Abstract
The envelope glycoprotein (Env) complex of human immunodeficiency virus type 1 has evolved a structure that is minimally immunogenic while retaining its natural function of receptor-mediated virus-cell fusion. The Env complex is trimeric; its six individual subunits (three gp120 and three gp41 subunits) are associated by relatively weak, noncovalent interactions. The induction of neutralizing antibodies after vaccination with individual Env subunits has proven very difficult, probably because they are inadequate mimics of the native complex. Our hypothesis is that a stable form of the Env complex, perhaps with additional modifications to rationally alter its antigenic structure, may be a better immunogen than the individual subunits. A soluble form of Env, SOS gp140, can be made that has gp120 stably linked to the gp41 ectodomain by an intermolecular disulfide bond. This protein is fully cleaved at the proteolysis site between gp120 and gp41. However, the gp41-gp41 interactions in SOS gp140 are too weak to maintain the protein in a trimeric configuration. Consequently, purified SOS gp140 is a monomer (N. Schülke, M. S. Vesanen, R. W. Sanders, P. Zhu, D. J. Anselma, A. R. Villa, P. W. H. I. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson, J. Virol. 76:7760-7776, 2002). Here we describe modifications of SOS gp140 that increase its trimer stability. A variant SOS gp140, designated SOSIP gp140, contains an isoleucine-to-proline substitution at position 559 in the N-terminal heptad repeat region of gp41. This protein is fully cleaved, has favorable antigenic properties, and is predominantly trimeric. SOSIP gp140 trimers are noncovalently associated and can be partially purified by gel filtration chromatography. These gp140 trimers are dissociated into monomers by anionic detergents or heat but are relatively resistant to nonionic detergents, high salt concentrations, or exposure to a mildly acidic pH. SOSIP gp140 should be a useful reagent for structural and immunogenicity studies.
The envelope glycoprotein (Env) complex of human immunodeficiency virus type 1 (HIV-1) mediates viral entry into CD4+ cells. The sequential binding of the surface subunit gp120 to the CD4 receptor and a coreceptor, usually CCR5 or CXCR4, induces conformational changes in the Env complex. These alterations in protein structure eventually enable the insertion of the hydrophobic fusion peptide of the transmembrane subunit, gp41, into the cell membrane. Subsequently, the viral and cell membranes fuse, allowing the release of the viral core into the cytoplasm and the initiation of a new cycle of infection (for reviews, see references 17, 24, 25, 31, 61, and 99). gp120 and gp41 are synthesized as a gp160 precursor that is cleaved within the cell to yield the native, prefusion form of the Env complex (39, 55, 63). This is generally considered to be a trimeric structure, containing three gp120 and three gp41 moieties held together by noncovalent interactions (31, 73, 99). The native Env complex is unstable, because the noncovalent intersubunit interactions that hold gp120 to gp41 are weak, as are the intermolecular interactions between the gp41 moieties (31, 73, 99). This instability is probably essential for receptor-triggered conformational changes to occur, but it does cause a problem for attempts to express the native complex as a recombinant protein (5).
One reason to prepare recombinant forms of the native Env complex is for structural studies. At present, structural information on HIV-1 Env is limited to core fragments of gp120 in the CD4-associated configuration and the six-helix bundle form of the gp41 core, which represent its terminal, most stable configuration (10, 16, 45, 46, 51, 54, 89, 94). Although the six-helix bundle is often referred to as the fusogenic form of gp41, this term can be misleading, because it is the formation and not the mere presence of the six-helix bundle that drives membrane fusion (25, 35, 56). Hence, antibodies to the six-helix bundle cannot interfere with fusion and are nonneutralizing (42, 59, 65, 71, 90). We therefore use the term “postfusion form” of gp41 when referring to the six-helix bundle, to reflect its persistence on infected cells as a major immunogen after the fusion process is complete and on virions when conformational changes in Env leading to gp120 shedding have occurred prematurely or abortively (25). Most antibodies to gp41 in HIV-1-infected individuals recognize this postfusion conformation (36, 59, 71, 78, 90, 100).
