Abstract
In many respects, picornaviruses are well suited for their proposed use as immunization vectors. However, their inherent genetic instability hinders application for prophylactic purposes. We demonstrate the improved expression and stability of a heterologous insert through a novel vector design strategy that partially replaces noncoding regulatory sequences with coding sequences for foreign gene products.
Vast numbers of different viruses have been proposed as candidates for the generation of immunization vectors against human immunodeficiency virus (reviewed in references 12 and 27). Among these viruses, picornaviruses have played a central role. Attempts to harness picornaviruses for vaccination purposes have included enteroviruses (e.g., poliovirus [1, 3, 5, 8] and coxsackieviruses [14]), rhinoviruses (6), and cardioviruses (e.g., mengovirus [4]).
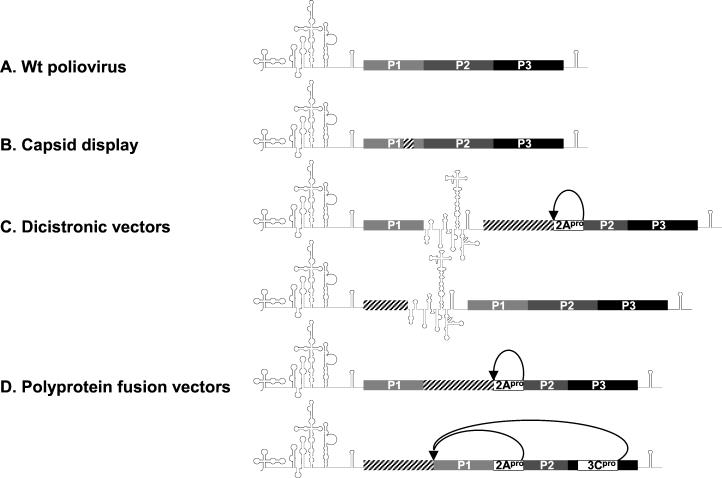
Various strategies have been employed to engineer picornavirus-based expression vectors (Fig. 1). These strategies were designed to conform to the known principles governing picornavirus gene expression. All picornaviruses lacking a 5′-end cap structure (21) rely on translational initiation through internal ribosomal entry (15, 16, 22, 23). The internal ribosomal entry site (IRES), a complex cis-acting genetic element with extensive secondary structure, mediates cap-independent translation of the viral genome. Translation of the single viral open reading frame (ORF) produces a large viral polyprotein that is subsequently processed to yield individual viral gene products (17, 28). Minimal inserts into the coding region for the viral capsid proteins (P1) gave rise to heterogeneous capsids which displayed foreign immunogenic peptides on the particle exterior (Fig. 1B) (6, 14). Dicistronic vectors that express foreign ORFs under the control of an intercistronic heterologous IRES element were generated (Fig. 1C) (2). Most recently, polyprotein fusion vectors were created by inserting foreign ORFs fused N terminally to the polyprotein or between P1 and P2 (Fig. 1D) (5, 8).
FIG. 1.
Genetic structure of proposed picornavirus-based expression vectors. (A) Wild-type (Wt) poliovirus. (B) Capsid display vectors with foreign peptides (hatched box) incorporated into the viral capsid (6, 14). (C) Dicistronic vectors expressing a foreign insert (hatched box) driven by a secondary encephalomyocarditis virus (EMCV) IRES (2). The intercistronic EMCV IRES may be placed between P1 and P2 or between the insert and P1. (D) Polyprotein fusion vectors (5, 8). A foreign insert is fused to the viral polyprotein either in between P1 and P2 or at the N terminus. Proteolytic processing of the fusion polyprotein occurs through artificial cleavage sites for the viral 2Apro or 3Cpro protease, respectively (5, 8).
