Abstract
Human immunodeficiency virus type 1 envelope glycoprotein gp120 interacts with CD4 and the CCR5 coreceptor in order to mediate viral entry. A CD4-induced surface on gp120, primarily composed of residues in the V3 loop and the C4 domain, interacts with CCR5. In the present study, we generated envelope glycoproteins comprising chimeric V3 loops and/or V3 loops with deletions and studied their binding to CCR5 amino-terminal domain (Nt)-based sulfopeptides and cell surface CCR5, as well as their ability to mediate viral entry. We thus delineated two functionally distinct domains of the V3 loop, the V3 stem and the V3 crown. The V3 stem alone mediates soluble gp120 binding to the CCR5 Nt. In contrast, both the V3 stem and crown are required for soluble gp120 binding to cell surface CCR5. Within the context of a virion, however, the V3 crown alone determines coreceptor usage. Our data support a two-site gp120-CCR5 binding model wherein the V3 crown and stem interact with distinct regions of CCR5 in order to mediate viral entry.
Entry of human immunodeficiency virus type 1 (HIV-1) R5 isolates into target cells is mediated by the successive interaction of the envelope glycoprotein gp120 with CD4 and the CCR5 coreceptor (for a review see reference 1). A cluster of negatively charged and sulfotyrosine residues within the CCR5 amino-terminal domain (Nt) are essential for CCR5-mediated fusion and entry of most HIV-1 R5 isolates (for a review see reference 10). Usage of other residues in the CCR5 Nt occurs in an isolate-dependent manner (10). Recently, it has been shown that HIV-1JRCSF can adapt to use CCR5 lacking the Nt (22). The mutant HIV-1JRCSF isolate has an increased affinity for the second extracellular loop (ECL2) of CCR5. Studies with anti-CCR5 monoclonal antibodies (MAbs) and low-molecular-weight inhibitors indicate that ECL2 epitopes are important for gp120 binding to CCR5 and for viral entry (7, 10). We have recently identified a cluster of ECL2 residues that are essential for the entry of non-clade B isolates (28). Residues dispersed throughout ECL1 and ECL3 of CCR5 have also been implicated in HIV-1 entry into target cells (10).
The binding of gp120 to the CD4 receptor creates and/or exposes a coreceptor binding site characterized by a hydrophobic core surrounded by a positively charged periphery (for a review see reference 23; 18). The coreceptor binding sites on envelope glycoproteins derived from R5 and X4 isolates are principally composed of conserved residues that are organized into nearly identical tertiary structures (17). Nonetheless, there is exquisite specificity for CCR5 and CXCR4 usage by R5 and X4 isolates, respectively. Soluble gp120-CD4 complexes wherein gp120 is derived from an R5 isolate but not from an X4 isolate bind specifically to CCR5 Nt-based sulfopeptides as well as to cell surface CCR5 (8, 9, 13). The interaction between soluble gp120-CD4 complexes and CCR5 Nt-based sulfopeptides involves residues located primarily in the V3 stem and the C4 region of gp120 (8, 9). Residues in the V3 crown, including the GPG motif, are important for soluble gp120-CD4 complex binding to cell surface CCR5 (9). The determinants of X4 gp120-CXCR4 interactions are isolate dependent and therefore far less clearly defined (for a review see reference 10).
Many studies have previously demonstrated that the gp120 V3 loop is a major determinant of HIV-1 tropism and coreceptor usage (for a review see reference 15). The V3 loop is composed of approximately 35 residues and has a global positive charge that can vary from +2 to +10. Sequence comparisons of V3 loops from different HIV-1 isolates show that the N-terminal segment, the GPG crest, and the C-terminal segment are conserved, whereas variability occurs in the regions flanking the crest (5). Structural analyses of V3 loop-based peptides predict an N-terminal loop and a C-terminal alpha helix (5, 25, 29). The GPG crest forms a beta turn, and the variable flanking regions form the two strands of an antiparallel beta sheet (5, 25, 29). A single amino acid change in V3 can switch viral coreceptor usage (4, 16, 27, 30). An increase in the positive charge of the V3 loop is often associated with CXCR4 usage (15, 20). Sequence changes in the GPG motif can modulate coreceptor usage (16, 27). Furthermore, variable residues flanking the GPG motif can alter the stability of the beta sheet and/or alter the surface accessibility of this element, thereby influencing coreceptor usage (5, 6, 29).