A second reason to make the native Env complex is to study its immunogenicity and determine its suitability as a vaccine antigen. The few monoclonal antibodies (MAbs) that potently neutralize HIV-1 all recognize epitopes exposed on the native Env complex and may well have been induced by such a complex (9, 34, 58, 69-71, 73, 81). In contrast, nonneutralizing MAbs do not bind to the native complex and probably represent immune responses to nonnative forms of Env, such as uncleaved gp160 precursors, dissociated gp120 subunits, or the six-helix bundle, postfusion form of gp41 (9, 59, 69, 70, 73).
Eliciting neutralizing antibodies by vaccination with any form of Env is problematic, because of the mechanisms that the native Env complex has evolved to shield its most critical sites and to limit its overall immunogenicity. Thus, conserved regions of gp120 involved in receptor binding are shielded by variable loops and by extensive glycosylation. The CD4 binding site (CD4BS) is recessed, and the coreceptor binding site is only formed or exposed for a short period after CD4 has already bound, thereby limiting the time and space available for antibody interference (57, 58, 66, 97). Whether such defense mechanisms can be overcome by vaccine-induced antibodies remains uncertain (59, 71). Our approach to the problem has been to try to make a stabilized native Env complex which may then have to be further modified to improve its immunogenicity.
The lability of the noncovalent interaction between gp120 and the gp41 ectodomain (gp41ECTO) is an obstacle to the production of stable, fully processed HIV-1 Env trimers. It was previously reported that the association between gp120 and gp41ECTO can be stabilized by the introduction of a correctly positioned intermolecular disulfide bond to make a soluble form of Env, SOS gp140 (5, 79). In the presence of cotransfected furin, the peptide bond linking gp120 to gp41ECTO is cleaved, allowing the production of properly processed gp140 (5, 79). It was initially reported that oligomeric proteins were present in supernatants from 293T cells transiently expressing SOS gp140 (5). However, these oligomers were not abundant, and they did not survive purification: purified SOS gp140 is a monomeric protein (83). We now describe a way to further stabilize the gp41-gp41 interactions in SOS gp140. We have introduced amino acid substitutions into the N-terminal heptad repeat region of gp41 that we hypothesize destabilize the postfusion form of the protein and thereby render the transition to this configuration less likely. One such protein, SOS I559P gp140 (designated SOSIP gp140), is properly folded, proteolytically cleaved, substantially trimeric, and has appropriate receptor binding and antigenic properties. The SOSIP gp140 trimer can be converted to the monomeric form by heat or anionic detergents but is partially resistant to nonionic detergents.
MATERIALS AND METHODS
Env expression.
Various forms of virus strain JR-FL gp140 were expressed in 293T cells from the pPPI4 vector and furin was expressed from pcDNA3.1-Furin as described previously (5, 79). Uncleaved JR-FL gp140 (gp140UNC) with amino acid substitutions to prevent its proteolytic processing has been described elsewhere (5). Specific mutations were made by using a QuickChange mutagenesis kit (Stratagene, La Jolla, Calif.). Random mutations were generated with primers that could contain any nucleotide at the relevant positions. Numbering is based on the virus strain HXB2 Env sequence. Recombinant JR-FL gp120 has been described elsewhere (92).
Antibody binding assays.
The procedures and MAbs used to perform radioimmunoprecipitation assays have all been described elsewhere (1, 5, 60, 80, 93). MAbs were provided by James Robinson (17b and 2.2B), George Lewis (B12), Hermann Katinger (2F5, 2G12, 4D4, and 4E10), and Dennis Burton (b12).
SDS-PAGE, BN-PAGE, and Western blot analyses.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), blue native (BN)-PAGE, and Western blot analyses were performed as described previously (5, 79, 83). Culture supernatants from transiently transfected 293T cells were concentrated 10-fold before gel electrophoresis by using Ultrafree-15 concentrators (Millipore, Bedford, Mass.).
Gel filtration analysis of envelope glycoproteins.
Supernatants from 293T cells, transfected with pPPI4-gp140 plus pcDNA3.1-Furin, were concentrated 100-fold and then size fractionated by using an analytical Superdex 200 HR 10/30 column equilibrated with 100 mM NaCl-50 mM sodium phosphate (pH 7.0) (phosphate-buffered saline [PBS]) (Amersham-Pharmacia, Piscataway, N.J.). Fractions (300 μl) were analyzed by using SDS-PAGE and BN-PAGE; in both cases, Western blotting was used to detect Env. The column was calibrated by using protein standards of known sizes (HMW-standard; Amersham-Pharmacia).
