due to loss of the checkpoint. It is equally possible that Mad2 may play an additional role in higher eukaryotes.
Through a screen of a HeLa cell cDNA library by the yeast two-hybrid system with MAD2 as bait, we have identified a novel human protein, designated CMT2 (Caught by MAD Two). Here we show that formation of the CMT2–MAD2 complex coincides with dissociation of MAD2 from the target, p55CDC, and overexpression of CMT2 abolishes the function of the spindle checkpoint. Depletion of CMT2 causes a transient cell cycle delay followed by cell death. These results indicate that CMT2 is required for a cell cycle event in mitosis, particularly at or beyond the point where the spindle checkpoint is satisfied.
Results
Structure of CMT2
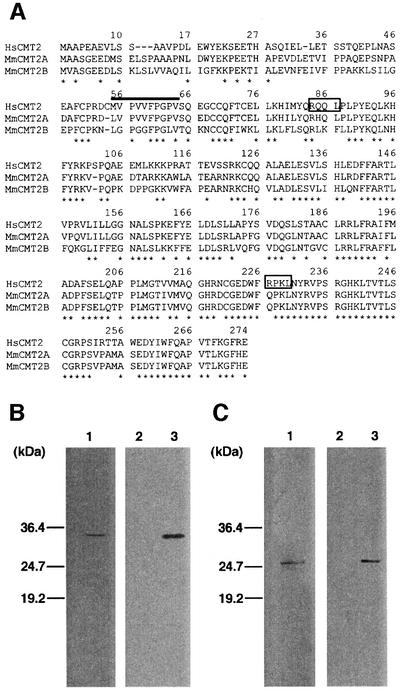
A yeast two-hybrid screen (Feilotter et al., 1994) with human MAD2 as bait allowed identification of two genes, one encoding human MAD1, which was previously shown to interact with MAD2 (Jin et al., 1998), and another encoding a novel protein, termed CMT2. Through database searches, we found two mouse cDNA clones homologous to human CMT2 (Figure 1A). We failed to identify any significant homologs in the entire budding yeast genome. The isolated cDNA most likely encodes the full-length CMT2 protein, whose calculated molecular weight is 31 kDa. By comparison of the human CMT2 to the mouse counterparts, we found the first methionine at analogous positions (Figure 1A). We also performed 5′-RACE and only obtained products that contained a short sequence upstream from the putative first methionine (see Materials and methods for details). With bacterially expressed CMT2, we raised an antibody to CMT2. An affinity-purified antibody to CMT2 detected a 34 kDa protein as a single band (Figure 1B, lane 1). When the cDNA isolated through the yeast two-hybrid screen was transiently expressed, this band became more intense (Figure 1B, lane 3) with no appearance of additional bands. We also raised an antibody to MAD2 and tested its activity (Figure 1C).
Fig. 1. Structure of CMT2. (A) Human CMT2 gene and mouse homologs. The amino acid sequences of the human CMT2 gene (HsCMT2; DDBJ/EMBL/GenBank accession No. NM_014628) and two mouse homologous genes (MmCMT2A and B; accession Nos NP_079925 and BAB27350, respectively) are aligned. The asterisks indicate the amino acids conserved in the three genes. The two putative destruction boxes are boxed. (B) Antibody to CMT2. One hundred micrograms (lane 1) of HeLa cell extracts were run on SDS–PAGE and processed for western blotting with the antibody to CMT2. One microgram of extracts prepared from HeLa cells transiently transfected with a plasmid for a transient expression of CMT2 (lane 3) or a negative control vector (lane 2) were processed as lane 1. (C) Antibody to MAD2. Same as (B) except that a plasmid for overexpression of MAD2 was used for lane 3 and the antibody to MAD2 was used as a probe.
With the yeast two-hybrid assay, we found that a domain containing 34 amino acids (between positions 45 and 78) of MAD2 is necessary for interaction with CMT2 (Supplementary figure 1, available at The EMBO Journal Online).
Expression of CMT2 in the cell cycle
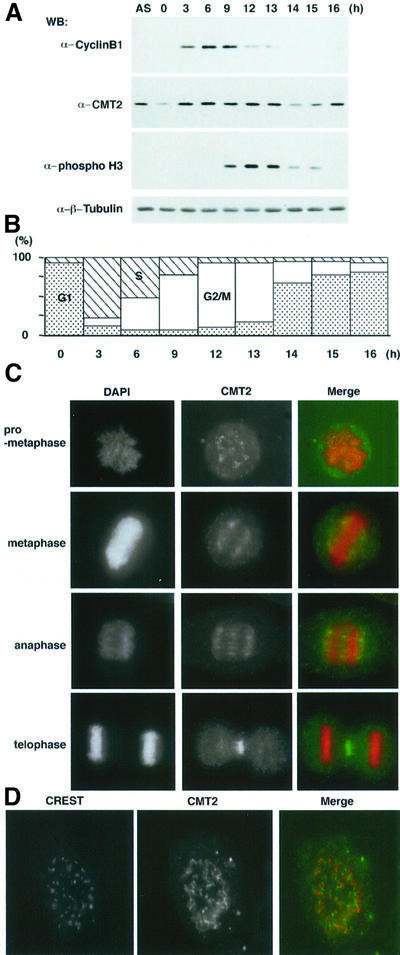
We examined the expression level of CMT2 in the cell cycle. HeLa cell culture was synchronized by double thymidine block and release, and cell extracts were prepared for western blotting. The levels of cyclin B1 and phosphorylated histone H3 were also measured to illustrate the synchronization. As shown in Figure 2A, the level of cyclin B1 reached a peak at 9 h after release and dropped in the following 3 h. The level of phosphorylated histone H3, which serves as a marker for mitosis (Mahadevan et al., 1991), reached a peak at 12–13 h after release and decreased in the following 2 h. The results, along with flow cytometry (FACS) analysis (Figure 2B), indicated that the culture was highly synchronized and most of the cells were in mitosis 9–13 h after release. The level of CMT2, which was low at time 0, increased and remained constant until late mitosis. Fourteen hours after release, it suddenly dropped; CMT2 may be destroyed after late mitosis. Although we did not address the biological significance, CMT2 contains two motifs similar to a destruction box, a signature for proteins to be destroyed at or after the onset of anaphase.