This report studied the function of the crown and stem of the V3 loop. We previously showed that a number of residues in the V3 stem were required for soluble R5 gp120-CD4 binding to CCR5 Nt-based sulfopeptides (9). Additional residues in the V3 crown were required for binding to cell surface CCR5 (9). We generated a number of novel alanine substitutions in the gp120JR-FL V3 loop in order to determine precisely the N- and C-terminal junctions of the V3 stem and crown (8, 9). Briefly, mutant gp120JR-FL proteins were generated with the QuickChange kit (Stratagene, San Diego, Calif.), and nucleotide sequencing was performed to ascertain the presence of alanine substitutions in gp120. Supernatants containing soluble, mutant gp120 proteins were harvested 24 h after calcium phosphate transfection of 293T cells, and gp120 in the supernatants was quantified and adjusted to a final concentration of 20 nM as described previously (2, 9, 26). Enzyme-linked immunosorbent assay plates were coated with the Nt sulfopeptide comprising residues 2 to 18 of CCR5 and sulfotyrosines in positions 10 and 14. We note that peptides with a full complement of tyrosine sulfates were not used since we showed previously that sulfation of Y3 had no effect on gp120-peptide binding and that a Y-to-F substitution at position 15 had no effect on viral entry, indicating that sulfation of Tyr-15 was also dispensable. Different dilutions of gp120-CD4-immunoglobulin G2 (IgG2) were added to the plates and horseradish peroxidase-conjugated goat anti-human IgG was used to detect the presence of bound CD4-IgG2 as described previously (9). The percentage of gp120-CD4-IgG2 binding to the sulfopeptide comprising residues 2 to 18 was calculated by the formula (optical density at 450 nm [OD450] with mutant gp120/OD450 with wild-type gp120) × 100%. Calculations were performed on data where the binding of wild-type gp120 to the sulfopeptide was half-maximal. CD4-IgG2 binding to native and mutant gp120 also was measured by enzyme-linked immunosorbent assay as described previously (9). All of the mutants exhibited similar patterns of binding to CD4-IgG2 (data not shown), consistent with the CD4 binding site on gp120 lying outside the V3 loop. We found that alanine substitutions for V3 loop N-terminal residues 297 to 303 decreased soluble gp120-CD4 binding to the CCR5 Nt sulfopeptide by approximately 10-fold (Table 1). Replacements of residues R304 and K305 caused a twofold decrease in binding. Mutagenesis of V3 loop residues 306 to 320 had little to no effect on gp120-CD4 complex binding to the Nt sulfopeptide. Finally, alanine substitutions for V3 loop C-terminal residues 321 to 326 decreased binding to the sulfopeptide anywhere from 4- to 10-fold. Based on these findings, amino acids 306 to 320 were designated the V3 crown and residues 296 to 305 as well as 321 to 330 were designated the N- and C-terminal strands of the V3 stem.
TABLE 1.
Binding of gp120JR-FL alanine mutants to the CCR5 Nt sulfopeptidea
| V3 residue | % gp120-CD4 binding to CCR5 Nt sulfopeptide |
|---|---|
| C 296 | — |
| T 297 | — |
| R 298* | 11.6 |
| P 299 | — |
| N 300 | — |
| N 301* | 11.9 |
| N 302 | 16 |
| T 303* | 11.08 |
| R 304* | 47 |
| K 305 | 49 |
| S 306* | 87.5 |
| I 307 | — |
| H 308 | — |
| I 309 | — |
| G 310* | 83.1 |
| P 311* | 118.7 |
| G 312* | 118.7 |
| R 313* | 98.2 |
| A 314 | — |
| F 315 | 95 |
| Y 316* | 62.6 |
| T 317 | — |
| T 318 | 97.4 |
| G 319 | — |
| E 320* | 96 |
| I 321 | 15 |
| I 322* | 25.2 |
| G 323 | 15 |
| D 324* | 11.2 |
| I 325* | 14.3 |
| R 326* | 18.9 |
| Q 327 | — |
| A 328 | — |
| H 329 | — |
| C 330 | — |
The biotinylated CCR5 Nt sulfopeptide comprising residues 2 to 18 was immobilized on streptavidin-coated plates and incubated with soluble gp120-CD4 complexes. The mutated amino acids and their locations in gp120JR-FL are in the left column. Mutant gp120JR-FL-CD4-IgG2 binding to the sulfopeptide is expressed as a percentage of wild-type gp120JR-FL-CD4-IgG2. The values shown are averages of three independent experiments, and standard deviations were no more than ±20%. —, residue not studied. Residues within the crown of the V3 loop are in boldface. Highly conserved (>80%) residues are in italics. Asterisk, residue previously characterized for binding to the sulfopeptide (9).