A major obstacle common to the proposed replicating picornavirus expression vectors is their inherent genetic instability. All proposed expression vectors share the tendency to revert to wild-type sequences with maximal propagation potential. This tendency may be simply explained by the deleterious effect of the insertion of foreign sequences on virus replication efficiency, triggering adaptation to a faster growing phenotype (9). These adaptation events invariably lead to the elimination of parts or all of the inserted foreign sequences. Genetic instability of picornavirus expression vectors greatly limits their usefulness for vaccination purposes.
We have adopted a novel strategy to engineer picornavirus-based expression vectors. This strategy is based on forcing picornaviruses to retain foreign sequences that confer a replicative advantage to the virus. This was achieved by replacing conserved viral noncoding IRES sequences with foreign sequences designed to assume secondary structure favored by replicating virus.
Vector design.
We based our expression vectors on a nonpathogenic chimera, known as PVS-RIPO (11), containing the IRES element of human rhinovirus type 2 (HRV2) in a poliovirus type 1 (Sabin) background. This chimera is characterized by highly attenuated neurovirulence and has been proposed for use as an oncolytic agent against malignant glioma (13).
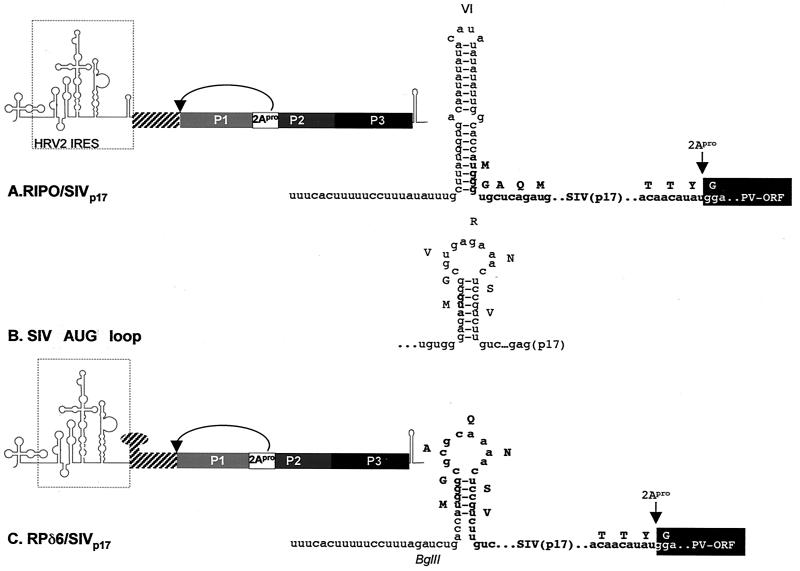
We produced two RIPO constructs designed to express sequences of simian immunodeficiency virus (SIVmac239) encoding the matrix protein (p17). First, replicating the conventional polyprotein fusion vector strategy of Andino et al. (5), SIVp17 was incorporated between the HRV2 IRES and the viral ORF to yield RIPO/SIVp17. The authentic AUG of the HRV2 IRES was used to drive translation of the fusion polyprotein, and proteolytic release of the foreign gene product occurred via an engineered cleavage site for the viral proteinase 2Apro (Fig. 2A).
FIG. 2.
Construction of recombinant IRES expression vectors. Both vector types feature artificial cleavage sites for 2Apro for the release of foreign polypeptides from the fusion polyprotein. Inserted SIVp17 sequences are indicated by hatched boxes. (A) Genetic structure of a conventional fusion polyprotein expression vector based on PVS-RIPO. The rhinoviral IRES element is shown. The enlarged sequence detail depicts HRV2 IRES stem-loop domain VI and the genetic structure of the insert region. Initiation of translation of the fusion polyprotein occurs from the authentic HRV2 initiation codon, preceding the N-terminal 4 amino acids of the poliovirus polyprotein and the ORF for SIVp17 (boldface letters). (B) Sequence and proposed secondary structure of the SIV AUG loop (7). The initiating AUG of SIVgag (boldface letters) is in a position similar to that of Y(n)X(m)AUG in the HRV2 IRES, forming the base of stem-loop domain VI [for the position of the Y(n)X(m)AUG motif, compare with Fig. 3A]. (C) Genetic structure of RPδ6/SIVp17. Sequences shown indicate the SIV AUG loop which has been used to replace HRV2 IRES stem-loop domain VI. Initiation occurs at Y(n)X(m)AUG, which had been placed in Kozak context (ACCAUGG). X(m) has been altered to contain a BglII restriction site. SIVp17 sequences are shown in boldface.