Fig. 2. Expression and localization of CMT2. (A) HeLa cells were arrested by double thymidine block and released at time 0. Cell extracts were prepared at indicated time points after the release. Extracts prepared from an asynchronous culture (AS) were also examined. (B) Synchronization of the culture used in (A) was confirmed by FACS analysis. (C) Mitotic HeLa cells were stained with DAPI and the antibody to CMT2. In the merged figures, DAPI is shown as red and immunofluorescence from CMT2 as green. (D) Mitotic HeLa cells were stained with the CREST antibody (red) and the antibody to CMT2 (green).
Cellular localization of CMT2 was examined by indirect immunofluorescent staining in HeLa cells. At an early stage of mitosis, such as prophase and prometaphase, it was found unevenly distributed throughout the nucleoplasm (Figure 2C). A fraction of CMT2 seemed to concentrate as speckles, some of which localized near the kinetochore-specific antigen recognized by CREST antibody (Figure 2D). While the CREST antibody stained its antigen as sharp dots, the fluorescent signal from the antibody to CMT2 was more diffused. We noticed that the CMT2 speckles increased as mitosis progressed. From metaphase to anaphase, CMT2 was concentrated on the spindle. After the completion of chromosome segrega tion, the majority of CMT2 remained in the mid-zone (Figure 2C).
p55CDC–MAD2 complex versus CMT2–MAD2 complex
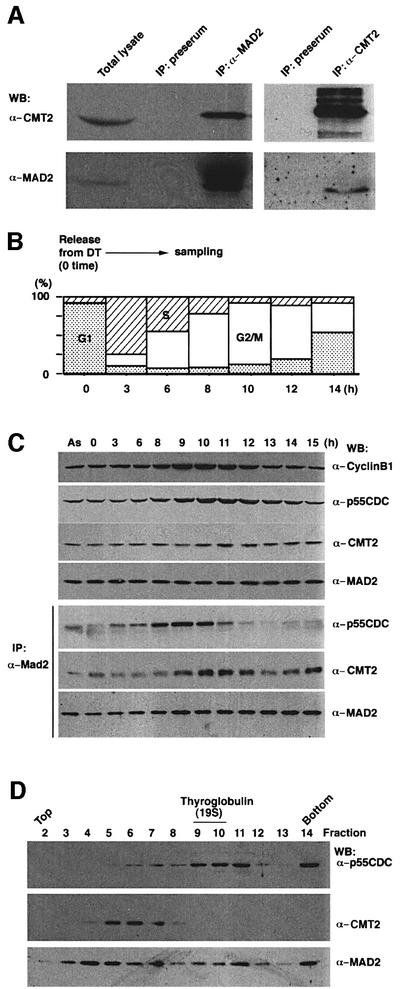
To examine physical interaction between CMT2 and MAD2 in HeLa cell extracts, immunoprecipitation was performed with the antibody to MAD2. As shown in Figure 3A, MAD2 antibody immunoprecipitates contained MAD2 as well as CMT2 (lane 3). A reciprocal experiment performed with the antibody to CMT2 also showed both proteins (lane 5). Under identical conditions, the precipitates by pre-immune serum did not contain either of the proteins (lanes 2 and 4).
Fig. 3. CMT2–MAD2 and p55CDC–MAD2 complexes. (A) Immuno precipitation was performed with the antibody to MAD2 (lane 3) or antibody to CMT2 (lane 5) with extracts prepared from an asynchronous HeLa cells culture. The precipitates were run on SDS–PAGE and analyzed by western blotting. Total lysates used for immunoprecipitation (lane 1) and precipitates by pre-immune serum (lanes 2 and 4) were also analyzed. (B) Cell culture, blocked by double thymidine (DT), was released at time 0 and sampled for (C). Synchronization of the culture was confirmed by FACS analysis. (C) Cell cycle analysis. Cell extracts were prepared at indicated time points after the release and processed for western blotting (upper three panels), or immunoprecipitation with the antibody to MAD2 followed by western blotting (lower three panels). Extracts prepared from an asynchronous culture (AS) were also examined in the same way. (D) Glycerol gradient centrifugation. Mitotic HeLa cell extracts were subjected to glycerol gradient centrifugation. Each fraction was run on SDS–PAGE and processed for western blotting.
We examined at which stage of the cell cycle CMT2 forms a complex with MAD2. Culture of HeLa cells was synchronized by double thymidine block and release, and cell extracts were prepared for immunoprecipitation with the antibody to MAD2. At early mitosis, the majority of MAD2 appeared to form a complex with p55CDC (Figure 3C; IP, α-MAD2 probed with α-p55CDC, lanes 8–10). A minor fraction of CMT2 was also found to form a complex with MAD2 at these time points. As the cell cycle progressed to mid-mitosis, MAD2 no longer formed the complex with p55CDC. Coincidentally, the great majority of MAD2 bound to CMT2 (Figure 3C; IP, α-MAD2 probed with α-CMT2, lanes 10–12). It should be noted that while the level of p55CDC measured by western blotting (Figure 3C, top panel) reached a peak around lanes 10–12, the level of p55CDC associated with MAD2 reached a peak earlier (Figure 3C; IP, α-MAD2 probed with α-p55CDC, lanes 8–10). The result indicates that p55CDC which is not in complex with MAD2 exists around mid-mitosis and that this form of p55CDC probably represents an active form responsible for promoting APC-dependent ubiquitylation. The level of p55CDC measured by western blotting decreased in late mitosis (lanes 13–15) due to probable destruction at or around anaphase. These results are consistent with previous reports (Weinstein, 1997; Wassmann and Benezra, 1998). The result also indicates that there are at least two different protein complexes containing MAD2. The MAD2–p55CDC complex is formed and disassembled prior to the formation of the MAD2–CMT2 complex that becomes prominent in mid-mitosis.