Figure 1 shows a comparison of V3 loop sequences from R5 HIV-1JR-FL (JR-FL) and X4 HIV-1LAI (LAI) (amino acid numbering is based on the JR-FL sequence). The JR-FL V3 loop has a net charge of +5, whereas the LAI V3 loop has a net charge of +9 (counting histidines). The N termini of the JR-FL and LAI V3 loops are identical. The majority of differences between the JR-FL and LAI V3 loops lie in the crown: the LAI V3 crown contains an RQ insertion between residues I309 and G310 and substitutions Y316V, T318I, and E320K. Finally, in the C terminus of the V3 loop, I322 is absent in LAI and residues D324 and I325 in JR-FL are replaced by N and M, respectively, in LAI. We generated gp120 V3 loop chimera wherein the crown and the stem of the JR-FL and LAI V3 loops were exchanged or deleted. The different gp120 chimeras are described according to their backbone, V3 stem, and V3 crown contents. A three-letter nomenclature was chosen wherein the first letter indicates the source of the backbone, the second letter indicates the source of the V3 stem (residues 296 to 305 and 321 to 330), and the third letter indicates the source of the V3 crown (residues 306 to 320). J stands for JR-FL and L stands for LAI. When deleted, the V3 crown was replaced by GAG. Soluble, mutant gp120 proteins were generated and quantified as described above. The binding of the gp120 V3 loop chimera to the CCR5 Nt sulfopeptide was tested as described above (Fig. 2). Chimeras JJL, LJJ, and LJL all bound to the CCR5 Nt peptide with efficiencies similar to that of the wild-type gp120JR-FL. Furthermore, deletion mutants JJΔ and LJΔ also bound to the sulfopeptide with nearly wild-type efficiency. Neither the chimeras and deletion mutants carrying the LAI stem nor wild-type LAI bound the CCR5 Nt sulfopeptide. From these results we conclude that the V3 stem alone determines soluble gp120 binding to the CCR5 Nt sulfopeptide.
FIG. 1.
Sequence comparisons of the JR-FL and LAI V3 loops. The amino acid sequences of the JR-FL and LAI V3 loops were aligned, and identical residues are indicated by vertical lines between the two sequences. Residues are numbered according to the gp120HxB2 sequence. Residues within the crowns of the V3 loops are in boldface.
FIG. 2.
gp120 chimera and deletion mutant binding to the CCR5 Nt sulfopeptide. The CCR5 Nt sulfopeptide comprising residues 2 to 18 was immobilized on streptavidin-coated plates and incubated with soluble gp120-CD4 complexes. The gp120 chimera and deletion mutants are on the x axis. Mutant gp120JR-FL-CD4-IgG2 binding to the sulfopeptide is expressed as a percentage of wild-type gp120JR-FL-CD4-IgG2 binding. The values shown are averages of three independent experiments.
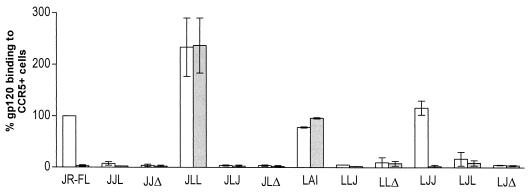
We then tested the ability of the gp120 V3 loop chimera and deletion mutants to bind to cell surface CCR5. L1.2 CCR5+ cells were incubated with gp120-containing supernatants and biotinylated CD4-IgG2 as described previously (9, 21). gp120-CD4-IgG2 binding to the cells was revealed by flow cytometry analysis of the mean fluorescence intensity (mfi) after addition of streptavidin-phycoerythrin (Pharmingen, San Diego, Calif.). Binding was calculated with the formula: (mfi for gp120 mutants/mfi for gp120 wild type) × 100%. Both wild-type gp120JR-FL and gp120LAI bound to the CCR5+ cells; however, only gp120JR-FL binding was completely inhibited by anti-CCR5 MAb PA14 (Fig. 3). The binding of gp120LAI was not inhibited by anti-CXCR4 MAb 12G5 or SDF-1, which indicated that it was not mediated by CXCR4 (E. G. Cormier, unpublished results). Only chimeras LJJ and JLL bound to the CCR5+ cells; however, only LJJ binding was inhibited by PA14. We therefore conclude that gp120 binding to cell surface-expressed CCR5 necessitates both the stem and crown of the V3 loop. The V3 crown alone or the V3 stem alone, regardless of gp120 background, is insufficient to mediate binding to CCR5.
