Abstract
The nuclear function of the heterodimeric NF-κB transcription factor is regulated in part through reversible acetylation of its RelA subunit. We now demonstrate that the p300 and CBP acetyltransferases play a major role in the in vivo acetylation of RelA, principally targeting lysines 218, 221 and 310 for modification. Analysis of the functional properties of hypoacetylated RelA mutants containing lysine-to-arginine substitutions at these sites and of wild-type RelA co-expressed in the presence of a dominantly interfering mutant of p300 reveals that acetylation at lysine 221 in RelA enhances DNA binding and impairs assembly with IκBα. Conversely, acetylation of lysine 310 is required for full transcriptional activity of RelA in the absence of effects on DNA binding and IκBα assembly. Together, these findings highlight how site-specific acetylation of RelA differentially regulates distinct biological activities of the NF-κB transcription factor complex.
Keywords: acetylation/deacetylation/IκBα/p300/RelA
Introduction
The inducible NF-κB transcription factor complex plays a central role in regulating the inflammatory, immune and anti-apoptotic responses in mammals (Baldwin, 1996; Ghosh et al., 1998). The prototypical NF-κB complex corresponds to a heterodimer of p50 and RelA subunits, which is chiefly sequestered in the cytoplasm through its assembly with a family of inhibitory proteins termed the IκBs (Baldwin, 1996). Stimulus-induced phosphorylation of two N-terminal serines in the IκBs mediated by the macromolecular IκB kinase complex (IKK) (Karin, 1999) triggers the rapid ubiquitylation and subsequent degradation of this inhibitor by the 26S proteasome complex. The liberated NF-κB heterodimer rapidly translocates into the nucleus where it engages cognate κB enhancer elements and activates gene expression (Baldwin, 1996; Ghosh et al., 1998). One of the cellular genes induced by NF-κB is IκBα (Beg et al., 1993; Brown et al., 1993; Sun et al., 1993). Newly synthesized IκBα proteins shuttle between the cytoplasm and the nucleus and can remove NF-κB from DNA, promoting return of the now inactive NF-κB–IκBα complex to the cytoplasm. These events lead to the termination of the NF-κB transcriptional response (Arenzana-Seisdedos et al., 1995, 1997).
The NF-κB signaling pathway is evolutionarily conserved. In mammals, five Rel family members have been identified: RelA/p65, RelB, c-RelA, p50/p105 and p52/p100 (Baldwin, 1996; Ghosh et al., 1998). All of these proteins contain an N-terminal Rel homology domain (RHD) consisting of ∼300 amino acids. The N-terminal portion of the RHD mediates both backbone and sequence-specific major groove contacts with DNA, while the C-terminal portion of the RHD is responsible for backbone contacts as well as dimerization with other Rel family members and interaction with IκBα (Baeuerle, 1998; Ghosh et al., 1998). While all of the Rel proteins bind DNA, only RelA, c-Rel and RelB contain C-terminal transcriptional activation domains (TADs) (Ghosh et al., 1998). These domains regulate the interaction of NF-κB with various components of the basal transcription apparatus, including TATA-binding protein, through these C-terminal domains (Xu et al., 1993; Schmitz et al., 1995) and TFIIB (Blair et al., 1994). RelA also associates with the p300/CBP transcriptional co-activators through its RHD and C-terminal transactivation domain; overexpression of p300/CBP enhances the transactivation potential of NF-κB (Gerritsen et al., 1997; Perkins et al., 1997; Sheppard et al., 1999). Nuclear receptor co-activators from the p160 family, including SRC-1/N-CoA-1, TIF2/GRIP-1 and SRC-3/Rac3/ACTR, also function as co-activators with NF-κB (Na et al., 1998; Sheppard et al., 1999; Werbajh et al., 2000). RelA also interacts with various transcriptional co-repressors, such as histone deacetylase 1 (HDAC1), HDAC2 and HDAC3 (Ashburner et al., 2001; Chen,L. et al., 2001).
The mechanism by which p300/CBP enhances NF-κB transcriptional activity is likely multi-factorial. Both p300 and CBP contain a histone acetyltransferase (HAT) enzymatic activity that regulates gene expression in part through acetylation of the N-terminal tails of histones. Acetylated histones are associated with transcriptionally active segments of chromatin, whereas deacetylated histones accumulate in transcriptionally repressed regions of chromatin (Imhof and Wolffe, 1998; Kuo and Allis, 1998; Sterner and Berger, 2000). In addition to modifying histones, p300/CBP also directly acetylates several transcription factors, including p53, GATA-1, MyoD, TFIIEβ and E2F (Berger, 1999; Bannister and Miska, 2000; Chen,H. et al., 2001). Acetylation of these factors leads to changes in their biological activity, such as alterations in DNA binding affinity, transcriptional activity, interaction with other proteins and intracellular protein stability (Berger, 1999; Bannister and Miska, 2000; Chen,H. et al., 2001).
We have recently described stimulus-coupled acetylation of the RelA subunit of NF-κB in vivo and additionally have shown that overexpression of p300/CBP promotes RelA acetylation in vivo. Our studies further indicate that acetylated RelA is subject to deacetylation by histone deacetylase 3 (HDAC3). This deacetylation reaction enhances IκBα binding and leads in turn to IκBα-dependent nuclear export of the NF-κB complex (Chen,L. et al., 2001).
We now describe our studies aimed at identifying the endogenous acetyltransferase(s) mediating RelA acetylation, the site(s) where such acetylation occurs and the functional consequences of this post-translational modification.
Results
p300 plays a key role in RelA acetylation
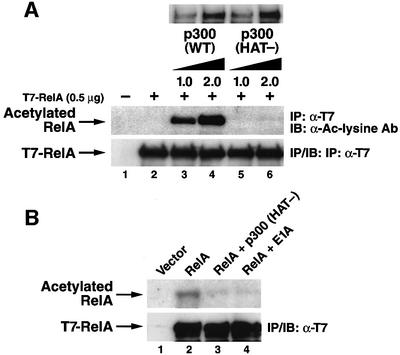
Having shown that RelA is acetylated in vivo in [3H]acetate radiolabeling assays (Chen,L. et al., 2001), we next sought to identify HATs mediating this modification. Overexpression of p300 or CBP effectively induces acetylation of RelA (Chen,L. et al., 2001). To further explore the potential involvement of the p300/CBP in RelA acetylation, we compared the ability of wild-type p300 and a p300 mutant lacking a functional HAT domain to promote RelA acetylation. This p300 (HAT-) mutant contains six point mutations in the HAT domain and lacks HAT activity (Kraus et al., 1999). 293T cells were co-transfected with expression vectors encoding RelA and either wide-type p300 or the p300 (HAT-) mutant. When the level of RelA acetylation was assessed by immunoblotting with anti-acetylated lysine antibodies, RelA was acetylated in a dose-related manner by wild-type p300 (Figure 1A, middle panel, lanes 3 and 4) but not by the p300 (HAT-) mutant (lanes 5 and 6). These results indicate that acetylation of RelA by p300 requires the HAT activity of p300.
