Abstract
We previously constructed a series of simian immunodeficiency virus (SIV) mutants containing deletions within a 97-nucleotide region of the SIVmac239 untranslated region or leader sequence. However, as is common with live attenuated viruses, several of the mutants exhibited a moderate propensity for reversion. Since the M184V mutation in human immunodeficiency virus type 1 reverse transcriptase is associated with diminished fitness as well as lamivudine resistance, we introduced this substitution into several of our deletion mutants to determine its effects on viral replication and compensatory reversion. Our results indicate that M184V impaired viral fitness in pair-wise comparisons of mutants that contained or lacked this substitution. We also observed that M184V significantly impaired the potential for both compensatory mutagenesis and reversion in these mutants both in cell lines and in peripheral blood mononuclear cells.
Genetic variation in human immunodeficiency virus type 1 (HIV-1) and other retroviral lineages has been associated with multiple factors, which include viral recombination, reverse transcriptase (RT) infidelity, and both viral and cellular factors that can influence mutation rates (10, 15, 21, 25, 33). The generation of viral quasispecies as a result of RT infidelity is largely due to the lack within viral RT of a 3′-to-5′ proofreading ability, combined with high rates of viral replication (8, 31).
The catalytic domains of the RTs of HIV-1 and its simian counterpart, simian immunodeficiency virus (SIV), both include a highly conserved YMDD motif. This domain is common throughout the polymerase family of enzymes (26), and mutations within this region are commonly lethal. However, viruses resistant to antiretroviral nucleoside 2′,3′-dideoxy-3′-thiacytidine (3TC) harbor a single M184V substitution within the aforementioned motif (32). This same substitution in RT is associated with resistance to 3TC in the case of SIV (7).
In HIV-1, it is well documented that the M184V mutation also confers a deficit in fitness that is most apparent in primary cell lines (2). The reasons for this are multifaceted and include the fact that RT enzymes containing M184V are associated with diminished processivity (2, 3, 23, 28), diminished nucleotide primer unblocking (11), and diminished ability to initiate reverse transcription (M. Gotte, X. Wei, K. Diallo, B. Marchand, A. Schaffer, and M. A. Wainberg, 5th Int. Workshop HIV Drug Resist. Treat. Strateg., abstr. 46, 2001). These events are also modulated by intracellular deoxynucleoside triphosphate substrate availability (3).
In the aftermath of deletion mutagenesis, leading to attenuation of replication, genetic variation requires passage through the constraints of an artificially produced bottleneck. Under these conditions, the spectrum of compensatory mutations is likely to be restricted (29, 30). In this study, we demonstrate that the M184V substitution can impair the viral capacity for reversion in the context of specific deletions within the 5′ leader regions of a series of attenuated SIVmac239 constructs. As well, the presence of the M184V substitution may affect the process of compensatory mutagenesis in regard to codon change.
(This work was performed by J. B. Whitney in partial fulfillment of the requirements for a Ph.D. degree in the Faculty of Graduate Studies, McGill University, Montreal, Canada, 2002.)
Several of our viral deletion mutants were previously shown to display moderate reversion kinetics over serial passage (12, 13), i.e., constructs SD2, SD5, and SD6. The M184V mutation was introduced into the RTs of these constructs by site-directed mutagenesis of the pCRII vector containing 1.7 kb of the SIV RT coding region as described previously (7). The coding sequence for the recovered M184V-containing RT fragment was then inserted between the NarI and BamHI sites in the full-length wild-type (WT) and mutant SIV proviral clones. All recombinant viruses were confirmed by sequencing.
After transfection of COS-7 cells with appropriate plasmid DNA using lipofectamine (GIBCO, Burlington, Ontario, Canada), viral supernatants were recovered and the concentration of p27 antigen in these stocks was quantified with a Coulter SIV core antigen assay kit (Immunotech Inc., Westbrook, Maine) as described previously (12).
Viral replication assays.