FIG. 3.
gp120 chimera and deletion mutant binding to CCR5+ cells. L12 CCR5+ cells (106) were incubated with soluble gp120-CD4 complexes (20 nM gp120 and 5 nM CD4-IgG2) in the absence (white bars) or presence (gray bars) of MAb PA14 (10 μg/ml), and binding was detected by flow cytometry analysis. The gp120 chimeras and deletion mutants are indicated along the x axis. Mutant gp120JR-FL-CD4-IgG2 binding to the cells is expressed as a percentage of wild-type JR-FL-CD4-IgG2 binding. The values shown are averages of three independent experiments.
The gp120 chimera and deletion mutants also were tested for their ability to mediate viral entry. The mutant gp120 coding sequences were subcloned into a gp160JR-FL or a gp160LAI SV7D expression vector and NL luc+ env− pseudotyped particles were generated as described previously (11). U87 CD4+ cells expressing CCR5 or CXCR4 were infected with the pseudotyped reporter viruses, and luciferase activity (relative light units [rlu]) was assayed in cell lysates 48 h postinfection with a standard kit (Promega, Madison, Wis.) (11). The LJJ chimera was able to mediate entry into CCR5+ cells to the same extent as wild-type gp160JR-FL, and viral entry was completely inhibited by MAb PA14 (Table 2). CCR5-dependent entry also was mediated by the LLJ and JLJ chimeras, albeit ∼10- and ∼100-fold less efficiently than entry mediated by the wild-type JR-FL envelope glycoproteins. The LJL chimera mediated entry into CXCR4+ cells as efficiently as wild-type gp160LAI. JLL and JJL also mediated CXCR4-dependent entry ∼10-fold less efficiently than the wild-type gp120LAI envelope glycoproteins. CXCR4-dependent entry of JJL, JLL, and LJL was completely inhibited by MAb 12G5. These results indicate that, in the context of a virion, coreceptor specificity depends solely on the V3 crown. We note that, even though LAI binding to cells appears nonspecific, viral entry mediated by both LAI and the JLL chimera is clearly CXCR4 dependent. Finally, our results show that the overall positive charge of the V3 loop does not determine coreceptor usage. Wild-type JR-FL and LAI V3 loops have net charges of +5 and +9, respectively, yet chimeras JLJ and LLJ, in which the V3 loops have a net charge of +8, use CCR5 exclusively. Similarly, the V3 loops of JJL and LJL have a net charge of +6 yet exclusively mediate CXCR4-dependent entry. Interestingly, none of the chimeras exhibited dual tropism.
TABLE 2.
Viral entry into CCR5+ and CXCR4+ target cells mediated by mutant envelope glycoproteinsa
| Protein | Entry into:
|
|||
|---|---|---|---|---|
| CCR5+ cells
|
CXCR4+ cells
|
|||
| −PA14 | +PA14 | −12G5 | +12G5 | |
| JR-FL | +++ | − | − | |
| JJL | − | ++ | − | |
| JJΔ | − | − | ||
| JLL | − | ++ | − | |
| JLJ | + | − | − | |
| JLΔ | − | − | ||
| LAI | − | +++ | − | |
| LLJ | ++ | − | − | |
| LLΔ | − | − | ||
| LJJ | +++ | − | − | |
| LJL | − | +++ | − | |
| LJΔ | − | − | ||
U87 CD4+ cells expressing CCR5 or CXCR4 were infected with NL luc+ env− virions (100 to 200 ng of p24/ml) pseudotyped with mutant envelope glycoproteins and expressing the luciferase gene. Infections of CCR5+ cells were carried out in the presence or absence of MAb PA14 (10 μg/ml), whereas infections of CXCR4+ cells were carried out in the presence or absence of MAb 12G5 (10 μg/ml). −, rlu >150; +, rlu between 5 × 103 and 1 × 104; ++, rlu between 104 and 105; +++, rlu > 105.