Fig. 1. p300 acetylates RelA in vivo. (A) The HAT activity of p300 is required for acetylation of RelA. 293T cells were co-transfected with the indicated amounts of T7-RelA and wild-type p300 or p300 (HAT-) mutant expression vector DNA. Acetylation was detected by immunoblotting of anti-T7 immunoprecipitates with anti-acetylated lysine antibodies (Cell Signaling) (middle panel). Levels of p300, p300 (HAT-) and T7-RelA are shown in the upper and lower panels, respectively. (B) p300 (HAT-) inhibits the acetylation of RelA. COS-7 cells were co-transfected with expression plasmid DNA encoding T7-RelA alone (5 µg) or in combination with p300 (HAT-) (10 µg) or adenovirus E1A (10 µg). Acetylation levels of RelA (upper panel) were assessed by [3H]sodium acetate labeling as described previously (Chen,L. et al., 2001). Levels of T7-RelA expression in each sample are shown in the lower panel.
To determine whether endogenous p300/CBP is involved in RelA acetylation, we examined whether the p300 (HAT-) mutant dominantly interfered with acetylation of RelA by endogenous cellular HATs. When expressed in COS-7 cells, RelA was readily acetylated in vivo as detected by [3H]acetate radiolabeling (Figure 1B, lane 2) in agreement with our previous results (Chen,L. et al., 2001). However, co-expression of the p300 (HAT-) mutant inhibited this acetylation (lane 3), most likely by interfering with the action of endogenous p300. Both wild-type p300 (Supplementary figure 2, available at The EMBO Journal Online) and the p300 (HAT-) mutant (data not shown) physically interacted with RelA at comparable levels. In addition, co-expression of E1A, an adenovirus protein that interacts with the HAT domain of both p300 and CBP and inhibits their acetyltransferase activities (Chakravarti et al., 1999; Hamamori et al., 1999), significantly reduced the levels of acetylation of RelA (lane 4). Together, these results suggest that endogenous p300 plays an important role in the acetylation of RelA observed in vivo. However, these findings do not exclude the possible involvement of other cellular acetyltransferases in this modification.
Acetylation of RelA occurs at multiple sites
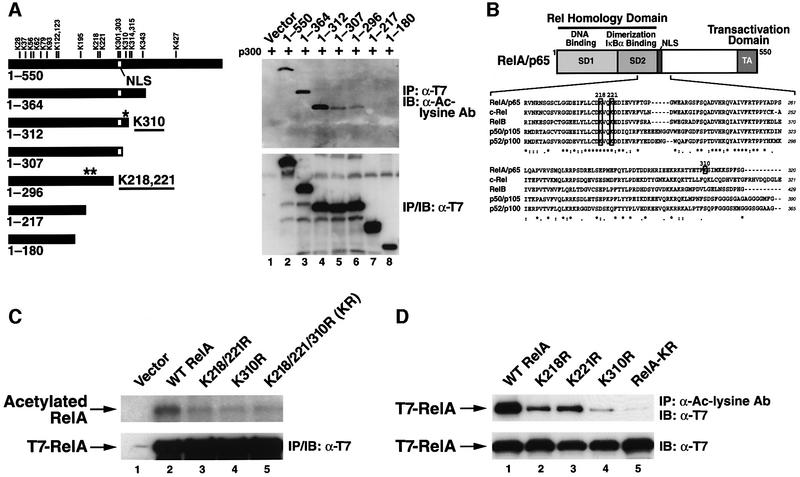
We next investigated the site(s) of acetylation in RelA. Inspection of its coding sequence revealed 18 lysine residues, any of which could form a potential site of acetylation. As a first step to identify the specific lysine residues involved, we generated a series of C-terminal deletion mutants of RelA (Figure 2A, left panel) and tested each in in vivo acetylation assays in 293T cells co-expressing p300 (Figure 2A, right panel). Anti-acetylated lysine antibody immunoblotting revealed that RelA 1–312 was acetylated to the same extent as with full-length RelA, suggesting that the lysine residues located C-terminal of amino acid 312 did not represent major targets for acetylation. However, further truncation to amino acid 307 sharply diminished RelA acetylation (Figure 2A, right panel, lane 5), despite the fact that both RelA 1–312 and RelA 1–307 interacted equivalently with p300 (Supplementary figure 1). Lysine 310 is the only potential acetylation site that is present in RelA 1–312 and is absent in RelA 1–307. Analysis of additional deletion mutants revealed further loss of the acetylation signal when RelA was deleted from amino acids 296 to 217 (Figure 2A, right panel, lanes 6 and 7). Two lysine residues at positions 218 and 221 are removed by this deletion, and one or both could form additional sites of acetylation. Similar results were obtained when CBP was substituted for p300 in the analysis of these deletion mutants (data not shown).
Fig. 2. Lysines 218, 221 and 310 in RelA correspond to major sites of p300-mediated acetylation. (A) Left: schematic depiction of the various C-terminal deletion mutants of RelA tested and the position of each of the 18 lysines that form potential acetylation sites. Asterisks indicate the relative positions of lysines 218, 221 and 310. Right: p300-mediated acetylation of the RelA deletion mutants. 293T cells were co-transfected with expression vector DNA (0.5 µg) encoding either full-length RelA or the indicated deletion mutants with p300 expression plasmid DNA (2 µg). Acetylation levels of each RelA deletion mutant (upper panel) were detected as in Figure 1A. Levels of expression of each of the RelA deletion mutants are shown in the lower panel. All these deletion mutants retain the ability to bind to p300 at comparable levels (see Supplementary figure 1). (B) Schematic depiction of the domain structure of RelA and the protein sequence alignment of RelA (amino acids 198–320), c-Rel, RelB, p50/p105 and p52/p100. Note that lysines 218 and 221 are highly conserved while lysine 310 is uniquely present in RelA. (C) In vivo acetylation assay of K-to-R substitution mutations in the context of full-length RelA. Mutants corresponding to K218/221R, K310R and RelA-K218/221/310R (designated RelA-KR) were transfected into COS-7 cells. Acetylation levels of wild-type RelA and the various substitution mutants were assessed by [3H]sodium acetate labeling as in Figure 1B (upper panel). Levels of expression of wild-type RelA and the various substitution mutants are shown in the lower panel. (D) 293T cells were transfected with the various lysine-to-arginine substitution mutants of RelA (0.5 µg) together with expression vector encoding p300 (2 µg). The level of acetylation of wild-type RelA and the various mutants was tested by immunoprecipitation with an anti-acetylated lysine antibody (Cell Signaling) followed by immunoblotting with anti-T7 antibodies (levels of expression of wild-type RelA and each mutant are shown in the lower panel).