Trimer stability experiments.
Eluates from gel filtration columns, containing SOSIP gp140 trimers, were incubated with various reagents or under different conditions and then analyzed by BN-PAGE and Western blotting. The following detergents were obtained from the following suppliers: SDS, Sigma, St. Louis, Mo.; t-octylphenoxypolyethoxyethanol (Triton X-100), Sigma; polyoxyethylene sorbitan monolaurate (Tween 20), Sigma; ethylphenyl-polyethylene glycol (Nonidet P-40), United States Biochemicals, Cleveland, Ohio; n-octyl β-d-glucopyranoside, Sigma; and Empigen BB, 30% solution (Empigen), Calbiochem, La Jolla, Calif.
Peptide production.
Plasmid pN36/C34JR-FL, encoding the HIV-1JR-FL N36(L6)C34 model peptide (which consists of the N36 and C34 peptides connected via a short peptide linker that replaces the disulfide-bonded loop region of gp41ECTO), was derived from pN36/C34HXB2 (51). Amino acid substitutions were introduced into the N36 segment of pN36/34JR-FL by using the method of Kunkel et al. (44) and then verified by DNA sequencing. All recombinant peptides were expressed in Escherichia coli strain BL21(DE3)/pLysS (Novagen, Madison, Wis.). The bacteria were grown at 37°C in Luria-Bertani medium to an optical density at 600 nm of 0.8 and induced with isopropylthio-β-d-galactoside for 3 to 4 h. Cells were lysed at 0°C with glacial acetic acid. The bacterial lysate was centrifuged (35,000 × g for 30 min) to separate the soluble fraction from inclusion bodies. The soluble fraction, containing denatured peptides, was dialyzed in 5% acetic acid overnight at room temperature. Peptides were purified from the soluble fraction to homogeneity by reverse-phase high-performance liquid chromatography (Waters, Milford, Mass.) with a C18 preparative column (Vydac, Hesperia, Calif.) and a water-acetonitrile gradient in the presence of 0.1% trifluoroacetic acid and then lyophilized. The molecular weight of each peptide was confirmed by using matrix-assisted laser desorption ionization-time of flight mass spectrometry (PerSeptive Biosystems, Framingham, Mass.). The concentration of each peptide was determined at 280 nm after solubilization in 6 M guanidinium chloride (32).
CD spectroscopy.
High-performance liquid chromatography-purified peptides were solubilized in 6 M guanidinium chloride-10 mM Tris-HCl (pH 7.0) and refolded by dilution in PBS at a neutral pH. The single-point-substituted variant peptides were named according to the position of the substitution. Circular dichroism (CD) experiments were performed by using an Aviv 62A DS CD spectrometer. The wavelength dependence of molar ellipticity, θ, was monitored at 4°C by using a 10 μM peptide solution in PBS. Helix content was calculated by the method of Chen et al. (22). Thermal stability was determined by monitoring the change in the CD signal at 222 nm (θ222) as a function of temperature. Thermal melts were performed in 2°C increments with an equilibration time of 2 min at the desired temperature and an integration time of 30 s. All melts were reversible. Superimposable folding and unfolding curves were observed, and >90% of the signal was regained upon cooling. The melting temperatures, or midpoints of the cooperative thermal unfolding transitions (Tm), were determined from the maximum of the first derivative, with respect to the reciprocal of the temperature, of the θ222 values (12). The error in the estimation of Tm was ±0.5°C.
Sedimentation equilibrium analysis.
An XL-A analytical ultracentrifuge equipped with an An-60 Ti rotor (Beckman Coulter, Fullerton, Calif.) was used for sedimentation equilibrium analysis. Peptide solutions were dialyzed overnight against PBS, loaded at initial concentrations of 10, 30, and 100 μM, and analyzed at rotor speeds of 20,000 and 23,000 rpm at 20°C. Data sets were fitted simultaneously to a single-species model of ln(absorbance) versus (radial distances)2 by using the program NONLIN (43). Protein partial specific volume and solvent density were calculated as described by Laue et al. (48).