Viral inocula for each construct, equivalent to 10 ng of p27 CA antigen, were treated with DNase I and used to infect CEMx174 cells (12). RT assays were used as a surrogate for viral replication and revealed that the presence of M184V together with the various deletions in the 5′ leader resulted in an additional impairment in viral replication compared to when M184V was not present. These results were observed consistently in replicate experiments, regardless of which leader mutant was studied. The impairment for each mutant virus containing M184V was further amplified by the presence of 8 μM 3TC, which further constrained viral replication by an additional 2 to 4 days (results not shown). This may have been due to additional selective pressure by 3TC to maintain the M184V mutation and prevent the outgrowth of revertant viruses.
To establish the potential for viral reversion over protracted periods, we performed serial passage or “forced evolution” of our mutant constructs using the CEMx174 cell line. Typically, aliquots of viral supernatants were taken at the observed peak of infection, and these samples were then used to infect fresh CEMx174 cells at doses equivalent to 10 ng of viral p27 antigen. During each successive passage, viruses containing deletions in leader sequences plus M184V showed delays in growth kinetics and reduced replicative capacity as assessed by RT assay (results not shown).
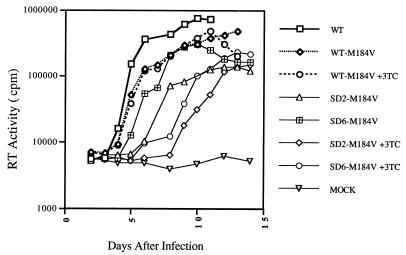
Subsequent PCR and sequencing analysis at each passage of these M184V-containing variants indicated that the original leader sequence deletions were retained in all instances. As well, each of the three leader mutants that encoded the M184V mutation retained this mutation over at least three consecutive passages. However, both SD5-M184V and SD6-M184V lost the M184V substitution by the fourth round of passage in the absence of 3TC. This sequence alteration was commensurate with a measurable increase in viral replication (Fig. 1; only SD2-M184V and SD6-M184V are shown). In contrast, the SD2-M184V variant retained M184V through at least four passages. The last variant also showed decreased RT activity and replication rates in comparison to either the SD5 or SD6 mutant at the same passage (only SD2-M184V and SD6-M184V are shown). A similar number of clones from experiments performed under the pressure of 3TC were also sequenced and showed no loss of either the leader or M184V in four passages. As well, we carried out equivalent experiments with only the leader mutations under 3TC pressure, and these cultures showed undetectable levels of viral replication during 2 months of passage as assessed by RT assay.
FIG. 1.
Delay of phenotypic reversion of the SD2-M184V mutant after long-term culture in CEMx174 cells. Shown are growth curves of viruses over extended culture. Note that the scale of the ordinate is logarithmic. Equivalent amounts of virus from transfected COS-7 cells were used to infect CEMx174 cells based on levels of p27 antigen (10 ng per 106 cells). Infected cells were grown over protracted periods, and culture fluids were monitored by RT assay. Mock infection denotes exposure of cells to heat-inactivated WT virus as a negative control. A representative example of the SD2-M184V and SD6-M184V variants at the fourth passage is shown (the experiment was performed three times with similar results).
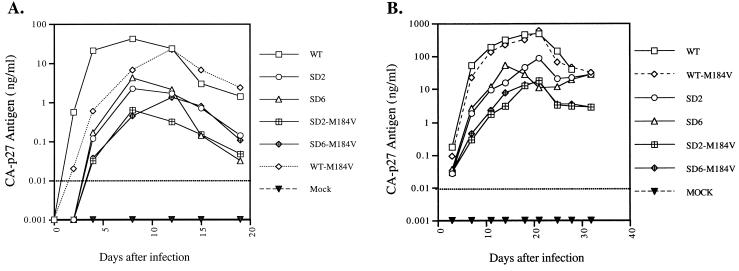
To evaluate viral replication under more-relevant cellular conditions, peripheral blood mononuclear cells (PBMCs) were obtained from two healthy, simian type D retrovirus 1-negative rhesus macaques. These cells were stimulated with phytohemagglutinin for 72 h prior to infection as described previously (13). Infection and viral antigen production were assessed by using complete RPMI 1640 culture medium supplemented with 10% heat-inactivated fetal bovine serum and 20 U of interleukin-2 (GIBCO)/ml throughout the monitored period. Viral replication for donors A and B was assessed in 10- and 2-ml total culture volumes, respectively, and fresh stimulated PBMCs were added to the cultures at weekly intervals. The results show that all the mutant viruses grew at diminished levels relative to WT (Fig. 2A and B). Furthermore, the addition of M184V to the SD2, SD5, and SD6 variants resulted in further reductions in p27 antigen production (SD2 and SD6 are shown).