In this study, we have precisely delineated two functional domains of the gp120 V3 loop: the V3 stem, which spans amino acid residues 296 to 305 and 321 to 330, and the V3 crown, which spans residues 306 to 320. Structural analyses of V3 loop-based peptides predict that these functional domains are characterized by distinct physical properties. The V3 stem comprises an N-terminal loop and a C-terminal alpha helix, whereas the V3 crown is an antiparallel beta sheet (5, 25, 29). In the context of soluble envelope glycoproteins, we find that the V3 stem (along with residues in C4 [9]) is responsible for gp120 binding to the CCR5 Nt (Table 3). Both the V3 crown and stem are required for soluble gp120 binding to cell surface CCR5 (Table 3). We interpret this to mean that the gp120-binding site on CCR5 has several components and propose that the V3 stem/C4 binds the Nt whereas the V3 crown interacts with residues in the extracellular loops of CCR5, most likely ECL2 (21, 22, 28). Interactions between the different domains of CCR5 and gp120 could occur either simultaneously or consecutively.
TABLE 3.
Chimeric gp120 binding and entry dataa
| Protein | gp120-CD4 binding to:
|
Viral entry
|
||
|---|---|---|---|---|
| CCR5 Nt sulfopeptide | Cell surface CCR5 | CCR5 mediated | CXCR4 mediated | |
| JR-FL | +++ | +++ | +++ | − |
| JJL | ++ | − | − | ++ |
| JJΔ | +++ | − | − | − |
| JLL | − | − | − | ++ |
| JLJ | − | − | + | − |
| JLΔ | − | − | − | − |
| LAI | − | − | − | +++ |
| LLJ | − | − | ++ | − |
| LLΔ | − | − | − | − |
| LJJ | ++ | +++ | +++ | − |
| LJL | ++ | − | − | +++ |
| LJΔ | ++ | − | − | − |
Simplified summary of binding to CCR5 Nt sulfopeptides, binding to cell surface CCR5, and entry into CCR5- or CXCR4-expressing cells mediated by the chimeric envelope glycoproteins used in this study. The extent of binding and entry is indicated by + and − signs.
In the context of a virion, however, the V3 crown alone is necessary and sufficient to direct exclusive usage of CCR5 or CXCR4 (Table 3). The V3 stem, despite being able to mediate specific binding to CCR5 Nt sulfopeptides, is not the main determinant of coreceptor usage. Thus, the LLJ chimera mediated entry into CCR5+ cells, even though we could not detect binding of the soluble protein to cell surface CCR5, and the JJL chimera mediated entry into CXCR4+ cells (Table 3). We propose that the oligomeric state of gp120 on the virion surface leads to multiple V3 crown-ECL2 interactions, which may override the need for a specific and/or strong V3 stem-Nt interaction and allow entry to occur, albeit less efficiently than for wild-type envelope glycoproteins. The recent observation that HIV-1 can adapt to use CCR5 with a truncated Nt further supports the notion that the V3 stem-CCR5 Nt interaction can be circumvented (22). Certain X4 isolates are able to enter cells using a CXCR4 with a truncated Nt, indicating that X4 gp120 V3 stem interactions with CXCR4 Nt also may be dispensable (3, 24, 31).
Several inhibitors of CCR5 coreceptor function, including anti-CCR5 MAbs and small molecules, have been shown to block gp120 binding to CCR5 without affecting the gp120-Nt interaction (12, 19, 21). Furthermore, MAbs that recognize epitopes in ECL2 of CCR5 are far more potent inhibitors of viral entry then MAbs that recognize epitope in the CCR5 Nt (19, 21). We propose that these MAbs inhibit the binding of the V3 crown to ECL2. By binding to a transmembrane pocket in CCR5, small-molecule inhibitors of R5 entry may destroy tertiary structures in ECL2 that are important for interactions with the V3 crown (T. Dragic, unpublished results; 12). Presumably, a similar mechanism of action is responsible for the action of small-molecule inhibitors of CXCR4 coreceptor function (14). The results from this study, together with our previous data, therefore, imply that the most potent inhibitors of HIV-1 entry target the V3 crown-ECL2 interaction.
Acknowledgments
We thank William Olson of Progenics Pharmaceuticals Inc. (Tarrytown, N.Y.) for generously providing us with CD4-IgG2, the PA14 MAb, and the biotinylated CCR5 Nt sulfopeptide.
This work was supported by NIH grant AI43847 to T.D.