Sequence alignment of all of the mammalian Rel proteins revealed that lysines 218 and 221 are highly conserved, while lysine 310 is uniquely present in RelA (Figure 2B). To confirm that lysines 221, 218 and 310 are acetylated by p300 in the context of the full-length RelA protein, arginine was substituted for these lysines (K to R) both singly and in combination. This strategy conserves the basic charge at the site while precluding acetylation. Combined mutation of all three lysines markedly reduced but did not completely eliminate the acetylation signal measured in [3H]acetate radiolabeling assays (Figure 2C, lane 5). Mutation of lysines 218 and 221 (lane 3) or lysine 310 (lane 4) also decreased acetylation compared with wild-type RelA (lane 2). When each of these RelA mutants was co-expressed with p300 and the lysates were immunoprecipitated with anti-acetylated lysine antibodies followed by immunoblotting with anti-T7 antibodies, the contribution of lysines 218, 221 and 310 to acetylation of the full-length RelA protein was confirmed (Figure 2D). These changes in acetylation were not due to impaired binding of p300, since each of the RelA mutants bound equivalently to p300 (Supplementary figure 2). Together, these findings indicate that lysines 218, 221 and 310 represent major acetylation sites in RelA in vivo. However, our findings do not exclude the involvement of other lysine residues, since a low level of acetylation remained after mutation of all three of these lysine residues (Figure 2C and D, lanes 5).
RelA-KR mutant displays impaired transactivation of RelA and does not functionally cooperate with p300/CBP
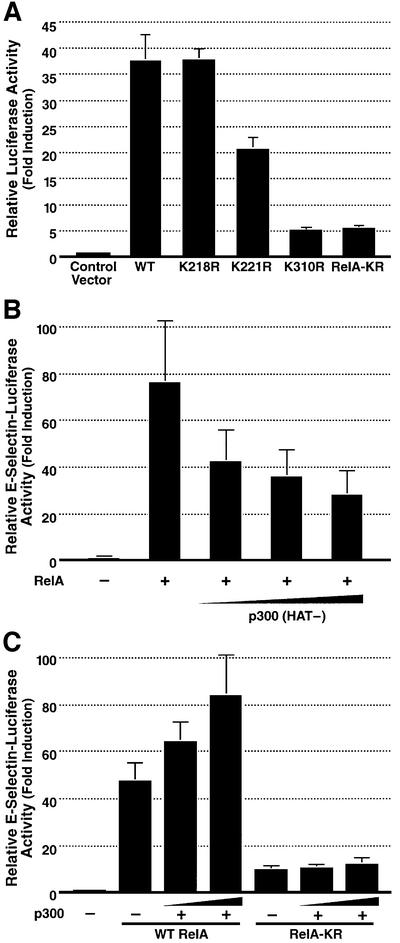
We next tested the functional significance of acetylation at these three sites in RelA using the lysine-to-arginine substitution mutants of RelA. We first examined whether mutation of these lysines altered the transcriptional activity of RelA. 293T cells were co-transfected with either wild-type RelA or the site-specific arginine substitution mutants together with an E-selectin–luciferase reporter plasmid containing a κB enhancer (Madge and Pober, 2000). Transactivation by the RelA K218R mutant proved comparable to that by wild-type RelA (Figure 3A). Conversely, both the K310R and triple KR RelA mutants were greatly impaired in transactivation function, while the K221R mutant exhibited intermediate activity. Together, these findings raise the possibility that acetylation at lysine 310, and to a lesser extent at lysine 221, is required for the full transactivation function of RelA. To further confirm that these findings were the result of diminished acetylation rather than of changes in the primary sequence of RelA introduced by the point mutations, transactivation activity of RelA was tested in the presence of the p300 (HAT-) mutant. This mutant induced dose-related inhibition of RelA-mediated transactivation (Figure 3B). Together, these results suggest that acetylation of RelA plays an important role in regulating RelA transcriptional activity and that modification of K310 is particularly important in this response.
Fig. 3. RelA K221R, K310R and RelA-KR display impaired transcriptional activation properties. (A) 293T cells were transfected with E-selectin–luciferase reporter plasmid DNA (0.1 µg) and the various lysine-to-arginine substitution mutants of RelA (1 ng). Luciferase activity was measured as described previously (Chen,L. et al., 2001). Results represent the average of three independent experiments ± SD. (B) The HAT-deficient mutant of p300 inhibits RelA-mediated transactivation of RelA. 293T cells were transfected with E-selectin–luciferase reporter plasmid DNA (0.1 µg) and expression plasmid DNA encoding RelA (5 ng) alone or with graded amounts of p300 (HAT-) expression plasmid DNA (100 ng, 200 ng and 400 ng). Luciferase activity was measured as in (A). (C) Lack of cooperative transactivation of RelA-KR and p300/CBP. 293T cells were co-transfected with E-selectin–luciferase reporter plasmid DNA (0.1 µg) and either wild-type RelA or RelA-KR expression plasmid DNA (1 ng), alone or in combination with increasing amounts of p300 expression vector DNA (100 ng and 200 ng). Luciferase activity was measured as in (A).
As noted, p300/CBP has been proposed to function as a transcriptional co-activator of RelA (Gerritsen et al., 1997; Sheppard et al., 1999), and this activity depends on its acetyltransferase function (Figure 3B). To further explore whether the decreased transactivation activity of the RelA-KR mutant stems from its hypoacetylation, we examined whether the residual transcription activity of the RelA-KR mutant is enhanced in the presence of co-expressed p300. While the transcriptional activity of wild-type RelA was enhanced in a dose-dependent manner following co-expression of p300 (Figure 3C), the activity of the RelA-KR mutant was not (Figure 3C). Similar findings were obtained when CBP was substituted for p300 (data not shown). These findings suggest that p300/CBP enhances RelA-mediated transcription at least in part by promoting direct acetylation of this NF-κB subunit.
Acetylation of RelA at lysine 221 alters its κB enhancer binding properties
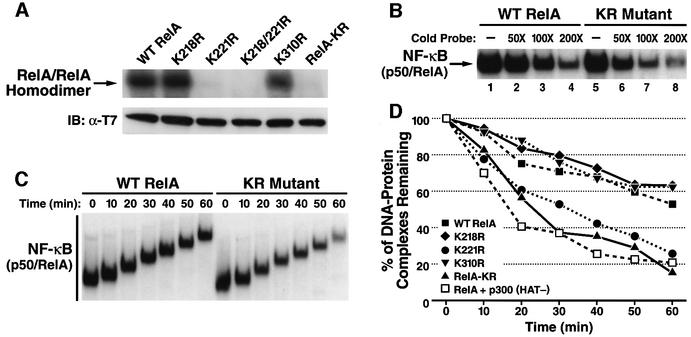
We next investigated whether mutation of these lysines alters the DNA binding activity of RelA. 293T cells were co-transfected with expression vectors encoding wild-type RelA or the various RelA mutants and p300. Whole-cell extracts were isolated, and electrophoretic mobility shift assays (EMSAs) were performed with a 32P-radio labeled κB enhancer oligonucleotide. The RelA K221R and RelA-KR mutants displayed sharply diminished DNA binding activity as homodimers, while the K218R and K310R RelA mutants displayed normal activity (Figure 4A, upper panel). All of the RelA mutants were expressed at levels comparable to those of wild-type RelA in these extracts (Figure 4A, lower panel) and retained the ability to form homodimers with efficiencies indistinguishable from that of wild-type RelA (Supplementary figure 3).