RESULTS
Stabilization of the SOS gp140 trimer by mutagenesis.
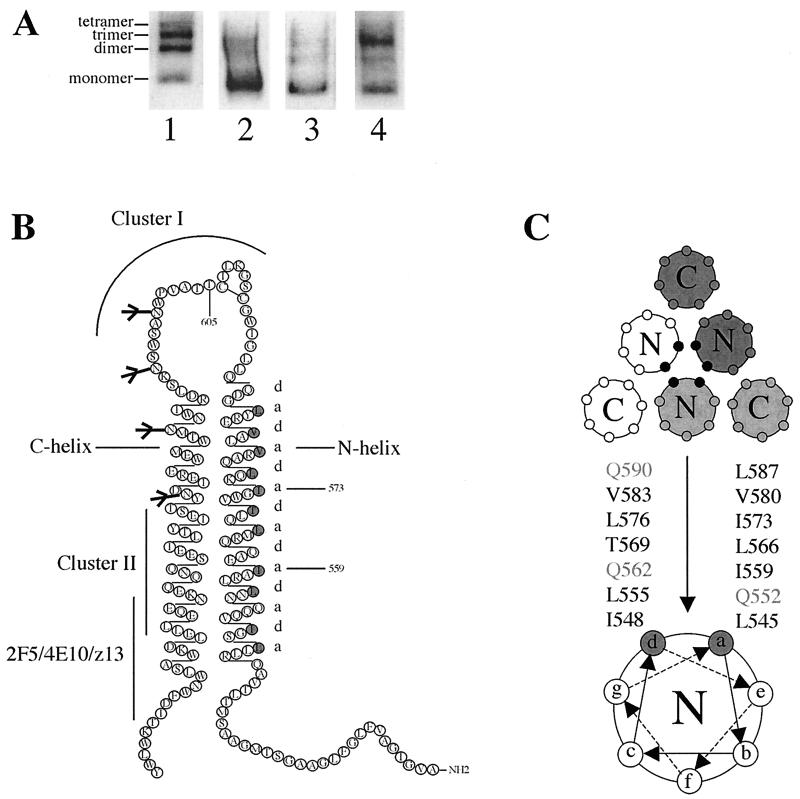
Although purified, cleaved SOS gp140 is a monomer, we have observed oligomeric forms of this protein in freshly prepared supernatants from transiently transfected 293T cells (5, 83). An example of SOS gp140 oligomers, together with the experiment-to-experiment variation in their abundance, was provided by a BN-PAGE analysis of four different 293T cell transfections (Fig. 1A). In some preparations, a significant fraction of SOS gp140 is oligomeric (Fig. 1A, lanes 1 and 4); in others, a negligible proportion is (lanes 2 and 3).
FIG. 1.
Instability of SOS gp140 and a mutagenesis strategy for gp41ECTO. (A) BN-PAGE analysis of SOS gp140 present in culture supernatants derived from four individual experiments each involving transient transfection of 293T cells with the same Env and furin expression plasmids. (B) The residues at positions a and d in the N-terminal heptad repeat of gp41ECTO, depicted in gray in the secondary structure, were substituted in this study. Major MAb epitopes are indicated (26, 68, 100, 106). (C) Schematic representation of a cross-section of the six-helix bundle, postfusion form of gp41ECTO and a helical-wheel representation of one N-terminal helix. The residues at positions a and d of the N-terminal heptad repeat (black symbols) form the trimer interface.