In order to demonstrate further the presence of the two different protein complexes containing MAD2, we analyzed mitotic HeLa cell extracts by glycerol density gradient centrifugation. Each fraction was examined by western blotting for the presence of p55CDC, CMT2 and MAD2. The great majority of p55CDC was enriched in the fractions around a 19S marker, thyroglobulin (Figure 3D, lanes 9–11). These fractions would represent p55CDC that associates with the 20S APC/cyclosome. In contrast, CMT2 was found in significantly lighter fractions (lanes 5–7). Distribution of MAD2 was relatively broad. However, two peaks, one coincident with p55CDC and the other with CMT2, were noticeable. We performed immunoprecipitation of each fraction and found that MAD2, at least in part, was in a complex either with p55CDC or CMT2 (Supplementary figure 2).
Effect of spindle poison on formation of CMT2–MAD2 complex
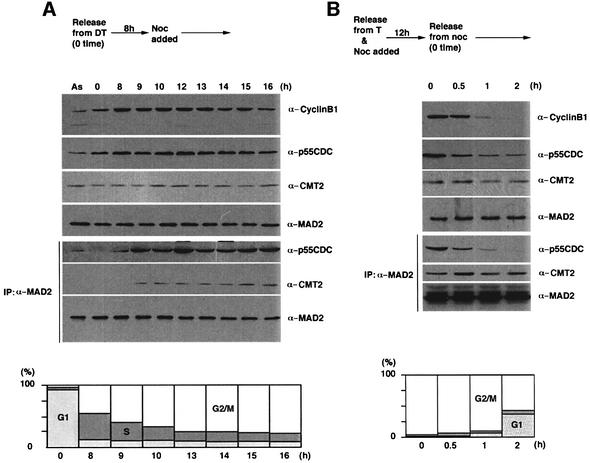
In as much as the CMT2–MAD2 complex appeared in mid-mitosis, we suspected that the complex is assembled depending on the progress of proper bipolar attachment of kinetochores to the spindle. To test this hypothesis, a synchronized culture at the G1/S boundary was released into media containing a spindle poison, nocodazole, and the formation of p55CDC–MAD2 and CMT2–MAD2 complexes was monitored by immunoprecipitation. As shown in Figure 4A, from 9 to 16 h after release, the p55CDC–MAD2 complex remained stable in the presence of the spindle poison. This distinctly contrasts with the result shown in Figure 3C, where in absence of the drug, the p55CDC–MAD2 complex was no longer detectable beyond 11 h after release. Although a small amount of the CMT2–MAD2 complex was detectable, it did not become prominent in the presence of the spindle poison (Figure 4A, Supplementary figure 3). We also examined the formation of the p55CDC–MAD2 and CMT2–MAD2 complexes after removal of the spindle poison. Within 1 h after the removal, the p55CDC–MAD2 complex was disassembled. In contrast, the CMT2–MAD2 complex became more prominent 30 min following removal of nocodazole (Figure 4B).
Fig. 4. Transition from p55CDC–MAD2 to CMT2–MAD2. (A) HeLa cells were arrested by double thymidine (DT) block and released at time 0. Nocodazole was added at 8 h after the release. Extracts were prepared at indicated time points and processed for western blotting (upper three panels), or immunoprecipitation with the antibody to MAD2 followed by western blotting (lower three panels). Extracts prepared from an asynchronous culture (AS) were also examined in the same way. FACS analysis was performed to monitor cell cycle progression. (B) HeLa cells were arrested by a single thymidine (T) block and released into the media containing nocodazole. After a 12 h incubation, the cells were released into the drug-free media (time 0). Cell extracts were prepared at indicated time points for western blotting (upper three panels), or immunoprecipitation with the antibody to MAD2 followed by western blotting (lower three panels). FACS analysis was performed to monitor cell cycle progression.
Abrogation of the spindle checkpoint by overexpression of CMT2
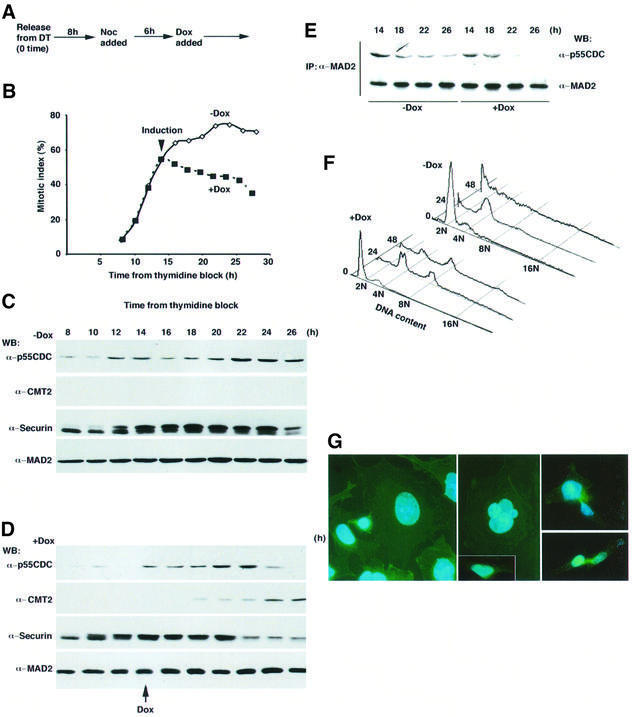
The CMT2–MAD2 complex became prominent when the checkpoint-induced arrest was released upon the completion of spindle formation (Figure 4B). This result suggested that formation of the CMT2–MAD2 complex was a result of the release of the spindle checkpoint, or that it caused the release of the checkpoint. If the latter explanation is true, then overexpression of CMT2 might compromise an arrest induced by the checkpoint. To test this possibility, CMT2 was overexpressed in HeLa cells that were arrested by the checkpoint. The CMT2 gene was placed under the control of an inducible promoter (CMV minimum promoter/tet operator) and integrated into the genome of HeLa cells expressing the tet repressor (TetR). Upon addition of doxycycline (Dox, a derivative of tetracycline), the system allowed overexpression of CMT2. Transfected HeLa cells were blocked by thymidine and released into media that contained nocodazole.