Fig. 4. The hypoacetylated RelA-KR mutant displays lower binding affinity for κB enhancer DNA. (A) Analyses of κB enhancer binding activity of wild-type RelA and the lysine-to-arginine mutants of RelA. Whole-cell extracts from 293T cells transfected with expression vectors encoding p300 and wild-type RelA or the indicated substitution mutants of RelA were prepared. EMSA was performed (upper panel) as described in Materials and methods. The levels of expression of RelA and each RelA mutant are shown in the lower panel (B) NF-κB heterodimers composed of p50 and wild-type RelA or the RelA-KR mutant display different sensitivity to unlabeled κB enhancer competition in EMSA. Whole-cell extracts from 293T cells co-transfected with expression vectors encoding wild-type RelA or the RelA-KR mutant together with p50 and p300 expression plasmids were isolated and tested in EMSA for binding to 32P-radiolabeled κB enhancer probes in the presence of increasing amounts of unlabeled κB enhancer oligonucleotides (cold probe). (C) Analyses of the off-rate of RelA binding to the κB enhancer. The off-rate of binding of wild-type RelA and RelA-KR to the κB enhancer was tested by EMSA as described in the Materials and methods. The results for wild-type RelA (left panel) and the RelA-KR mutant (right panel) are shown. (D) Measurement of the off-rate of binding of wild-type RelA in the presence of p300 (HAT-) mutant and wild-type RelA and the collection of lysine-to-arginine substitution mutants in the presence of p300 and p50. Binding to 32P-radiolabeled κB enhancer was measured as in (B) followed by assessment of the EMSA results by radiodensitometry. The results presented are an average of two independent experiments.
We next examined the DNA binding activity of these RelA mutants in the presence of p300 when assembled with p50, thus forming the prototypical NF-κB complex. In contrast with the marked defect in DNA binding activity displayed by the RelA-KR mutant when tested as a homodimer, complexes of p50/RelA-KR exhibited levels of steady-state DNA binding similar to that found with p50/wild-type RelA (Figure 4B, lanes 1 and 5). However, when increasing amounts of unlabeled κB enhancer oligonucleotide were added to these reaction mixes, p50/RelA-KR complexes competed more effectively than the p50/wild-type RelA complexes (compare lanes 2–4 with lanes 6–8). These results raised the possibility that acetylation of RelA might diminish the overall affinity of these NF-κB complexes for the κB enhancer. To further assess this possibility, we measured the kinetics of dissociation of these complexes from the κB enhancer (Figure 4C). As shown in Figure 4D, p50/RelA-KR complexes dissociated more rapidly than p50/wild-type RelA complexes. Quantification of these results revealed that p50/RelA displayed a T1/2 for dissociation of ∼64 min, while NF-κB complexes containing the RelA-KR mutant exhibited a T1/2 of ∼22 min (Figure 4D). Analysis of the individual point mutants of RelA revealed that the K221R mutant displayed accelerated kinetics of dissociation like the KR mutant. Conversely, the RelA K218R and K310R mutants exhibited DNA binding properties indistinguishable from those of wild-type RelA (Figure 4D). Further supporting a role for acetylation in regulating the DNA binding properties of RelA, overexpression of the p300 (HAT-) mutant produced a more rapid off-rate of binding of NF-κB to the κB enhancer (Figure 4D). Overexpression of the p300 (HAT-) mutant also inhibited RelA DNA binding activity as a homodimer (data not shown). Using biotinylated forms of the κB enhancer, we further found that acetylated forms of RelA do directly engage this enhancer (Supplementary figure 4). Together, these results suggest that the acetylation of RelA at lysine 221 enhances the binding affinity of the NF-κB complex for the κB enhancer.
Acetylation of lysine 221 and possibly lysine 218 in RelA regulates assembly with IκBα
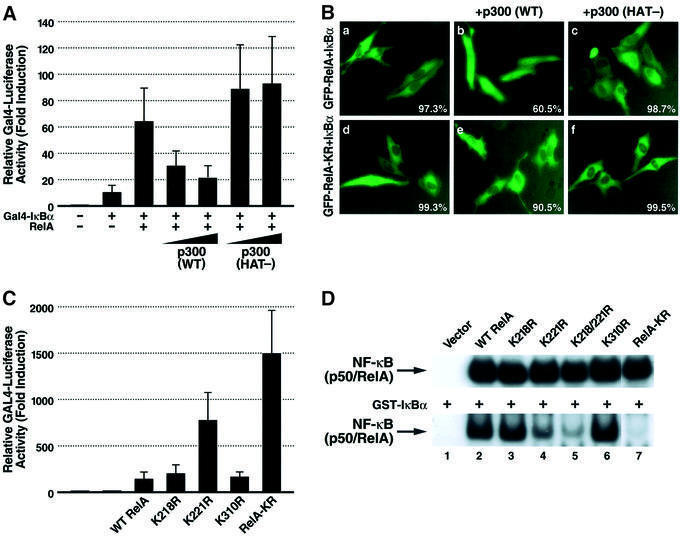
Our previous studies indicated that deacetylation of RelA by HDAC3 promotes its assembly with IκBα. In contrast, acetylated RelA binds weakly, if at all, to IκBα (Chen,L. et al., 2001). To further explore whether acetylation regulated this interaction, we employed a mammalian one-hybrid system to examine the IκBα binding properties of RelA in the presence of either wild-type p300 or the p300 (HAT-) mutant. For these assays, full-length IκBα (pFA-IκBα) was expressed in 293T cells as a GAL-4 fusion protein in the presence of wild-type RelA. Effective binding of RelA to IκBα leads to transcriptional activation of a co-transfected GAL-4 responsive reporter (Figure 5A). Analysis of the interplay of RelA and IκBα in this system revealed that co-expression of p300 produced a dose-dependent decrease in luciferase activity. Conversely, co-expression of the p300 (HAT-) mutant did not diminish the luciferase activity. These findings are consistent with the notion that acetylation of RelA blocks its interaction with IκBα.
Fig. 5. Acetylation of lysine 221 in RelA plays a key role in regulating assembly with IκBα. (A) p300 inhibits the interaction of RelA with IκBα in a mammalian one-hybrid assay. Expression plasmids encoding wild-type RelA (0.1 µg) were co-transfected into 293T cells with plasmids encoding the GAL4 DNA binding domain fused to IκBα (pFA-IκBα) (1 ng) together with a GAL4 enhancer–luciferase reporter (0.1 µg) (pFR-luc) (Stratagene) in the presence of increasing amounts of either p300 or p300 (HAT-) mutant expression plasmids (100 ng and 200 ng, respectively). Luciferase activity was measured as in Figure 3A. (B) p300 prevents the cytoplasmic sequestration of wild-type RelA but not RelA-KR induced by co-expression of IκBα. HeLa cells were co-transfected with expression vectors encoding GFP–RelA (0.2 µg) or GFP–RelA-KR and IκBα (50 ng) together with wild-type p300 expression vector (1 µg). Note that in the presence of p300, the majority of GFP–RelA localizes in the nucleus while GFP–RelA-KR localizes predominantly in the cytoplasm. The percentage of cells displaying the depicted phenotype derived from inspection of at least 500 cells present in multiple fields in two independent experiments is presented in the bottom right corner of each panel. (C) The RelA-KR mutant displays a stronger interaction with IκBα in vivo. Interaction of IκBα with wild-type RelA or the lysine-to-arginine substitution mutants was tested in the mammalian one-hybrid assay system as in (A). (D) Whole-cell extracts from 293T cells co-transfected with expression vectors encoding wild-type RelA or the various RelA substitution mutants as indicated together with expression vectors encoding p50 and p300 were prepared and tested in EMSA. Binding of the nuclear complexes to DNA in the presence (lower panel) or absence (upper panel) of added recombinant of GST–IκBα (50 ng) is shown.