The above observations suggest that the oligomeric form of SOS gp140, although clearly unstable, might not be too far from stability. We therefore adopted a mutagenesis strategy to try to increase the stability of cleaved, oligomeric SOS gp140. We focused on altering the gp41ECTO sequence, because of the clear preponderance of evidence that interactions between gp41ECTO subunits are responsible for the oligomerization of gp140 (29, 85, 99). However, no structural or other data are available on the likely points of contact between the gp41 moieties in the native, prefusion form of the envelope glycoprotein trimer. We therefore adopted a more theoretical approach, based on the premise that destabilization of the postfusion state of gp41 might stabilize its prefusion configuration, by shifting the conformational equilibrium in favor of the prefusion state (40, 41, 49-51). A plethora of structural and genetic data on the postfusion, six-helix bundle structure of gp41 was available to guide our mutagenesis strategy (3, 10, 13, 16, 20, 21, 39, 49, 51, 54, 74, 89, 94-96)
The trimeric stability of gp41 in the six-helix bundle form of the protein is determined by the residues at positions a and d of the N-terminal heptad repeat region (40, 41, 52) (Fig. 1B and C). Most of these amino acids are absolutely conserved. Hydrophobic Val, Leu, and Ile residues form the apolar interface of the three N-terminal helices; these residues are critical determinants of the folding and thermal stability of the six-helix bundle (84). We assumed that making nonconservative substitutions at these conserved, hydrophobic residues would destabilize the six-helix bundle and so, by reducing the probability of its formation, cause the gp41ECTO subunits to remain in their desired, prefusion configuration. The hydrophilic glutamine residues at positions 552, 562, and 590 were initially left unaltered, since buried polar interactions involving these residues confer structural specificity for the formation of the N-terminal coiled-coil trimer at the expense of its thermal stability (41) (Fig. 1B and C). Substitutions at the Thr-569 residue were also evaluated.
Because we could not predict which amino acids would be tolerated in the prefusion configuration of gp41ECTO, we performed random mutagenesis. Hence, various amino acids with different biochemical properties were introduced in place of the targeted Val, Ile, and Leu residues. An emphasis of our mutagenesis approach was to alter the Ile-559 and Ile-573 residues, because core isoleucines are known to confer the greatest stability to trimeric coiled coils (also known as isoleucine zippers) (38). In most simian immunodeficiency virus (SIV) strains, Val and Thr residues are found, respectively, at these positions, where they may serve to destabilize the postfusion, coiled-coil structure (40, 49, 50). Of note is that SIV gp140UNC has a greater tendency to be trimeric than the corresponding HIV-1 protein (15, 19).
The use of BN-PAGE to monitor the oligomeric state of HIV-1 gp140 has been described elsewhere (83). This technique served as a screening assay to identify more stable SOS gp140 variants, i.e., ones that remained trimeric under conditions in which unmodified SOS gp140 ran mainly as a monomer. The SOS gp140 variants were expressed in transiently transfected 293T cells, in the presence of cotransfected furin to facilitate gp120-gp41 cleavage. The effects of various single-residue substitutions on the expression and trimer stability of these SOS gp140 variants are summarized in Table 1. An example of how BN-PAGE was used to derive this information is shown in Fig. 2A.
TABLE 1.
Summary of characteristics of SOS gp140 variants
| Residue | Mutation in SOS gp140 | Expressiona | Trimer stabilityb | Cleavagec |
|---|---|---|---|---|
| L545 | F | ± | ± | |
| N | ++ | ± | ||
| P | ++ | ± | ||
| G | ++ | ± | ||
| I548 | V | ++ | ± | |
| L | ++ | ± | ||
| H | ++ | ± | ||
| N | ++ | ± | ||
| S | ++ | ± | ||
| G | ++ | ± | ||
| R | ++ | ± | ||
| L555 | V | − | ND | |
| W | − | ND | ||
| Y | − | ND | ||
| S | − | ND | ||
| P | − | ND | ||
| I559 | V | + | ± | |
| F | ± | ++ | ||
| N | − | ND | ||
| P | ++ | ++ | ++ | |
| G | + | ++ | ++ | |
| R | + | ++ | ||
| L566 | V | + | + | ++ |
| I | ± | ± | ||
| N | + | + | ||
| T | + | + | ||
| P | + | ± | ||
| K | ± | ± | ||
| T569 | S | + | ± | |
| P | + | + | ++ | |
| K | + | + | ||
| E | − | ND | ||
| I573 | L | ++ | ± | |
| F | ++ | ± | ||
| Y | ++ | ± | ||
| Q | ++ | ± | ||
| N | ++ | ± | ||
| T | ++ | ± | ||
| P | ++ | ± | ||
| G | ++ | ± | ||
| K | ++ | ± | ||
| L576 | V | ± | ± | |
| F | ± | ± | ||
| Y | ± | ± | ||
| Q | ± | ± | ||
| N | ± | ± | ||
| G | ± | ± | ||
| K | ± | ± | ||
| V580 | L | ++ | ± | |
| H | ++ | ± | ||
| T | ++ | ± | ||
| P | ++ | ± | ||
| G | ++ | ± | ||
| V583 | L | ++ | ± | |
| Q | ++ | ± | ||
| N | ++ | ± | ||
| S | ++ | ± | ||
| P | ++ | ± | ||
| R | ++ | ± | ||
| K | ++ | ± | ||
| L587 | A | ± | ± | |
| P | ± | ± | ||
| R | ± | ± | ||
| D | ± | ± | ||
| E | ± | ± |
Relative scale: −, no expression; ±, minimal expression; +, expression level lower than that of wild-type SOS gp140; ++, expression level equivalent to that of wild-type SOS gp140; +++, expression level greater than that of wild-type SOS gp140. All proteins were expressed in the presence of cotransfected furin, which decreases gp140 expression (5, 6, 79). The data were derived from at least three independent transfections.