When Dox was not added (thus CMT2 was not overexpressed), the mitotic index, (percentage of the non-adherent pseudo-mitotic cells in population) was ∼60%, at 14 h after release from the thymidine block (Figure 5B). Continued incubation with nocodazole resulted in an increase of the mitotic index up to 80%. Examination of the level of human securin, which normally drops in mid-mitosis (Zou et al., 1999), remained stable under this condition, indicating that the cells were tightly arrested due to the effect of nocodazole (Figure 5C).
Fig. 5. Overexpression of CMT2. (A) HeLa cells in which CMT2 was inducible by Dox were arrested by double thymidine (DT) block and released at time 0. Nocodazole was added at 8 h after the release. Dox was added [+Dox for (D)] or not [–Dox for (C)] at 14 h after the release. (B) Mitotic index. (C and D) Extracts were prepared at each time point and processed for western blotting. For detection of p55CDC and CMT2, 10 µg of total proteins were run on SDS–PAGE. Note that the endogenous CMT2 was not detectable under this condition. Thirty micrograms of total proteins for detection of securin and 50 µg for MAD2 were used. (E) Extracts were prepared at each time point for immunoprecipitation with the antibody to MAD2, followed by western blotting. (F) FACS analysis. (G) The cells prepared as in (D) were grown for 28 h after the release from double thymidine block and stained with DAPI (blue, for DNA) and FITC-conjugated phalloidin (green, for cell shape). Induction of CMT2 in the cells arrested by nocodazole resulted in a forced exit from mitosis. The majority of them exhibited larger nuclei (left panel) or multiple nuclei (middle panel). A fraction of them showed a ‘cut’-like phenotype (right panels). If CMT2 was not induced, the majority of the cells exhibited condensed chromosomes with a rounded-up cell shape, a typical phenotype of prometaphase arrest (data not shown). Interphase nuclei of normal HeLa cells at late G2 are shown for comparison (middle panel, inset).
When Dox was added at 14 h after release from the thymidine block, the induced CMT2 became detectable within 3–4 h (Figure 5D). The level of the induced CMT2 was ∼10-fold of the endogenous CMT2. Upon the addition of Dox, the mitotic index dropped (Figure 5B). The levels of p55CDC and securin also dropped ∼8 h after the addition of Dox (Figure 5D). These results indicate that induction of CMT2 abrogates the function of the spindle checkpoint and overcomes the cell cycle arrest caused by the spindle poison. The overexpression of CMT2 most likely affects the spindle checkpoint in a direct manner. Immunoprecipitation by the antibody to MAD2 demonstrated that the p55CDC–MAD2 complex was disassembled after the induction of CMT2, while it persisted if CMT2 was not induced (Figure 5E). We also measured the DNA content of individual cells. If CMT2 was not induced, most of the cells had a 4N content 24 h after release from the thymidine block, indicating that they were arrested in G2 or mitosis (Figure 5F, 24 h, –Dox). Continued presence of nocodazole resulted in appearance of cells with <2N content (48 h, –Dox), probably reflecting cell death due to prolonged arrest by nocodazole. In contrast, the overexpression of CMT2 resulted in appearance of cells with 8N content (Figure 5F, 24 and 48 h, +Dox), indicating that the next round of DNA synthesis was completed without previous chromosome segregation. This phenotype is typical in cells lacking the functional spindle checkpoint (Taylor and McKeon, 1997; Cahill et al., 1998; Michel et al., 2001). It should be noted that overexpression of CMT2 prevents appearance of cells with <2N content, suggesting rescue of cells from cell death due to prolonged arrest by nocodazole. We examined 11 independent cell lines in which the inducible CMT2 construct was integrated and found that they all consistently exhibited this phenotype. In the absence of the spindle poison, the induction of CMT2 resulted in a slight reduction in the growth rate. Perhaps it partially abolishes the checkpoint function. A reduction in the level of MAD2 partially abrogates the checkpoint and affects cell cycle progression only when the checkpoint is challenged (Michel et al., 2001).
Cytological observation of the cells indicated that overexpression of CMT2 in cells arrested by nocodazole caused extensive abnormality in chromosome morphology. The great majority of cells (∼80%) exhibited nuclei that were larger than those of normal mitotic cells (Figure 5G, left panel). It was also noticeable that they occasionally contained multiple nuclei (Figure 5G, middle panel). In addition, a minor fraction of the cells (20%) showed partially and unequally segregating nuclear mass (Figure 5G, right panels), which may be analogous to the phenotype of fission yeast cut mutants (Yanagida, 1998). The cut-like phenotype may be because nocodazole was losing its efficacy over time and allowed partial segregation of chromosomes.