Studies were next performed to examine the effects of co-expression of wild-type p300 or p300 (HAT-) proteins on the subcellular localization of GFP–RelA in the presence of IκBα. When IκBα was co-transfected with GFP–RelA, GFP–RelA was chiefly detected in the cytoplasm (Figure 5B, panel a). However, IκBα failed to produce such cytoplasmic localization of GFP–RelA when wild-type p300 was co-expressed (panel b). In contrast, in the presence of the p300 (HAT-) mutant, the cytoplasmic pattern of GFP–RelA expression in the presence of IκBα was maintained (panel c). Of note, co-expression of p300 did not alter the cytoplasmic pattern of GFP–RelA-KR expression observed in the presence of IκBα (panel e). Together, these findings are consistent with a role for p300-mediated acetylation of RelA not only in preventing its assembly with IκBα but also in promoting its nuclear expression.
We next examined the IκBα binding properties of the various RelA mutants using the mammalian one-hybrid system described in Figure 5A. The K218R and K310R mutants of RelA only modestly activated the GAL4 reporter at levels similar to wild-type RelA, indicating relatively similar degrees of interaction of these RelA mutants with the GAL4–IκBα fusion protein. In contrast, the K221R mutant of RelA and the composite RelA-KR mutant displayed markedly greater activation of the GAL4 response reporter (Figure 5C). These results suggest that acetylation of lysine 221 most likely plays a key role in impairing the assembly of RelA with IκBα.
To further test this hypothesis, we performed EMSA with whole-cell extracts of 293T cells transfected with expression vectors encoding wild-type RelA or mutant forms of RelA, p50 and p300. Consistent with the results presented earlier in Figure 4C, wild-type RelA and each of the lysine mutants of RelA displayed similar steady-state levels of binding to the κB enhancer when tested in the presence of p50 (Figure 5D). However, the addition of recombinant GST–IκBα to the EMSA reaction mixtures produced markedly diminished DNA binding with the RelA K221R mutant compared with either wide-type RelA or the RelA K218R or K310R mutants (Figure 5D). Dual arginine substitution of lysine 218 and 221 or all three sites (RelA-KR) led to even greater levels of inhibition of DNA binding in the presence of GST–IκBα. Together, these findings support the notion that acetylation of RelA at lysine 221, alone or in combination with lysine 218, impairs assembly with IκBα.
The RelA-KR and K221R mutants predominantly localize in the cytoplasm
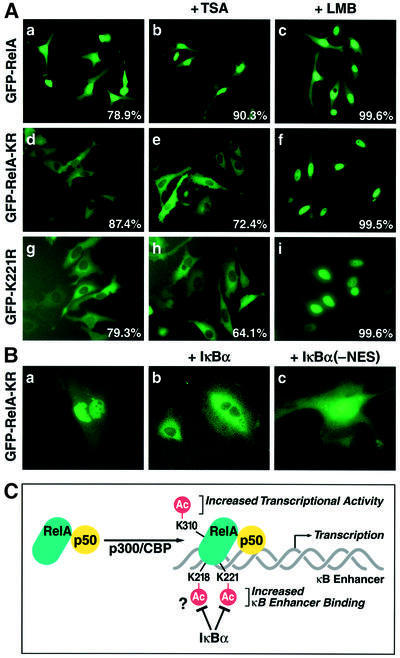
We have reported previously that HDAC3-mediated deacetylation of RelA promotes RelA export from the nucleus to the cytoplasm and that this export is dependent on IκBα (Chen,L. et al., 2001). A prediction of this model is that the RelA-KR and RelA-K221R mutants, as a consequence of their hypoacetylation and enhanced IκBα binding activities, would predominantly reside in the cytoplasm. GFP fusion proteins containing wild-type RelA, RelA-KR or RelA-K221R were expressed in HeLa cells. When studied by fluorescence microscopy, GFP–wild-type RelA and the GFP–RelA-K218R and GFP–RelA-K310R (data not shown) proteins were predominantly localized in the nucleus (Figure 6A, panel a). In contrast, both the GFP–RelA-KR and GFP–RelA- K221R mutants were principally localized in the cytoplasm (panels d and g). Both of these mutants remained in the cytoplasm in the presence of the HDAC inhibitor, trichostatin A (TSA) (panels e and h). However, both the GFP–RelA-KR and GFP–RelA-K221R mutants were apparently subject to limited nucleocytoplasmic shuttling since addition of leptomycin B, a specific inhibitor of CRM-1-dependent nuclear export, promoted nuclear accumulation of both of these fusion proteins (panels f and c).
Fig. 6. GFP–RelA-KR and K221R mutants are principally localized in the cytoplasm of expressing cells. (A) HeLa cells were transfected with GFP–RelA (2 µg) (panels g, h and i), GFP–RelA-KR (2 µg) (panels d, e and f) or GFP–RleA-K221R (panels g, h and i) expression plasmid DNA. Selected cultures of these cells were treated with TSA (400 nM, 5 h) (panels b, e and h) or with leptomycin B (20 nM, 2 h) (panels c, f and i). (B) GFP–RelA-KR is principally expressed in the nucleus of IκBα–/– MEFs. MEFs derived from IκBα–/– mice were transfected with GFP–RelA-KR (1 µg) (panel a) expression vector DNA. The IκBα–/– MEFs were also reconstituted with expression vectors encoding either wild-type IκBα (panel b) or an IκBα NES-deficient mutant (panel c). (C) A model summarizing how acetylation of different lysine residues regulates distinct functions of RelA. Acetylation of lysine 221 increases DNA binding affinity for the κB enhancer and prevents the association of RelA with IκBα; acetylation of lysine 310 likely controls the association of an unknown factor that is required for full transactivation by RelA.
We suspected that the difference between the subcellular patterns of localization obtained with wild-type RelA and the RelA-KR mutant was due to the fact that RelA-KR forms an excellent substrate for IκBα binding and this interaction leads to rapid export of the RelA KR mutant from the nucleus into the cytoplasm. To test this hypothesis, we expressed GFP–RelA-KR in murine embryo fibroblasts (MEFs) isolated from IκBα–/– mice. As predicted, the GFP–RelA-KR fusion protein was predominantly localized in the nucleus of these IκBα–/– cells (Figure 6B, panel a). When these MEFs were reconstituted with wild-type IκBα by cDNA transfection, GFP–RelA- KR was detected in the cytoplasm (panel b). However, reconstitution with an IκBα mutant containing a mutation in its nuclear export signal (NES) (Johnson et al., 1999) produced a nuclear predominant pattern of expression (panel c). Taken together, these findings are consistent with the hypothesis that the lack of the acetylation of RelA-KR mutant, specifically at lysine 221, leads to enhanced interaction with IκBα and nuclear export of the hypoacetylated RelA in a manner that is principally dependent on the NES present in IκBα.