In our second vector, we followed a new strategy, attempting to partially replace IRES structures with foreign coding sequences. This design utilized a highly conserved sequence element within picornavirus IRESs known as the Y(n)X(m)AUG motif (24, 28). In polio- and rhinoviruses (containing type 1 IRESs [reviewed in reference 28]), the AUG triplet contained within this motif located at the base of stem-loop domain VI (Fig. 3A) is not in Kozak context and does not serve as an initiation codon. Instead, an AUG triplet in Kozak context located 43 nucleotides (nt) (HRV2 [Fig. 3A]) or 153 nt (poliovirus [25]) downstream of Y(n)X(m)AUG serves as the initiation codon (19, 26). However, the cryptic AUG sequence within Y(n)X(m)AUG has been reported to serve in initiation in the poliovirus IRES if placed in proper Kozak context (25). Upon shifting the initiation codon to Y(n)X(m)AUG and deleting stem-loop domain VI of the rhinovirus IRES in PVS-RIPO (Fig. 3B), we generated a viable deletion recombinant (RPδ6) with replication kinetics similar to those of full-length PVS-RIPO (Fig. 3C).
FIG. 3.
Position and structure of the Y(n)X(m)AUG motif within type 1 (enterovirus and rhinovirus) IRES elements. The polypyrimidine tract [Y(n)], spacer [X(m)], and cryptic AUG (asterisks) in the intact HRV2 IRES (A) and in a stem-loop domain VI deletion mutant (B) are indicated. The sequence of X(m) was altered in panel B to put the adjacent cryptic AUG into Kozak context (CUUAUGG to ACCAUGG). (C) Growth characteristics of PVS-RIPO (open squares) and RPδ6 (filled diamonds) in HeLa cells. The IRES deletion construct gave rise to viable virus that grew only slightly less efficiently than did PVS-RIPO in HeLa cells. p.i., postinfection.
The proposed secondary structure for the SIV leader RNA predicts the formation of a stem-loop surrounding the initiation codon of SIVgag (the AUG loop [Fig. 2B] [7]). The AUG loop contains coding sequences for N-terminal SIVp17 and was used to replace stem-loop domain VI in RPδ6 to yield vector RPδ6/SIVp17 (Fig. 2C). Manipulations necessary to insert the SIVp17 ORF into RPδ6 were designed to maintain the overall structure of the AUG loop. To ensure proper processing of the viral fusion polypeptide, the authentic N terminus of the wild-type poliovirus polyprotein (MGAQ) was placed at the N-terminal junction of the expression cassette. These changes altered the N terminus of SIVp17 from MGVRNSVL to MGAQNSVL. Proteolytic release of SIVp17 was again provided through an engineered site for 2Apro. Thus, foreign coding sequences partially mimicked cognate IRES structure and the initiation of translation occurred at Y(n)X(m)AUG (Fig. 2C).
Virus passaging and emergence of an enlarged variant.
In vitro-transcribed RNA of these constructs was transfected into HeLa cells to derive virus for subsequent passaging for assessment of genetic stability. Each passage was incubated for 24 h at 37°C and processed to isolate total RNA for reverse transcription (RT)-PCR analysis of the insert region (Fig. 4).
FIG.4.