Inactivation of CMT2
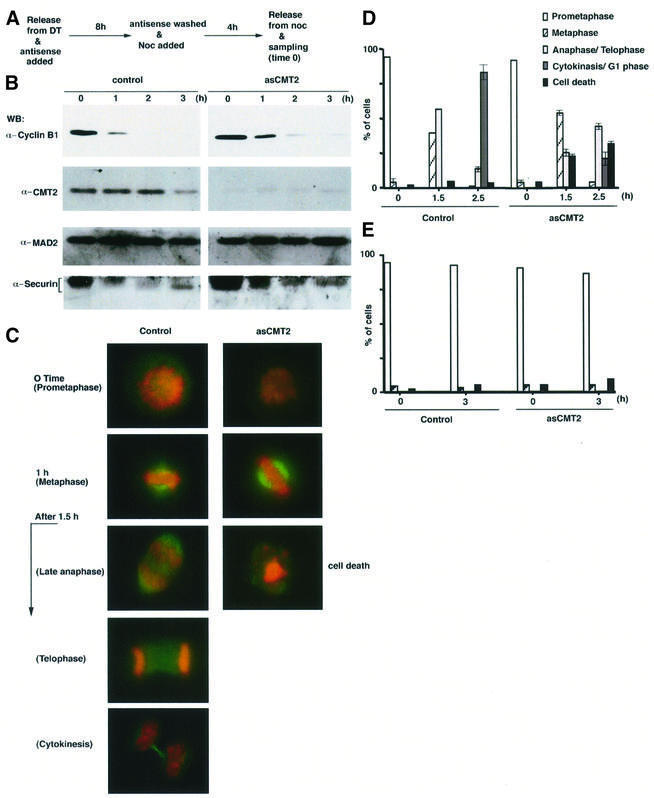
We finally examined the effect of CMT2 depletion by the antisense oligonucleotide-based technology that has been utilized successfully in similar studies (Li et al., 1999; Yao et al., 2000). Since CMT2 and MAD2 interact with each other around mid-mitosis, we were particularly interested in testing whether the cell cycle would proceed normally after mid-mitosis under conditions of low CMT2 expression. Following thymidine block and release, antisense oligonucleotides for CMT2 (hereafter termed asCMT2) were transfected into HeLa cells for 8 h. The transfected cells were subsequently treated with nocodazole for 4 h, and released into drug-free media (Figure 6A). As a control experiment, another group of HeLa cells was treated in the same way, but with random oligonucleotides. Western blotting with the antibody to CMT2 demonstrated that the treatment with asCMT2 was effective. In cell extracts prepared from time 0 through the following 3 h, the level of CMT2 was <10% of that of the control group (Figure 6B). Both the cells treated with asCMT2 and the cells in the control group were arrested in early mitosis at time 0 (Figure 6C and D). Between 1 and 1.5 h after release into drug-free media, condensed chromosomes were found near the spindle equator (Figure 6C). Thus, the treatment with asCMT2 did not affect progression of the cell cycle up to metaphase. Further incubation after the release, however, revealed a dramatic difference. The percentage of the cells which remained at metaphase was higher in the asCMT2-treated cells (53.2 versus 39.4% at 1.5 h after release, Figure 6D). While 56.4% of the control group entered into anaphase/telophase at 1.5 h, only 24.7% did so when treated with asCMT2. Thus, loss of CMT2 caused a delay in the transition from metaphase to anaphase. Examination of the levels of cyclin B1 and securin also demonstrated a delay in their destruction in the asCMT2-treated cells (Figure 6B). In addition, the asCMT2-treated cells exhibited an abnormality in chromosome morphology (22.1% at 1.5 h and 31.5% at 2.5 h). In these cells, fragmentation of nuclear mass, which might be an indication of apoptotic cell death, was apparent (Figure 6C and D). Two and a half hours after release, 82.8% of the control cells exited from mitosis, whereas only 20.7% of the asCMT2-treated cells did so (Figure 6D). The cell death observed in the asCMT2-treated cells was a post-metaphase event. If nocodazole was not removed at time 0, both the control and the asCMT2-treated cells remained arrested at prometaphase (Figure 6E).
Fig. 6. Inactivation of CMT2. (A) HeLa cells were released from double thymidine (DT) block and transfected with asCMT2 for 8 h. After asCMT2 was removed, they were grown in media containing nocodazole for 4 h and released into drug-free media at time 0. The samples were taken at indicated time points and processed for western blotting for (B) or stained with DAPI (red, for DNA) and the antibody to β-tubulin conjugated with FITC (green, for spindle) for (C). (D) The mitotic stage was determined by examining the cells after staining with DAPI. For each time point, 100 cells were scored. The average percentages of four independent experiments are shown with error bars. (E) The cells were prepared in the same way as (A) except that nocodazole was not removed at time 0. The mitotic stage was determined as in (D).
Discussion
CMT2–MAD2 complex
Previous studies (Fang et al., 1998; Kallio et al., 1998; Wassmann and Benezra, 1998) have demonstrated that MAD2 forms a complex with p55CDC and suggested a role for MAD2 in preventing p55CDC from promoting APC-dependent ubiquitylation (Li et al., 1997; Fang et al., 1998). A recent study (Sudakin et al., 2001) has suggested that MAD2 is a component of the MCC along with BubR1, Bub3 and p55CDC. Results from other laboratories (Wu et al., 2000; Skoufias et al., 2001; Tang et al., 2001) suggest that p55CDC–MAD2 and p55CDC–BubR1 exist in separate complexes that inhibit APC. We have shown by two means that the great majority of CMT2 does not interact with the p55CDC–MAD2 complex to form a ternary complex. First, the timing of formation of the CMT2–MAD2 complex coincides with that of disassembly of the p55CDC–MAD2 complex. Secondly, the two complexes, MAD2–p55CDC and CMT2–MAD2, can be physically separated by glycerol gradient centrifugation. CMT2 forms a stable complex with MAD2 in the absence of p55CDC. Therefore, we speculate that CMT2 is not required for a process in which MAD2 inhibits the activity of p55CDC. However, it should be stressed that we do not exclude the possibility that CMT2 transiently interacts with the p55CDC–MAD2 complex.