Discussion
In this study, we demonstrate that p300/CBP participates in the acetylation of RelA in vivo. We further identify lysines 218, 221 and 310 as major sites of acetylation within RelA. Finally, we show that acetylation of these different lysines regulates different biological properties of RelA. Specifically, acetylation of lysine 221 both enhances DNA binding and impairs IκBα assembly, while acetylation of lysine 310 is required for full transcriptional activity of NF-κB in the absence of effects on DNA and IκBα binding.
It is now well recognized that p300/CBP and other HATs acetylate a variety of non-histone substrates both in vitro and in vivo (Gu and Roeder, 1997; Boyes et al., 1998; Martínez-Balbás et al., 2000; Marzio et al., 2000). We find that p300 acetylates RelA in vivo and that the HAT activity of p300 is essential for this modification. CBP can also mediate the acetylation of RelA in vivo, in agreement with the prior finding that CBP cooperatively stimulates RelA-mediated transcription (Gerritsen et al., 1997; Sheppard et al., 1999; Vanden Berghe et al., 1999). However, our data do not exclude the involvement of other acetyltransferases in the acetylation of RelA in vivo. In addition to p300/CBP, NF-κB interacts with members of the p160 nuclear receptor co-activator family, which can function as co-activators of NF-κB-induced gene expression (Na et al., 1998; Sheppard et al., 1998; De Bosscher et al., 2000; Werbajh et al., 2000). However, the mechanism by which these co-activators cooperate with RelA remains unclear. One possibility is that they, like p300/CBP, may directly acetylate RelA, since many of these proteins contain HAT activity (Leo and Chen, 2000; Sterner and Berger, 2000; Werbajh et al., 2000). It is also possible that their effect on RelA depends on a preceding action of p300/CBP. Most of these co-activators are in fact found in complexes with p300/CBP (Leo and Chen, 2000; Sterner and Berger, 2000). Alternatively, these enzymes may act in a more indirect manner, perhaps by modifying histones and altering chromatin structure.
Acetylation alters the transcriptional activity of many transcription factors. For example, acetylation of E2F by PCAF enhances its transactivation potential (Martínez-Balbás et al., 2000). Substitution of arginine for lysine 310 in RelA markedly impairs its transcriptional activity. Conversely, substitution to a glutamine residue at this site, which may approximate the physical changes associated with acetylation, enhances the transcriptional activity of RelA (data not shown). Lysine 310 is located immediately C-terminal of the RHD, and mutation of this site affects neither the DNA binding properties of RelA nor its assembly with IκBα (Figure 4C). These results raise the question of how acetylation at this site promotes increased transcriptional activity of RelA. Acetylated lysine 310 might form a platform for the binding of a bromodomain-containing protein (Dhalluin et al., 1999; Polesskaya et al., 2001) that is required for the full transcriptional activity of RelA. However, the identity of the putative factor binding to acetylated lysine 310 remains unknown. This factor does not appear to be p300 since wild-type RelA and RelA K310R bind similar amounts of p300 in co-immunoprecipitation assays (Supplementary figure 2).
Acetylation of transcription factors may also alter their intrinsic DNA binding properties. Acetylation of p53 or GATA-1 enhances their affinity for DNA (Gu and Roeder, 1997; Boyes et al., 1998) while acetylation of CDP/cut or HMGI (Y) impairs their DNA binding (Munshi et al., 1998; Li et al., 2000). Arginine substitution of lysine 221, but not lysine 218 or 310, sharply impairs the DNA binding activity of RelA homodimers (Figure 4A), and when tested as a heterodimer with p50 the RelA-K221R mutant leads to a 3-fold faster rate of dissociation from the κB enhancer (T1/2, 22 min versus 64 min). The kinetics of NF-κB dissociation from the κB enhancer also increases in the absence of p300 when wild-type RelA or the RelA-K218R or K310R mutant is tested. However, the absence of p300 does not accelerate the already rapid kinetics of dissociation observed with NF-κB complexes formed with RelA-KR or RelA-K221R. The further finding that co-expression of a dominantly interfering HAT mutant of p300 recapitulates these more rapid kinetics of dissociation observed with wild-type p50/RelA complexes (Figure 4D) supports the conclusion that acetylation at lysine 221, rather than a change in amino acid sequence at this position, underlies the alteration in DNA binding.
Analysis of the crystal structure of the RHDs of RelA and p50 complexed with DNA indicates that lysine 221 forms a direct contact with the DNA backbone (Chen et al., 1998). Thus, acetylation of lysine 221 might be expected to impair DNA binding, since this modification neutralizes the positive charge on the lysine residue. However, our observation suggests that acetylation of lysine 221 in fact enhances the affinity of RelA for DNA. It is possible that acetylation of lysine 221 produces this effect by causing a conformational change within the protein.
Acetylation also regulates protein–protein interactions. For example, acetylation of MyoD enhances its interaction with p300/CBP (Polesskaya et al., 2001), while acetylation of Drosophila T cell factor by Drosophila CBP in vitro decreases its affinity for β-catenin/Armadillo (Waltzer and Bienz, 1998). Arginine substitution of lysine 221 alone or in combination with lysine 218 strengthens the interaction of RelA with IκBα. Acetylation at this site may thus impair the interaction of RelA with IκBα. We have shown previously that p300-mediated acetylation of RelA reduces IκBα binding, while HDAC3-mediated deacetylation of RelA promotes IκBα assembly (Chen,L. et al., 2001). Our studies now implicate the acetylation of lysine 221, alone or in combination with lysine 218, as a key modification that controls RelA assembly with IκBα. Lysine 221 of RelA directly interacts with methionine 279 located within the sixth ankyrin repeat of IκBα. Thus, it is not surprising that acetylation of this residue impairs the interaction of RelA with IκBα. The ability of the lysine 218 mutant to enhance the effect of the lysine 221 mutant, in the context of the double mutation (Figure 5B, lower panel, lane 5), may reflect involvement of the IκBα PEST sequence in RelA binding. The PEST sequence of IκBα is targeted for phosphorylation in vivo, and this phosphorylation is required for IκBα-mediated inhibition of DNA binding (Ernst et al., 1995; Chu et al., 1996; Lin et al., 1996; Schwarz et al., 1996). Therefore, if the phosphorylated PEST sequence interacts with lysine 218 of RelA, then acetylation of lysine 218 may interfere with this interaction. In contrast, acetylation of lysine 310 affects neither DNA binding activity nor IκBα assembly. This finding is consistent with the fact that residues immediately flanking lysine 310 interact with the cap region of IκBα and thus make little or no contribution to the assembly of RelA with IκBα (Huxford et al., 1998; Jacobs and Harrison, 1998).