As assessed by BN-PAGE. The trimer stability of SOS gp140 was set at ± (some trimers were present in some transfections). The maximum amount of trimers (++) was found only in gp140 variants containing substitutions at residue Ile-559 and ranged from 40 to >90% of total Env expression in independent transfections. The data were derived from at least three independent transfections. ND, not determined (no expression).
Data were derived from Fig. 2C. Symbols are as defined in footnote a.
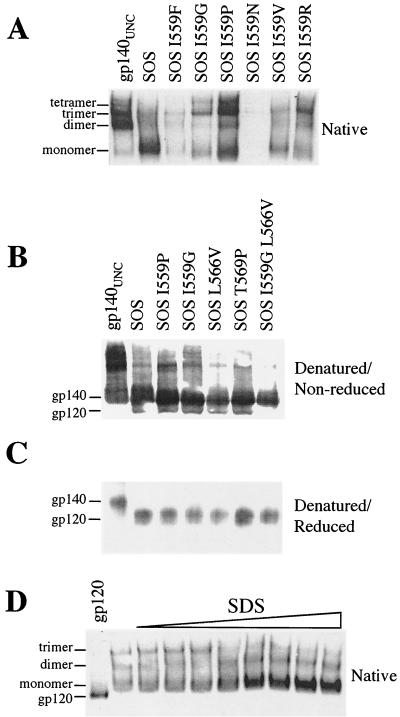
FIG. 2.
PAGE analysis of SOS gp140. (A) BN-PAGE of SOS gp140 containing changes at position 559. gp140UNC was included for comparison. (B and C) SDS-PAGE of SOS gp140 variants under nonreducing (B) or reducing (C) conditions. (D) BN-PAGE analysis after treatment with increasing SDS concentrations. SOSIG gp140 was treated with SDS for 1 h at 25°C. The SDS concentrations used were 0, 0.005, 0.01, 0.015, 0.02, 0.025, 0.03, 0.035, and 0.04%, increasing from left to right. BN-PAGE analysis was then performed. Monomeric gp120 (20 ng) served as a molecular weight standard.
Many of the randomly generated SOS gp140 mutants were not expressed or were expressed poorly, particularly those with substitutions at positions 555, 576, and 587. This result probably occurred because amino acid changes at these positions have adverse effects on protein folding. However, other conserved residues (e.g., those at positions 548, 573, 580, and 583) were more tolerant of substitutions, in that the SOS gp140 mutants were still expressed efficiently. The effects on SOS gp140 expression of substitutions at other positions, notably, 545, 559, 566, and 569, were dependent on the identity of the amino acid introduced. Thus, at position 545, a Leu-to-Phe change (L545F) reduced SOS gp140 expression, whereas the introduction of Asn (L545N), Pro (L545P), or Gly (L545G) at this position had little effect. Similarly, Ile-to-Phe or Ile-to-Asn substitutions at position 559 (I559F or I559N, respectively) severely diminished SOS gp140 expression (Fig. 2A, third and sixth lanes). In contrast, the introduction of Val (I559V), Gly (I559G), or Arg (I559R) residues at position 559 had a lesser effect on SOS gp140 expression, and a Pro (I559P) substitution had no adverse effect at all (Fig. 2A, fourth, seventh, and eighth lanes). Other examples of how the introduced residue can have a variable effect on SOS gp140 expression include changes at residues 566 and 569 (Table 1).