Is CMT2 a silencer of the spindle checkpoint?
It has been well documented that sister chromatids separate, with a lag-time of 20–30 min, after the last kinetochore is attached to the spindle (Rieder et al., 1994, 1995). The separation is highly synchronous, suggesting that the attachment of the spindle to the last kinetochore triggers a process that promotes separation of all the sister chromatids at once. Perhaps a diffusible element is responsible for triggering such a process (Rieder et al., 1997). Prior to sister chromatid separation, the spindle checkpoint must be silenced so that Cdc20/Slp1/p55CDC can promote the proteolysis mediated by APC. A previous report (Wassmann and Benezra, 1998) and our results (Figure 3C) indicate that MAD2 dissociates from p55CDC around the transition from metaphase to anaphase. Dissociation of MAD2 from p55CDC would therefore be a central event in the silencing process of the spindle checkpoint.
The CMT2–MAD2 complex becomes prominent in mid-mitosis. When a spindle poison interferes with spindle formation, the p55CDC–MAD2 complex remains stable. Removal of the drug triggers the disassembly of the p55CDC–MAD2 complex as well as formation of the CMT2–MAD2 complex. These results suggest that, upon the completion of spindle formation, MAD2 switches its binding partner from p55CDC to CMT2. In this scenario, CMT2 may play an active role, that is, to remove MAD2 from p55CDC and tether MAD2 for silencing the checkpoint. Consistent with this role, we have also shown that overexpression of CMT2 in cells arrested by nocodazole results in abrogation of the arrest maintained by the spindle checkpoint. This is accompanied by disappearance of the p55CDC–MAD2 complex.
Cell death by depletion of CMT2
Although it is an attractive hypothesis that CMT2 is a silencer of the spindle checkpoint, it is equally possible that CMT2 may play another (or an additional) role in mitosis.
If CMT2 is a silencer of the spindle checkpoint, loss of CMT2 would cause a prolonged activation of the checkpoint, even when spindle formation is completed. This would result in a cell cycle arrest at metaphase. In fact, the spindle checkpoint can be activated chronically without damage to the spindle in budding and fission yeasts. Overexpression of a component of the checkpoint [Mps1 (Hardwick et al., 1996) or MAD2 (He et al., 1997)] results in a cell cycle arrest at metaphase. Depletion of CMT2 results in cell death following the very transient delay in the onset of anaphase. The discrepancy between yeasts and HeLa cells remains to be clarified experimentally, and we interpret this result as follows.
In HeLa cells lacking CMT2, mitosis proceeds normally up to metaphase. When nocodazole is added for inhibition of spindle formation, loss of CMT2 does not induce cell death. These results indicate that the cell death due to loss of CMT2 is a post-metaphase event. CMT2 is localized on the spindle from metaphase to anaphase. It is found in the mid-zone in later mitosis. Localization of CMT2 is reminiscent of that of ‘chromosome passenger proteins’, and suggests that CMT2 may play a role with these proteins. It should be noted that inactivation of survivin, one of the chromosome passenger proteins, also results in cell death (Li et al., 1999).
Despite the fact that most of the components of the spindle checkpoint are conserved from yeast to human, there is no apparent homolog of CMT2 in the yeast genome. Absence of CMT2 in yeasts may suggest that CMT2 plays a role in events specific to higher eukaryotes. As shown in Figure 5F, HeLa cells tend to die when treated with nocodazole for a long time (typically >24 h). Interestingly, overexpression of CMT2 significantly reduces cell death (Figure 5F). Therefore, CMT2 may suppress cell death induced by a chronic activation of the spindle checkpoint.
These possibilities are mutually inclusive and CMT2 may play multiple roles in mitosis. The results presented here suggest that CMT2 (most likely with MAD2) is required for concerted progression of the cell cycle when, and after, the spindle checkpoint is satisfied.
Materials and methods
Cell culture and transfection
HeLa cells were grown under standard conditions in DMEM supplemented with fetal bovine serum and antibiotics. HeLa T-Rex cells (Invitrogen) were grown according to the manufacturer’s protocol. For cell cycle synchronization, a standard protocol for double thymidine block and release was followed (first arrested with 2.5 mM thymidine for 18 h, released into thymidine-free medium for 8 h, and incubated in 2.5 mM thymidine for 18 h for the second arrest). To arrest in M phase, cells were incubated with 200 ng/ml nocodazole for 12 h, after a single thymidine block.
Cells were transfected with Effecten Transfection Reagent (Qiagen) according to the manufacturer’s protocol. HeLa T-Rex cells that express CMT2 protein were generated by transfecting them with pcDNA 4 (Invitrogen) plasmid carrying CMT2 gene. Zeocin-resistant foci were selected and purified using the dilution method. They were maintained in 0.2 µg/ml zeocin. To induce CMT2, cells were incubated with 1 µg/ml Dox. To arrest CMT2-inducible cells in M phase, 500 ng/ml nocodazole was used.
Antibodies
Fusion protein of glutathione S-transferase (GST) and the full-length CMT2 (GST–CMT2) was used to raise the antibody to CMT2 at Covance. The antibody to MAD2 was prepared at Bethyl Laboratory with His6-MAD2. The antibodies were affinity-purified with His6-CMT2 or His6-MAD2 immobilized on nitrocellulose membrane.