Mutation of lysine 221 causes both an increase in the off-rate of DNA binding (Figure 4C and D) and an increase in RelA interaction with IκBα. The increased off-rate we observed in the DNA binding experiments could reflect the greater ability of the RelA K221R mutant to interact with endogenous IκBα present in the cells, thereby promoting faster dissociation from the κB enhancer. However, because RelA K221R continues to display poor κB enhancer binding activity in cellular extracts treated with deoxycholate (data not shown), which promotes IκBα dissociation (Baeuerle and Baltimore, 1988), it seems likely that the K221R mutant is intrinsically impaired for DNA binding activity independently of its enhanced ability to bind IκBα. Of note, the compromised DNA binding of RelA K221R is not due to any impairment in its ability to homodimerize.
Acetylation of lysine residues within the nuclear localization signals of CIIAT and HNF-4 leads to an increased nuclear accumulation of both factors (Spilianakis et al., 2000; Soutoglou et al., 2001), likely reflecting impaired nuclear export. We now show that acetylation at lysine 221 plays a major role in determining the subcellular localization of RelA chiefly by preventing its assembly with IκBα. While cytoplasmic in normal cells, the RelA-KR protein expressed in IκBα–/– MEF cells displays a predominantly nuclear pattern of expression. Cytoplasmic localization of the RelA-KR protein is restored in these cells by transfection of wild-type IκBα expression vector DNA. In contrast, introduction of a nuclear export-defective mutant of IκBα fails to alter the nuclear pattern of RelA-KR expression occurring in these IκBα–/– MEF cells. These findings underscore the key role played by IκBα in promoting nuclear export of hypoacetylated forms of RelA. As noted above, IκBα assembly with RelA is enhanced by deacetylation of lysine 221. Thus, deacetylation of this residue in RelA functions as an intranuclear molecular switch that serves to terminate the NF-κB transcriptional response by promoting IκBα assembly and nuclear export of the NF-κB complex.
In summary, our studies demonstrate that acetylation of RelA at distinct sites differentially regulates various biological functions of NF-κB (Figure 6C). Acetylation of lysine 310 of RelA is required for full transactivation by the NF-κB complex, most likely by recruiting an unidentified cofactor. Acetylation of lysine 221 enhances RelA binding to the κB enhancer, while acetylation of lysine 221 alone or in combination with lysine 218 impairs the assembly of RelA with IκBα. Lysines 218 and 221 are highly conserved within all Rel family members, including Dorsal from Drosophila. The possibility that these evolutionarily conserved lysine residues are targets for reversible acetylation and contribute to the regulation of the biological functions of other Rel factors remains an intriguing possibility.
Materials and methods
Plasmid constructs
pcDNA3.1-p300 and pcDNA3.1-p300 (HAT-) were prepared by sub-cloning p300 and p300AT2Mut fragments from pBS-p300 and pBS-p300AT2Mut (Kraus et al., 1999) into HindIII and NotI sites of the pcDNA3.1 vector (Invitrogen). Various T7-tagged C-terminal deletion mutants of RelA were prepared by PCR and subcloned into the BamHI and XbaI sites of a modified pET3a vector. Lysine-to-arginine substitution mutants of RelA were generated by site-directed mutagenesis (Stratagene). Different GFP RelA mutants were prepared by subcloning fragments of these mutants from their T7-tagged expression vectors into BamHI and XbaI sites of the eGFP–C1 vector (Clontech). pFA-IκBα was produced by subcloning a PCR product of IκBα into BamHI and XbaI sites of pFA-CMV vector (Stratagene). A pCMX-IκBα NES-deficient mutant was prepared by deletion of coding sequence for GFP from an eGFP–IκBα double–NES mutant (I52A/L54A/L272A and L274A) (Johnson et al., 1999). Point mutations and sequences of all plasmids prepared by PCR were confirmed by DNA sequencing.
Immunoprecipitation and immunoblotting analyses
Immunoprecipitation and immunoblotting analyses were performed as described previously (Chen,L. et al., 2001).
Whole-cell extract preparation
293T cells were transfected with wild-type or mutant RelA (0.5 µg) together with p50 (0.5 µg) and p300 (2 µg) expression plasmid DNA. After 24–36 h, the cells were harvested and incubated in a buffer (20 mM HEPES pH 7.9, 0.4 M NaCl, 25% glycerol, 1 mM EDTA, 2.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail) for 30 min at 4°C. The cells were then freeze–thawed. After centrifugation at 4°C, the supernatant was isolated and used as the whole-cell extract.
EMSAs
The EMSAs were performed as described previously (Chen,L. et al., 2001). In the competition experiments, a 50-, 100- or 200-fold excess of unlabeled κB enhancer of oligonucleotide (cold probe) was added to each reaction. For the off-rate measurements, a 200-fold excess of cold probe was added 15 min after reaction of the extract with the 32P-radiolabeled κB probe. Aliquots of the reaction mixture were removed at 10 min intervals up to 60 min. The reactions were stopped by direct loading of the samples onto a running non-denaturing gel. The intensity of each band was measured with Scion Image 1.62. For the IκBα binding experiments, 50 ng of GST–IκBα was added to each reaction.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs T.Hope (University of Illinois), W.L.Kraus (Cornell University), J.Pober (Yale University) and J.F.Klement (Jefferson Medical College) for the gift of reagents; T.Huxford and G.Ghosh (University of California at San Diego) for assistance in RelA/IκBα structural modeling; J.Carroll and J.Hull for assistance in the preparation of the figures; and R.Givens for assistance in the preparation of the manuscript. This work was supported in part by funds provided by the J.David Gladstone Institutes, Pfizer, Inc., and benefited from core facilities provided by the University of California, San Francisco– Gladstone Institute of Virology and Immunology Center for AIDS Research (NIH P30 MH59037).