FIG. 2.
Replication capacity of wild-type and mutated viruses in monkey PBMCs. Equivalent amounts of virus were used to infect rhesus macaque PBMCs based on levels of p27 antigen, typically 10 ng of WT or mutant virus per 4 × 106 PBMCs. Viral replication was monitored by determining levels of SIV p27 antigen by enzyme-linked immunosorbent assay of culture fluids. Mock infection denotes exposure of cells to heat-inactivated wild-type virus as a negative control. The dotted line representing 0.01 ng of p27/ml indicates the threshold sensitivity of the assay. (A) Growth curves indicating antigen production in PBMCs from donor monkey A. (B) Growth curves for PBMCs from donor monkey B. p27 antigen results are the averages of duplicates. (C) Second passage of mutated viruses in 106 CEMx174 cells by using an inoculum of 10 pg of p27 antigen derived from the infected PBMCs of monkey B. Viral replication was monitored by RT assay of culture fluids. Shown is a representative replication curve of experiments conducted in duplicate. Mock infection denotes exposure of cells to heat-inactivated WT virus as a negative control. Note that the scales of the ordinates are logarithmic.
To determine the propensity for phenotypic reversion in PBMCs, additional passages were performed with supernatants taken at the peak levels of viral p27 antigen production, at 21 days after infection. These were used to infect fresh stimulated PBMCs from the same animal, i.e., donor B. This second passage of mutant virus harboring the M184V substitution exhibited levels of antigen production nearly identical to that seen during the first infection (not shown), suggesting that no increase in replication capacity occurred in this case.
We also used viral supernatants collected at the peak of the first passage in monkey PBMCs, again from donor B, to infect fresh CEMx174 cells. The results show that increased viral replication occurred in variants that lacked the M184V mutation (Fig. 2C). In contrast, replication in M184V-harboring species showed only minor increases compared to replication of clonal infectious stock.
RNA dimerization.
We also assessed the ability of the SD2 mutant virus to properly incorporate a mature RNA dimer. Nondenaturing Northern analysis of purified RNA preparations had indicated that deletion of the sequence between nucleotides +398 and +418 in SD2 completely eliminated viral RNA dimerization. The additional presence of the M184V mutation together with the leader mutation did not appear to outwardly affect RNA dimerization (unpublished data).
Those viruses that were continually passaged in the CEMx174 cell line were sequenced by PCR amplification of proviral DNA recovered from cells isolated at the peak of the fourth round of infection. The sequencing of the complete SD2-M184V untranslated region (UTR) and gag regions showed numerous point mutations in all clones. Despite this variability, one point mutation that corresponded to a G-to-A transition in MA, encoding a change from a threonine to isoleucine at residue 70 (T70I), was found in all six sequenced clones.
To assess the relevance of this mutation, site-directed mutagenesis was performed with the SD2-M184V clone to produce the variant termed SD2-MA-M184V. Infectious inocula were produced in COS-7 cells and were then used to infect CEMx174 cells in parallel with controls, which included two previously described SD2 reversion mutants (12, 14). The compensatory mutations that restored the SD2 virus to replication competence had been shown to be members of two distinct sets of mutations within the putative dimerization initiation site (DIS) loop (A423G) and within several different Gag proteins, i.e., NC (E18G and G31K) or CA (K197R) and p6 (E49K). These amino acid changes are responsible for restoration of viral RNA packaging and viral fitness (14). A similar situation has been observed for deletions within the SL1 region of HIV-1 (20).
Figure 3A also shows that the T70I mutation in MA was sufficient to confer a replicative advantage in the context of SD2-M184V. This codon change had no observable effect on a WT virus (not shown). MA is known to be involved in the targeting of both Gag and genomic RNA to the cell membrane and in the formation and stabilization of genomic RNA dimers (6, 9). The potential role of the T70I MA mutation in the rescue of viral replication is also suggested by recent studies on a role for upstream leader sequences and the MA coding sequence in formation of an extended RNA pseudoknot structure. Extended interactions involving a region of MA and the pol open reading frame have also been reported (24, 27). Additional in vitro evidence also supports a role for higher-order structures in the regulation of viral replication (16).