RT-PCR analysis of total RNA preparations obtained from serial passages of RIPO/SIVp17 and RPδ6/SIVp17 expression vectors. Lanes are marked according to the origin of template used in the diagnostic PCR: M, molecular mass marker; P, plasmid DNA; T, lysate of transfected cells; 1 through 5, individual passages. The schematic above indicates the positions of annealing sequences for primers used in the PCR. (A) Transfection of RIPO/SIVp17 RNA and subsequent passaging in HeLa cells resulted in the rapid deletion of inserted foreign sequences. (B) Transfection of RPδ6/SIVp17 RNA and subsequent passaging of virus resulted in a genetic adaptation event characterized by an insert enlarged by 114 nt (arrowhead). The enlarged insert was retained after five passages, without evidence for the occurrence of deletion variants.
In accordance with published reports on the poor genetic stability of conventional polyprotein fusion vectors (20), foreign inserts were rapidly eliminated from RIPO/SIVp17 (Fig. 4A). Sequencing of the amplified product obtained by RT-PCR analysis after five passages revealed either reversion to the parent PVS-RIPO genotype or retention of minimal insert residue (<25 nt).
Surprisingly, RT-PCR analysis of serially passaged RPδ6/SIVp17 revealed the occurrence of an enlarged insert after the second passage (Fig. 4B). Since picornavirus-based expression vectors usually eliminate insert sequences to restore efficient growth, this finding was highly unexpected. The enlarged insert was the sole product detected by RT-PCR analyses of subsequent passages, indicating a rapid eclipse of the RPδ6/SIVp17 parent by the emerging variant. Sequencing of the enlarged RPδ6/SIVp17 variant revealed the presence of an exact duplication of the Y(n)X(m)AUG motif, as well as 84 N-terminal nt of the SIVp17 coding region, which included the engineered IRES domain VI containing the SIV AUG loop (Fig. 5A). Notably, the entire SIVp17 coding region was retained. This variant exhibited a homogeneous plaque phenotype significantly larger than that of its parent (data not shown) and growth kinetics akin to those of PVS-RIPO (Fig. 5B). The duplication was in frame, enlarging the insert fragment from 420 to 534 nt (Fig. 5A).
FIG. 5.
Sequence and growth characteristics of variant RPδ6/SIVp17(21). (A) The amplified PCR product obtained through RT-PCR from total cellular RNA prepared from the fifth consecutive passage of RPδ6/SIVp17 in HeLa cells (Fig. 4B) was subjected to sequence analysis. The variant virus had acquired a duplication of IRES and insert sequences (duplicated sequences are boxed in gray). The duplication was in frame, containing two tandem AUG codons in Kozak context (capital letters). (B) RPδ6/SIVp17(21) virus (filled diamonds) exhibited propagation kinetics in HeLa cells that resembled those of PVS-RIPO (open squares). p.i., postinfection.
SIVp17 expression.
A larger plaque phenotype, robust replication in HeLa cells (Fig. 5B), and genetic stability with full retention of the insert after five passages (Fig. 4B) suggested that the acquisition of the duplicated sequences confers a replicative advantage to variant RPδ6/SIVp17 over its parent. To correlate this phenotype with the expression of inserted sequences, we analyzed the kinetics of expression of SIVp17 in the enlarged RPδ6/SIVp17 variant. To that end, we conducted parallel Western blot analyses of infected cell lysates by using monoclonal antibodies against poliovirus gene products 2C/2BC and simian anti-simian human immunodeficiency virus (α-SHIV) polyclonal antibody sera (Fig. 6). These analyses revealed the rate of synthesis and proteolytic processing of SIVp17 released from the fusion polyprotein to be in step with the kinetics and processing of cognate viral gene products (compare synthesis rates over time in Fig. 6A and B). The recombinant expression construct, by virtue of the duplicated sequence element, contained two tandem initiation codons in Kozak context (Fig. 5A). Simian α-SHIV sera recognized a protein of 21 kDa (Fig. 6B), in accordance with initiation at the 5′-most AUG. Proteolytic processing of the fusion polyprotein produced an SIVp17(21) variant enlarged by 38 amino acids encoded by the duplication, yielding a size increase of about 4 kDa (Fig. 6B).