References
- Arenzana-Seisdedos F., Thompson,J., Rodriguez,M.S., Bachelerie,F., Thomas,D. and Hay,R.T. (1995) Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol. Cell. Biol., 15, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenzana-Seisdedos F., Turpin,P., Rodriguez,M., Thomas,D., Hay,R.T., Virelizier,J.L. and Dargemont,C. (1997) Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J. Cell Sci., 110, 369–378. [DOI] [PubMed] [Google Scholar]
- Ashburner B.P., Westerheide,S.D. and Baldwin,A.S.,Jr (2001) The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol., 21, 7065–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P.A. (1998) IκB–NF-κB structures: at the interface of inflammation control. Cell, 95, 729–731. [DOI] [PubMed] [Google Scholar]
- Baeuerle P.A. and Baltimore,D. (1988) Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-κB transcription factor. Cell, 53, 211–217. [DOI] [PubMed] [Google Scholar]
- Baldwin A.S. Jr, (1996) The NF-κ B and IκB proteins: new discoveries and insights. Annu. Rev. Immunol., 14, 649–683. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Miska,E.A. (2000) Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci., 57, 1184–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg A.A., Finco,T.S., Nantermet,P.V. and Baldwin,A.S.,Jr (1993) Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: a mechanism for NF-κB activation. Mol. Cell. Biol., 13, 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L. (1999) Gene activation by histone and factor acetyltrans ferases. Curr. Opin. Cell Biol., 11, 336–341. [DOI] [PubMed] [Google Scholar]
- Blair W.S., Bogerd,H.P., Madore,S.J. and Cullen,B.R. (1994) Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol. Cell. Biol., 14, 7226–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- Brown K., Park,S., Kanno,T., Franzoso,G. and Siebenlist,U. (1993) Mutual regulation of the transcriptional activator NF-κ B and its inhibitor, IκBα. Proc. Natl Acad. Sci. USA, 90, 2532–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D., Ogryzko,V., Kao,H.Y., Nash,A., Chen,H., Nakatani,Y. and Evans,R.M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chen F.E., Huang,D.B., Chen,Y.Q. and Ghosh,G. (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature, 391, 410–413. [DOI] [PubMed] [Google Scholar]
- Chen H., Tini,M. and Evans,R.M. (2001) HATs on and beyond chromatin. Curr. Opin. Cell Biol., 13, 218–224. [DOI] [PubMed] [Google Scholar]
- Chen L., Fischle,W., Verdin,E. and Greene,W.C. (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science, 293, 1653–1657. [DOI] [PubMed] [Google Scholar]
- Chu Z.L., McKinsey,T.A., Liu,L., Qi,X. and Ballard,D.W. (1996) Basal phosphorylation of the PEST domain in the IκBβ regulates its functional interaction with the c-rel proto-oncogene product. Mol. Cell. Biol., 16, 5974–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bosscher K., Vanden Berghe,W., Vermeulen,L., Plaisance,S., Boone,E. and Haegeman,G. (2000) Glucocorticoids repress NF-κB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc. Natl Acad. Sci. USA, 97, 3919–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Ernst M.K., Dunn,L.L. and Rice,N.R. (1995) The PEST-like sequence of IκBα is responsible for inhibition of DNA binding but not for cytoplasmic retention of c-Rel or RelA homodimers. Mol. Cell. Biol., 15, 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsen M.E., Williams,A.J., Neish,A.S., Moore,S., Shi,Y. and Collins,T. (1997) CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl Acad. Sci. USA, 94, 2927–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., May,M.J. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Hamamori Y., Sartorelli,V., Ogryzko,V., Puri,P.L., Wu,H.Y., Wang,J.Y., Nakatani,Y. and Kedes,L. (1999) Regulation of histone acetyltrans ferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell, 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Huxford T., Huang,D.B., Malek,S. and Ghosh,G. (1998) The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell, 95, 759–770. [DOI] [PubMed] [Google Scholar]
- Imhof A. and Wolffe,A.P. (1998) Transcription: gene control by targeted histone acetylation. Curr. Biol., 8, R422–R424. [DOI] [PubMed] [Google Scholar]
- Jacobs M.D. and Harrison,S.C. (1998) Structure of an IκBα/NF-κB complex. Cell, 95, 749–758. [DOI] [PubMed] [Google Scholar]
- Johnson C., Van Antwerp,D. and Hope,T.J. (1999) An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J., 18, 6682–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. (1999) How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene, 18, 6867–6874. [DOI] [PubMed] [Google Scholar]
- Kraus W.L., Manning,E.T. and Kadonaga,J.T. (1999) Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol., 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Leo C. and Chen,J.D. (2000) The SRC family of nuclear receptor coactivators. Gene, 245, 1–11. [DOI] [PubMed] [Google Scholar]
- Li S., Aufiero,B., Schiltz,R.L. and Walsh,M.J. (2000) Regulation of the homeodomain CCAAT displacement/cut protein function by histone acetyltransferases p300/CREB-binding protein (CBP)-associated factor and CBP. Proc. Natl Acad. Sci. USA, 97, 7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Beauparlant,P., Makris,C., Meloche,S. and Hiscott,J. (1996) Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol. Cell. Biol., 16, 1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madge L.A. and Pober,J.S. (2000) A phosphatidylinositol 3-kinase/Akt pathway, activated by tumor necrosis factor or interleukin-1, inhibits apoptosis but does not activate NF-κB in human endothelial cells. J. Biol. Chem., 275, 15458–15465. [DOI] [PubMed] [Google Scholar]
- Martínez-Balbás M.A., Bauer,U.M., Nielsen,S.J., Brehm,A. and Kouzarides,T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G., Wagener,C., Gutierrez,M.I., Cartwright,P., Helin,K. and Giacca,M. (2000) E2F family members are differentially regulated by reversible acetylation. J. Biol. Chem., 275, 10887–10892. [DOI] [PubMed] [Google Scholar]
- Munshi N., Merika,M., Yie,J., Senger,K., Chen,G. and Thanos,D. (1998) Acetylation of HMG I(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol. Cell, 2, 457–467. [DOI] [PubMed] [Google Scholar]
- Na S.Y., Lee,S.K., Han,S.J., Choi,H.S., Im,S.Y. and Lee,J.W. (1998) Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor κB-mediated transactivations. J. Biol. Chem., 273, 10831–10834. [DOI] [PubMed] [Google Scholar]
- Perkins N.D., Felzien,L.K., Betts,J.C., Leung,K., Beach,D.H. and Nabel,G.J. (1997) Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science, 275, 523–527. [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Naguibneva,I., Duquet,A., Bengal,E., Robin,P. and Harel-Bellan,A. (2001) Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol., 21, 5312–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M.L., Stelzer,G., Altmann,H., Meisterernst,M. and Baeuerle,P.A. (1995) Interaction of the COOH-terminal transactivation domain of p65 NF-κB with TATA-binding protein, transcription factor IIB, and coactivators. J. Biol. Chem., 270, 7219–7226. [DOI] [PubMed] [Google Scholar]
- Schwarz E.M., Van Antwerp,D. and Verma,I.M. (1996) Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IκBα. Mol. Cell. Biol., 16, 3554–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard K.A., Phelps,K.M., Williams,A.J., Thanos,D., Glass,C.K., Rosenfeld,M.G., Gerritsen,M.E. and Collins,T. (1998) Nuclear integra tion of glucocorticoid receptor and nuclear factor-κB signaling by CREB-binding protein and steroid receptor coactivator-1. J. Biol. Chem., 273, 29291–29294. [DOI] [PubMed] [Google Scholar]
- Sheppard K.A. et al. (1999) Transcriptional activation by NF-κB requires multiple coactivators. Mol. Cell. Biol., 19, 6367–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E., Viollet,B., Vaxillaire,M., Yaniv,M., Pontoglio,M. and Talianidis,I. (2001) Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J., 20, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis C., Papamatheakis,J. and Kretsovali,A. (2000) Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol., 20, 8489–8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.C., Ganchi,P.A., Ballard,D.W. and Greene,W.C. (1993) NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science, 259, 1912–1915. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W., De Bosscher,K., Boone,E., Plaisance,S. and Haegeman,G. (1999) The nuclear factor-κB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J. Biol. Chem., 274, 32091–32098. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Werbajh S., Nojek,I., Lanz,R. and Costas,M.A. (2000) RAC-3 is a NF-κB coactivator. FEBS Lett., 485, 195–199. [DOI] [PubMed] [Google Scholar]
- Xu X., Prorock,C., Ishikawa,H., Maldonado,E., Ito,Y. and Gelinas,C. (1993) Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol. Cell. Biol., 13, 6733–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]