FIG. 3.
(A) Reversion of the SD2-M184V variant after replication in CEMx174 cells. Growth curves of reverted viruses in CEMx174 cells. Equivalent amounts of virus from transfected COS-7 cells were used to infect CEMx174cells based on levels of p27 antigen (10 ng per 106 cells). Viral replication was monitored by RT assay of culture fluids. Shown is a representative growth curve of experiments conducted in triplicate. Mock infection denotes exposure of cells to heat-inactivated WT virus as a negative control. (B) The M184V mutation restricts compensatory mutagenesis in the case of the SD2 variant. Viruses derived from COS-7 cells were standardized on the basis of p27 CA antigen and used to infect 106 CEMx174 cells. RT activity of culture fluids was used to monitor replication. Shown is a representative growth curve of experiments performed in triplicate. Mock infection denotes exposure of cells to heat-inactivated WT virus as a negative control. Note that the scales of the ordinates are logarithmic in both panels.
To further explore the notion that reversion of SD2-M184V virus was impaired specifically as a result of M184V, we inserted this substitution into the two aforementioned SD2 reversion mutants, termed SD2-DIS-NC1+2-M184V and SD2-DIS-CA-P6-M184V. Both these variants were impaired in replication ability in CEMx174 cells compared with equivalent constructs that lacked M184V (Fig. 3B).
In summary, the M184V mutation in RT adversely impacts the replicative fitness of a number of SIV constructs. Furthermore, SIVs containing both the M184V and DIS mutations are less able to effect repair through compensatory mutagenesis than are viruses containing a WT RT. Our results also show that viral species that harbored both the M184V mutation and deletions in the region of the DIS displayed reduced replication capacity over multiple passages. Similar results have been reported with HIV-1 viruses containing the M184V mutation in RT (17, 19). In nonhuman primate studies, M184V-containing SIV failed to revert to WT and may have been initially impaired in its ability to multiply to high titer. However, this replication deficit may have been corrected over time, as a consequence of a distinct compensatory mutation within RT (22).
We should point out that attenuation of the SD2-M184V variant may be partly attributable to synergy between the 5′ UTR and the Gag-Pol region in regard to both structure and function. HIV-1 RTs that harbor M184V suffer from diminished ability to initiate reverse transcription and to participate in the elongation phase of minus-strand DNA synthesis (5, 18). In addition, the UTRs of both HIV and SIV play key roles in RNA dimerization and strand transfer (1, 4, 18).
Finally, we have shown that both RT and UTR sequences are necessary for restoration of viral replication, and our experiments suggest that viral recombination is involved in the process of compensatory mutagenesis. Mutants that are presumed to lack this function in the process of reversion might be relegated to fixing adventitious mutations in an iterative fashion, likely imparting delays to restoration of a WT replication phenotype.
Acknowledgments
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: the p239SpSp5′ and p239SpE3′ plasmids, contributed by R. Desrosiers, and the CEMx174 cell line, provided by P. Cresswell.
This research was supported by the Canadian Institutes of Health Research and by the Canadian Network for Vaccines and Immunotherapeutics for Cancer and Chronic Viral Diseases. J.B.W. was supported by a fellowship from The Canadian Institutes for Health Research.