FIG. 6.
Western blot analysis of the kinetics of viral and foreign gene expression in HeLa cells infected with RPδ6/SIVp17(21). Lane M, molecular mass marker. (A) A monoclonal antibody against the poliovirus gene products 2C and its precursor 2BC was used in Western blot assays of cell lysates obtained at the indicated intervals postinfection (p.i.). Initial viral gene expression could be detected at 3 h p.i. and reached its peak at 6 h p.i. (B) Serum from an SHIV-infected rhesus macaque was used to sample infected cell lysates assayed in panel A for expression of SIVp17(21). In parallel with native viral gene expression, SIVp17(21) could be detected at 3 h p.i. Synthesis greatly increased until 6 h p.i., synchronous with viral gene expression. The gel migration rate of SIVp17(21) is in accordance with that of an enlarged gene product produced through initiation at the first Y(n)X(m)AUG motif in variant RPδ6/SIVp17(21).
Our findings for RPδ6/SIVp17(21) indicated significantly more efficient expression of insert sequences with regard to synthesis rate and stability than with conventional fusion polyprotein vectors containing full-length IRES elements (e.g., RIPO/SIVp17). All attempts to demonstrate SIVp17 expression with RIPO/SIVp17 under the experimental conditions used to demonstrate SIVp17(21) expression failed (data not shown). This is not surprising, considering the exceedingly poor growth of the parent construct and very rapid deletion of foreign sequences.
RPδ6/SIVp17(21) stability.
To evaluate the long-term genetic stability of RPδ6/SIVp17(21), we performed RT-PCR analyses from infected cell lysates collected over 20 passages (Fig. 7A). Genetic stability of the recombinant was finite, because after nine passages, two prominent deletion variants emerged. These deletion variants eventually eclipsed replication of the full-length recombinant (Fig. 7A). Western blot analyses of cell lysates from consecutive passages revealed solid expression of SIVp17(21) to overlap with the positive identification of the full-length recombinant construct by RT-PCR (Fig. 7B). However, expression of SIVp17(21) could be detected up to the 14th passage, where the full-length insert could no longer be amplified (compare Fig. 7A and B). This observation may be explained by the preferential amplification of shorter fragments in a mixture of different-sized cDNA templates present at later passages.
FIG. 7.
Genetic stability of variant RPδ6/SIVp17(21) after extended passages. (A) RT-PCR analysis of 20 passages of variant RPδ6/SIVp17(21) in HeLa cells. Methods and primers are those employed for Fig. 4A and B. The source of the template cDNA is indicated by the following lane designations: M, molecular mass marker; P, original RPδ6/SIVp17 plasmid DNA; T, lysate of cells transfected with RPδ6/SIVp17 transcript RNA; 1 through 20, individual passages; P−, RPδ6 plasmid DNA). Two distinct deletion variants emerged after the ninth passage, slowly replacing the full-length RPδ6/SIVp17(21) variant. Deletions were comparatively minor [fragments corresponding to partially deleted insert sequences are labeled (i) and (ii)]. The amplicon from the P− template represents the expected size of the fragment obtained from virus genomes devoid of any insert (compare with Fig. 4A). (B) Comparative analysis of SIVp17(21) expression by variant RPδ6/SIVp17(21) after transfection of transcript RNA and 20 subsequent passages in HeLa cells. No expression of SIVp17 was observed in transfected HeLa cells and the first passage under experimental conditions that readily revealed SIVp17(21) expression in later passages. (C) Genetic structure and sequence of two deletion variants (i and ii from panel A) isolated from variant RPδ6/SIVp17(21) after 20 passages in HeLa cells. Internal deletions of 114 and 240 nt, respectively, were flanked by identical complementary sequences. Duplicated sequences and engineered stem-loop structures were not affected by deletion events.