REFERENCES
- 1.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 2.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brelot, A., N. Heveker, O. Pleskoff, N. Sol, and M. Alizon. 1997. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J. Virol. 71:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs, D. R., D. L. Tuttle, J. W. Sleasman, and M. M. Goodenow. 2000. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages). AIDS 14:2937-2939. [DOI] [PubMed] [Google Scholar]
- 5.Catasti, P., J. D. Fontenot, E. M. Bradbury, and G. Gupta. 1995. Local and global structural properties of the HIV-MN V3 loop. J. Biol. Chem. 270:2224-2232. [DOI] [PubMed] [Google Scholar]
- 6.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormier, E. G., and T. Dragic. 2000. An overview of HIV-1 coreceptor function and its inhibitors, p. 19-34. In C. Kuiken, F. McCutchan, B. Foley, J. W. Mellors, B. Hahn, J. Mullins, P. Marx, and S. Wolinsky (ed.), HIV sequence compendium 2000. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 8.Cormier, E. G., M. Persuh, A. D. Thompson, S. W. Lin, T. P. Sakmar, W. C. Olson, and T. Dragic. 2000. Specific interaction of CCR5 amino-terminal domain peptides containing sulfo-tyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 97:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 11.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farzan, M., N. Vasilieva, C. E. Schnitzler, S. Chung, J. Robinson, N. P. Gerard, C. Gerard, H. Choe, and J. Sodroski. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275:33516-33521. [DOI] [PubMed] [Google Scholar]
- 14.Hatse, S., K. Princen, L. O. Gerlach, G. Bridger, G. Henson, E. De Clercq, T. W. Schwartz, and D. Schols. 2001. Mutation of Asp(171) and Asp(262) of the chemokine receptor CXCR4 impairs its coreceptor function for human immunodeficiency virus-1 entry and abrogates the antagonistic activity of AMD3100. Mol. Pharmacol. 60:164-173. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman, T. L., and R. W. Doms. 1999. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 16:57-65. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Q., J. O. Trent, G. D. Tomaras, Z. Wang, J. L. Murray, S. M. Conolly, J. M. Navenot, A. P. Barry, M. L. Greenberg, and S. C. Peiper. 2000. Identification of Env determinants in V3 that influence the molecular anatomy of CCR5 utilization. J. Mol. Biol. 302:359-375. [DOI] [PubMed] [Google Scholar]
- 17.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 18.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 20.Moulard, M., H. Lortat-Jacob, I. Mondor, G. Roca, R. Wyatt, J. Sodroski, L. Zhao, W. Olson, P. D. Kwong, and Q. J. Sattentau. 2000. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J. Virol. 74:1948-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Platt, E. J., S. E. Kuhmann, P. P. Rose, and D. Kabat. 2001. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J. Virol. 75:12266-12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 24.Reeves, J., N. Heveker, A. Brelot, M. Alizon, P. Clapham, and L. Picard. 1998. The second extracellular loop of CXCR4 is involved in CD4-independent entry of human immunodeficiency virus type 2. J. Gen. Virol. 79:1793-1799. [DOI] [PubMed] [Google Scholar]
- 25.Rini, J. M., R. L. Stanfield, E. A. Stura, P. A. Salinas, A. T. Profy, and I. A. Wilson. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc. Natl. Acad. Sci. USA 90:6325-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu, N., Y. Haraguchi, Y. Takeuchi, Y. Soda, K. Kanbe, and H. Hoshino. 1999. Changes in and discrepancies between cell tropisms and coreceptor uses of human immunodeficiency virus type 1 induced by single point mutations at the V3 tip of the env protein. Virology 259:324-333. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, D. A. D., E. C. Cormier, and T. Dragic. 2002. CCR5 and CXCR4 usage by non-clade B human immunodeficiency virus type 1 primary isolates. J. Virol. 76:3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tugarinov, V., A. Zvi, R. Levy, Y. Hayek, S. Matsushita, and J. Anglister. 2000. NMR structure of an anti-gp120 antibody complex with a V3 peptide reveals a surface important for co-receptor binding. Struct. Fold Des. 8:385-395. [DOI] [PubMed] [Google Scholar]
- 30.Verrier, F., A. M. Borman, D. Brand, and M. Girard. 1999. Role of the HIV type 1 glycoprotein 120 V3 loop in determining coreceptor usage. AIDS Res. Hum. Retrovir. 15:731-743. [DOI] [PubMed] [Google Scholar]
- 31.Willett, B. J., K. Adema, N. Heveker, A. Brelot, L. Picard, M. Alizon, J. D. Turner, J. A. Hoxie, S. Peiper, J. C. Neil, and M. J. Hosie. 1998. The second extracellular loop of CXCR4 determines its function as a receptor for feline immunodeficiency virus. J. Virol. 72:6475-6481. (Erratum, 72:8460.) [DOI] [PMC free article] [PubMed] [Google Scholar]