Western blotting and immunoprecipitation
Harvested cells were washed once with PBS (pH 7.2) without calcium and magnesium, and lysed in NP-40 lysis buffer [50 mM HEPES pH 7.4, 250 mM NaCl, 1% NP-40, 5 mM β-glycerophosphate, aprotinin (10 µg/ml), leupeptin (10 µg/ml), 1 mM PMSF, 2 mM Na3VO4, 0.2 mM EDTA, 10 mM NaF, 1 µM Microcystin-LR (Calbiochem) and 1 mM DTT]. Cell lysates were incubated at 0°C for 20 min and centrifuged at 13 000 g for 5 min. For immunoprecipitation, the supernatants were incubated with indicated antibody (10 µg/ml) for 4 h at 4°C. Then 20 µl of protein G–Sepharose beads were added (Pierce) and the supernatants were incubated for another 4 h. The immunoprecipitates were washed once by NP-40 lysis buffer, twice by NP-40 lysis buffer without NaCl, and subjected to western blotting. Antibodies to CMT2 or p55CDC (Biosource International; Weinstein, 1997) were used at a concentration of 1 µg/ml. The antibody to MAD2 (Covance) was used at the recommended dilution. Other antibodies, anti-β-tubulin (Sigma) and anti-cyclin B1 (Pharmingen), were used at a concentration of 1 µg/ml. The anti-phospho-histone H3 antibody (Upstate Biotechnology) was used at a concentration of 0.5 µg/ml.
5′-RACE
5′-rapid amplification of cDNA ends System (Gibco BRL) was performed following the manufacturer’s protocols. The CMT2-specific antisense primers were CM1 (5′-GACAGAACCTCCGCC-3′) and CM2 (5′-GGA CTTCTCATACCA-3′). Poly(A)+ RNAs were isolated from HeLa cells and used for analysis. The product of 5′-RACE contained only 15 nucleotides (GGCACGAGGGTCGTG) upstream from the putative first methionine of CMT2, in which no additional open reading frame could be assembled.
Glycerol gradient centrifugation
Extracts were prepared from HeLa cells that were treated with nocodazole for 12 h (arrested extracts) or the cells that were released from nocodazole block (released extracts). The two extracts were mixed equally and a total of 500 µg of proteins in 200 µl was applied to a 2.2 ml, 18–40% glycerol gradient in NP-40 lysis buffer (100 mM NaCl). After a spin at 50 000 r.p.m. for 12 h with Beckman STL 500 rotor, they were fractionated into 14 fractions. Each fraction was concentrated by trichloroacetic acid precipitation and neutralized by 2 M Tris solution prior to being run on SDS–PAGE. Gel-filtration standard (Bio-Rad) was used as the marker for S value.
Cell staining
Cells grown on poly-l-lysine-coated cover slips were washed with ice-cold PHEM buffer (60 mM PIPES, 25 mM HEPES pH 6.9, 10 mM EDTA, 4 mM MgCl2) and fixed with 4% formamide in PHEM for 15 min. The cells were washed twice with PHEM and permeated with 0.2% Triton X-100 in PHEM for 2 min. After two washes with PHEM, the cells were incubated with appropriate antibodies. When necessary, 50 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) (Roche) and 500 ng/ml fluorescein isothiocyanate (FITC)-conjugated phalloidin (Fluka) were also added for 5 min.
Antisense study
All antisense oligonucleotides were designed and synthesized by Sequitur (Natick, MA). Transfection was performed with OligofectinG (Sequitur) following the manufacturer’s protocols. The sequences of the oligonucleotides used in this study were TGTCAATCAAGGCG TTGGTCGCTTC (control) and ACCACTGGTACCATGCAGTCTC TTG (asCMT2). We confirmed that ∼80% of the treated cells incorporated the oligonucleotides, with fluorescent-labeled oligonucleotides.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors thank Drs Kirschner and Zou (Harvard Medical School, MA) for the antibody to securin, Dr Brinkley (Baylor College of Medicine, TX) for the CREST antibody and Dr Query (Albert Einstein College of Medicine, NY) for comments on the manuscript. This work was supported by grants from NIH (GM56305), United States Army (BC980290) and the Ministry of Education, Culture, Sports, Science and Technology of Japan (COE Research) to T.M. An initial stage of the work was supported by an award from Howard Hughes Medical Institute to T.M. T.H. was a recipient of a fellowship from United States Army. S.H.K. was a recipient of fellowships from New York State Department of Health and United States Army and is currently supported by the Ministry of Science and Technology, Korea (M1-0016-00-0015 and R01-2000-000-00089-0). The authors also thank Kyowa Hakko Kogyo Co. Ltd. (Tokyo, Japan) for support.