Sequencing of the insert region of both variants recovered from the 20th passage revealed deletions of 114 and 240 nt, (Fig. 7C). These deletions occurred in the ORF of SIVp17 and did not affect the engineered 3′ IRES stem-loop structure or duplicated sequences [the repeat Y(n)X(m)AUG motif and the AUG stem-loop structure that emerged upon passaging of the RPδ6/SIVp17 parent were fully retained] (Fig. 7C). Both deletion sites were flanked by identical complementary stretches of nucleotides (CAUGUU…AACAGG; complementary nucleotides shown in boldface), suggesting the possibility of a “loop-out” mechanism to account for the deletion event (Fig. 7C). Even after 20 passages, the replicating RPδ6/SIVp17(21) variant retained a foreign insert of either 417 or 291 nt in length (Fig. 7C). Considering the length of the original insert (without duplicated sequences) of 420 nt, these represent retention rates of 99 and 70%, respectively.
These experiments illustrate a new approach to rational picornavirus expression vector design. The genetic plasticity of the 3′ IRES element allows certain structural features to be reconstituted by heterologous sequences coding for foreign gene products. Although apparently dispensable for efficient replication in cell culture or in vitro translation assays (Fig. 3C) (10, 18, 25), stem-loop domain VI and adjacent sequences are highly conserved among picornaviruses and are therefore likely to confer an advantage to replicating virus. Reconstitution of this conserved structural motif with heterologous coding sequences appears to facilitate the significantly enhanced retention of foreign inserts when compared with conventional fusion vectors in which foreign sequences simply encumber optimal viral growth. The observed partial deletions in RPδ6/SIVp17(21) occurred outside the regulatory domains and may reflect the virus's preference for a shorter insert, reducing distance between the 3′ duplicate stem-loop and the viral ORF. Experiments to assess and refine the application of this vector design strategy with other foreign ORFs and in related candidate picornavirus vectors are ongoing.
The specific mechanisms underlying the enhanced stability of the RPδ6/SIVp17(21) vector remain unclear. We view the duplication of the 3′ IRES regulatory sequences, including the Y(n)X(m)AUG and AUG stem-loop, and their maintenance over 20 passages as indicative of their critical role in virus replication. In addition, since foreign insert sequences were readily recruited to replace native regulatory domains, the virus's preference for these motifs appears to involve secondary structure rather than specific RNA sequences. Experimentation to dissect the factors conferring distinct growth, expression, and stability advantages to RPδ6/SIVp17(21) is needed.
Acknowledgments
We are grateful to D. Montefiori (Duke University) and E. Wimmer (SUNY at Stony Brook) for providing α-SHIV macaque sera and 2C/2BC antibodies, respectively. A genomic SIVmac239 cDNA clone was kindly provided by L. Alexander (Yale University).
This research was supported by Public Health Service grant CA87537 from the National Institutes of Health. M.G. is the recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences.