REFERENCES
- 1.Anderson, J. A., R. J. Teufel II, P. D. Yin, and W. S. Hu. 1998. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J. Virol. 72:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 3.Back, N. K., and B. Berkhout. 1997. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 41:2484-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakrishnan, M., P. J. Fay, and R. A. Bamabara. 2001. The kissing hairpin sequence promotes recombination within the HIV-1 5′ leader sequence. J. Biol. Chem. 276:36482-36492. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., A. T. Das, and J. L. B. van Wamel. 1998. The native structure of the human immunodeficiency virus type 1 RNA genome is required for the first strand transfer of reverse transcription. Virology 249:211-218. [DOI] [PubMed] [Google Scholar]
- 6.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherry, E., M. Slater, H. Salomon, E. Rud, and M. A. Wainberg. 1997. Mutations at codon 184 in simian immunodeficiency virus reverse transcriptase confer resistance to the (−) enantiomer of 2′-3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 41:2763-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 9.Garbitt, R. A., J. A. Albert, M. D. Kessler, and L. J. Parent. 2001. trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 75:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goncalves, J., Y. Korin, J. Zack, and D. Gabuzda. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 70:8701-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan, Y., J. B. Whitney, K. Diallo, and M. A. Wainberg. 2000. Leader sequences downstream of the primer-binding site are important for efficient replication of simian immunodeficiency virus. J. Virol. 74:8854-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan, Y., J. B. Whitney, C. Liang, and M. A. Wainberg. 2001. Novel, live attenuated simian immunodeficiency virus constructs containing major deletions in leader RNA sequences. J. Virol. 75:2776-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan, Y., K. Diallo, M. Detorio, J. B. Whitney, C. Liang, and M. A. Wainberg. 2001. Partial restoration of replication of simian immunodeficiency virus by point mutations in either the dimerization initiation site (DIS) or Gag region after deletion mutagenesis within the DIS. J. Virol. 75:11920-11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huthoff, H., and B. Berkhout. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inouye, P., E. Cherry, M. Hsu, S. Zolla-Pazner, and M. A. Wainberg. 1998. Neutralizing antibodies directed against the V3 loop select for different escape variants in a virus with mutated reverse transcriptase (M184V) than in wild-type human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 14:735-740. [DOI] [PubMed] [Google Scholar]
- 18.Jones, J. S., R. W. Allan, and H. M. Temin. 1994. One retroviral RNA is sufficient for synthesis of viral DNA. J. Virol. 68:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y., Z. Zhang, J. K. Wakefield, S. M. Kang, and C. D. Morrow. 1997. Nucleotide substitutions within U5 are critical for efficient reverse transcription of human immunodeficiency virus type 1 with a primer binding site complementary to tRNAHis. J. Virol. 71:6315-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, C., L. Rong, Y. Quan, M. Laughrea, L. Kleiman, and M. A. Wainberg. 1999. Mutations within four distinct Gag proteins are required to restore replication of human immunodeficiency virus type 1 after deletion mutagenesis within the dimerization initiation site. J. Virol. 73:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newstein, M. C., and R. C. Desrosiers. 2001. Effects of reverse-transcriptase mutations M184V and E89G on simian immunodeficiency virus in rhesus monkeys. J. Infect. Dis. 184:1262-1267. [DOI] [PubMed] [Google Scholar]
- 23.Oude Essink, B. B., N. K. Back, and B. Berkhout. 1997. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 25:3212-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paillart, J. C., E. Skripkin, B. Ehresmann, C. Ehresmann, and R. Marquet. 2002. In vitro evidence for a long-range pseudoknot in the 5′ untranslated and matrix coding regions of human immunodeficiency virus type 1 (HIV-1) genomic RNA. J. Biol. Chem. 277:5995-6004. [DOI] [PubMed] [Google Scholar]
- 25.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 26.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purohit, P., S. Dupont, M. Stevenson, and M. R. Green. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro selection. RNA 7:576-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quan, Y., P. Inouye, C. Liang, L. Rong, M. Gotte, and M. A. Wainberg. 1998. Dominance of the E89G substitution in HIV-1 reverse transcriptase in regard to increased polymerase processivity and patterns of pausing. J. Biol. Chem. 273:21918-21925. [DOI] [PubMed] [Google Scholar]
- 29.Rouzine, I. M., and J. M. Coffin. 1999. Search for the mechanism of genetic variation in the pro gene of human immunodeficiency virus. J. Virol. 73:8167-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouzine, I. M., A. Rodrigo, and J. M. Coffin. 2001. Transition between stochastic evolution and deterministic evolution in the presence of selection: general theory and application to virology. Microbiol. Mol. Biol. Rev. 65:151-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skalka, A. M., and S. P. Goff. 1993. Reverse transcriptase. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 32.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G-A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]