References
- Abrieu A., Kahana,J.A., Wood,K.W. and Cleveland,D.W. (2000) CENP-E as an essential component of the mitotic checkpoint in vitro. Cell, 102, 817–826. [DOI] [PubMed] [Google Scholar]
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Basto R., Gomes,R. and Karess,R.E. (2000) Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat. Cell Biol., 2, 939–943. [DOI] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer,C., Yu,J., Riggins,G.J., Willson,J.K., Markowitz,S.D., Kinzler,K.W. and Vogelstein,B. (1998) Mutations of mitotic checkpoint genes in human cancers. Nature, 392, 300–303. [DOI] [PubMed] [Google Scholar]
- Chan G.K.T., Jablonski,S.A., Starr,D.A., Goldberg,M.L. and Yen,T.J. (2000) Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol., 2, 944–947. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Waters,J.C., Salmon,E.D. and Murray,A.W. (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science, 274, 242–246. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko,A., Mann,M. and Murray,A.W. (1998) Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol., 143, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Brady,D.M., Smith,D., Murray,A.W. and Hardwick,K.G. (1999) The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell, 10, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters,J.M., Kirschner,M.W. and Koshland,D. (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev., 10, 3081–3093. [DOI] [PubMed] [Google Scholar]
- Dobles M., Liberal,V., Scott,M.L., Benezra,R. and Sorger,P.K. (2000) Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell, 101, 635–645. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998) The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev., 12, 1871–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilotter H.E., Hannon,G.J., Ruddell,C.J. and Beach,D. (1994) Construction of an improved host strain for two hybrid screening. Nucleic Acids Res., 22, 1502–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Yamano,H., Kumada,K., Nagao,K., Hunt,T. and Yanagida,M. (1996) Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature, 381, 438–441. [DOI] [PubMed] [Google Scholar]
- Guacci V., Koshland,D. and Strunnikov,A. (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell, 91, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss,E., Luca,F.C., Winey,M. and Murray,A.W. (1996) Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science, 273, 953–956. [DOI] [PubMed] [Google Scholar]
- He X., Patterson,T.E. and Sazer,S. (1997) The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 94, 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Hoffman,D.B., Fang,G., Murray,A.W. and Salmon,E.D. (2000) Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol., 150, 1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Trotis,L. and Roberts,B.T. (1991) S.cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66, 507–517. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Ikui A.E., Furuya,K., Yanagida,M. and Matsumoto,T. (2002) Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci., 115, 1603–1610. [DOI] [PubMed] [Google Scholar]
- Jin D.Y., Spencer,F. and Jeang,K.T. (1998) Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell, 93, 81–91. [DOI] [PubMed] [Google Scholar]
- Kallio M., Weinstein,J., Daum,J.R., Burke,D.J. and Gobsky,G.J. (1998) Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol., 141, 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science, 279, 1045–1047. [DOI] [PubMed] [Google Scholar]
- Li F., Ackermann,E.J., Bennett,C.F., Rothermel,A.L., Plescia,J., Tognin,S., Villa,A., Marchisio,P.C. and Altieri,D.C. (1999) Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat. Cell Biol., 1, 461–466. [DOI] [PubMed] [Google Scholar]
- Li R. and Murray,A.W. (1991) Feedback control of mitosis in budding yeast. Cell, 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Li Y. and Benezra,R. (1996) Identification of a human mitotic checkpoint gene: hsMAD2. Science, 274, 246–248. [DOI] [PubMed] [Google Scholar]
- Li Y., Gorbea,C., Mahaffey,D., Rechsteiner,M. and Benezra,R. (1997) MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits activity. Proc. Natl Acad. Sci. USA, 94, 12431–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan L.C., Willis,A.C. and Barratt,M.J. (1991) Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell, 65, 775–783. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk,R. and Nasmyth,K. (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell, 91, 35–45. [DOI] [PubMed] [Google Scholar]
- Michel L.S. et al. (2001) MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature, 409, 355–359. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet., 35, 673–745. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Jan-Michael,P. and Uhlmann,F. (2000) Splitting the chromosome: cutting the ties that bind sister chromatids. Science, 288, 1379–1384. [DOI] [PubMed] [Google Scholar]
- Nicklas R.B. (1997) How cells get the right chromosomes. Science, 275, 632–637. [DOI] [PubMed] [Google Scholar]
- Peters J.M. (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell, 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Schultz,A., Cole,R.W. and Sluder,G. (1994) Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol., 127, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Cole,R.W., Khodjakov,A. and Sluder,G. (1995) The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol., 130, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Khodjakov,A., Paliulis,L.V., Fortier,T.M., Cole,R.W. and Sluder,G. (1997) Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc. Natl Acad. Sci. USA, 94, 5107–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Melixetian,M., Faretta,M., Prosperini,E., Helin,K. and Musacchio,A. (2001) Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J., 20, 6371–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli,M., Knapp,S., Antoni,A.D., Jeang,K.T. and Musacchio,A. (2002) Crystal structure of the tetrameric Mad1–Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J., 21, 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V. and Hieter,P. (1998) Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet., 32, 307–337. [DOI] [PubMed] [Google Scholar]
- Skoufias D.A., Andreassen,P.R., Lacroix,F.B., Wilson,L. and Margolis,R.L. (2001) Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl Acad. Sci. USA, 98, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan,G.K.T. and Yen,T.J. (2001) Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BubR1, Bub3, CDC20, and Mad2. J. Cell Biol., 154, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bhardwaj,R., Li,B. and Yu,H. (2001) Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell, 1, 227–237. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Tomonaga T. et al. (2000) Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev., 14, 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich,F. and Nasmyth,K. (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature, 400, 37–42. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Wernic,D., Poupart,M.A., Koonin,E.V. and Nasmyth,K. (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell, 103, 375–386. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz,S. and Amon,A. (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science, 278, 460–463. [DOI] [PubMed] [Google Scholar]
- Wassmann K. and Benezra,R. (1998) Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc. Natl Acad. Sci. USA, 95, 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J.C., Chen,R.H., Murray,A.W. and Salmon,E.D. (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol., 141, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J. (1997) Cell cycle-regulated expression, phosphorylation, and degradation of p55Cdc. J. Biol. Chem., 272, 28501–28511. [DOI] [PubMed] [Google Scholar]
- Weiss E. and Winey,M. (1996) The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol., 132, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Lan,Z., Li,W., Wu,S., Weinstein,J., Sakamoto,K.M. and Dai,W. (2000) p55CDC/hCdc20 is associated with BubR1 and may be a downstream target of the spindle checkpoint kinase. Oncogene, 19, 4557–4562. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Guacci,V. and Koshland,D. (1996) Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathways. J. Cell Biol., 133, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M. (1998) Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol., 8, 144–149. [DOI] [PubMed] [Google Scholar]
- Yao X., Abrieu,A., Zheng,Y., Sullivan,K.F. and Cleveland,D.W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol., 2, 484–491. [DOI] [PubMed] [Google Scholar]
- Zou H., McGarry,T.J., Bernal,T. and Kirschner,M.W. (1999) Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science, 285, 418–422. [DOI] [PubMed] [Google Scholar]