REFERENCES
- 1.Agol, V. I., E. V. Pilipenko, and O. R. Slobodskaya. 1996. Modification of translational control elements as a new approach to design of attenuated picornavirus strains. J. Biotechnol. 44:119-128. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, L., H. H. Lu, and E. Wimmer. 1994. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc. Natl. Acad. Sci. USA 91:1406-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almond, J. W., D. Stone, K. Burke, M. A. Skinner, A. J. Macadam, D. Wood, M. Ferguson, and P. D. Minor. 1993. Approaches to the construction of new candidate poliovirus type 3 vaccine strains. Dev. Biol. Stand. 78:161-169. [PubMed] [Google Scholar]
- 4.Altmeyer, R., N. Escriou, M. Girard, A. Palmenberg, and S. van der Werf. 1994. Attenuated Mengo virus as a vector for immunogenic human immunodeficiency virus type 1 glycoprotein 120. Proc. Natl. Acad. Sci. USA 91:9775-9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andino, R., D. S. D. Silvera, P. L. Suggett, C. J. Achacoso, C. J. Miller, D. Baltimore, and M. B. Feinberg. 1994. Engineering poliovirus as a vaccine vector for the expression of diverse antigens. Science 265:1448-1451. [DOI] [PubMed] [Google Scholar]
- 6.Arnold, G. F., D. A. Resnick, A. D. Smith, S. C. Geisler, A. K. Holmes, and E. Arnold. 1996. Chimeric rhinoviruses as tools for vaccine development and characterization of protein epitopes. Intervirology 39:72-78. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 8.Crotty, S., B. L. Lohman, F. X. Lu, S. Tang, C. J. Miller, and R. Andino. 1999. Mucosal immunization of cynomolgus macaques with two serotypes of live poliovirus vectors expressing simian immunodeficiency virus antigens: stimulation of humoral, mucosal, and cellular immunity. J. Virol. 73:9485-9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eigen, M., and C. Biebricher. 1988. Sequence space and quasispecies distribution, p. 211-245. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, 3rd ed. CRC Press, Boca Raton, Fla.
- 10.Gmyl, A. P., E. V. Pilipenko, S. V. Maslova, G. A. Belov, and V. I. Agol. 1993. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5′-untranslated region yield a variety of pseudorevertants. J. Virol. 67:6309-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gromeier, M., H. H. Lu, L. Alexander, and E. Wimmer. 1997. Attenuated poliovirus as live vector, p. 315-329. In M. M. Levine (ed.), New generation vaccines, 2nd ed. Marcel Dekker, New York, N.Y.
- 13.Gromeier, M., S. Lachmann, M. Rosenfeld, P. Gutin, and E. Wimmer. 2000. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. USA 97:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halim, S. S., D. N. Collin, and A. I. Ramsingh. 2000. A therapeutic HIV vaccine using coxsackie-HIV recombinants: a possible new strategy. AIDS Res. Hum. Retrovir. 16:1551-1558. [DOI] [PubMed] [Google Scholar]
- 15.Jang, S. K., H.-G. Kräusslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang, S. K., M. V. Davies, R. J. Kaufman, and E. Wimmer. 1989. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNS in vitro. J. Virol. 63:1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 18.Kuge, S., and A. Nomoto. 1987. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5′ noncoding sequence in viral replication. J. Virol. 61:1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meerovitch, K., R. Nicholson, and N. Sonenberg. 1991. In vitro mutational analysis of cis-acting RNA translational elements within the poliovirus type 2 5′ untranslated region. J. Virol. 65:5895-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller, S., and E. Wimmer. 1998. Expression of foreign proteins by poliovirus polyprotein fusion: analysis of genetic stability reveals rapid deletions and formation of cardioviruslike open reading frames. J. Virol. 72:20-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomoto, A., Y. F. Lee, and E. Wimmer. 1976. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc. Natl. Acad. Sci. USA 74:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of tranlation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 23.Pelletier, J., and N. Sonenberg. 1989. Internal binding of eukaryotic ribosomes on poliovirus RNA: translation in HeLa cell extracts. J. Virol. 63:441-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pestova, T. V., C. U. T. Hellen, and E. Wimmer. 1991. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J. Virol. 65:6194-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestova, T. V., C. U. Hellen, and E. Wimmer. 1994. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology 204:729-737. [DOI] [PubMed] [Google Scholar]
- 26.Pilipenko, E. V., A. P. Gmyl, S. V. Maslova, Y. V. Svitkin, A. N. Sinyakov, and V. I. Agol. 1992. Prokaryotic-like cis-elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell 68:119-131. [DOI] [PubMed] [Google Scholar]
- 27.Schnell, M. 2001. Viral vectors as potential HIV-1 vaccines. FEMS Microbiol. Lett. 200:123-129. [DOI] [PubMed] [Google Scholar]
- 28.Wimmer, E., C. T. Hellen, and X. M. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]