We used BN-PAGE to determine whether there were differences in oligomer stability among the SOS gp140 mutants that were efficiently expressed. Under native conditions, wild-type SOS gp140 migrates predominantly as a monomer, with some dimeric and trimeric species also present (Fig. 2A, second lane). The proportion of SOS gp140 that is oligomeric varies from experiment to experiment (Fig. 1A), but the gel shown in Fig. 2A is typical of what is most commonly observed. In contrast, gp140UNC migrates as oligomeric forms, with the dimer predominating (Fig. 2A, first lane).
Most of the amino acid substitutions consistent with the efficient expression of SOS gp140 had little effect on the extent of its oligomerization (Table 1). However, several substitutions at position 559 clearly altered the oligomerization state of SOS gp140 (Fig. 2A). Thus, when Ile-559 was replaced by a nonconservative Gly (I559G), Arg (I559R), or Pro (I559P) residue, the most abundant protein form was consistently the trimer (Fig. 2A, fourth, fifth, and eighth lanes). In contrast, the conservative Ile-to-Val substitution at position 559 had no effect, in that the I559V protein and wild-type SOS gp140 were indistinguishable (Fig. 2A, second and seventh lanes). Some substitutions at positions 566 and 569 (e.g., L566V and T569P) marginally increased the proportions of SOS gp140 variants that were trimeric (Table 1). Note that the percentage of trimers varied per transfection, but the amount of trimer in the SOS gp140 I559P preparation shown in Fig. 2A (see also Fig. 3) was typical. Sometimes, larger amounts of trimer, up to 90%, were observed.
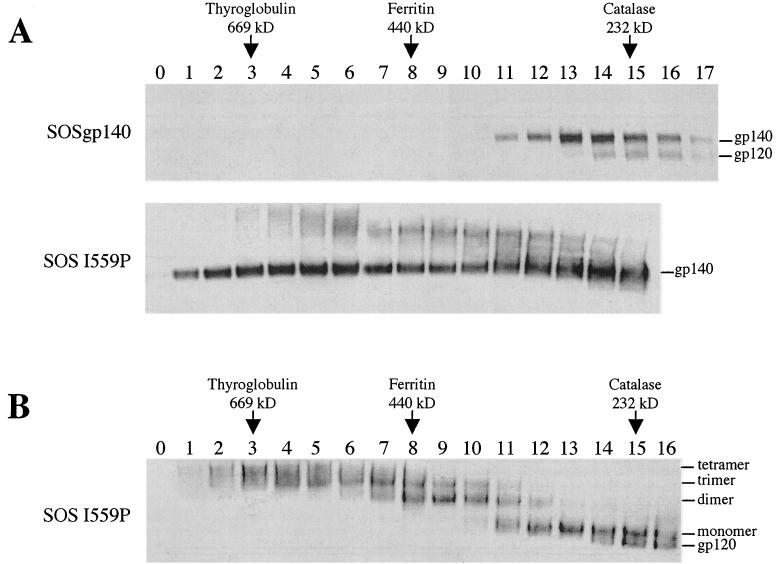
FIG. 3.
Gel filtration analysis of SOSIP gp140. (A) SOS gp140 and SOSIP gp140 (SOS I559P) were fractionated on a Superdex 200 column. The individual fractions were analyzed by SDS-PAGE and Western blotting. (B) The SOSIP gp140 fractions from panel A were analyzed by BN-PAGE and Western blotting. The elution positions (peak fractions) of standard proteins are indicated.
There appeared to be a correlation between the expression of some SOS gp140 mutants and the extent to which they were oligomeric. Thus, substitutions at positions 559, 566, and 569 affected both the expression and the oligomerization of SOS gp140 (Table 1 and Fig. 2A). Residues at these positions may be particularly important for the correct folding of the trimeric, prefusion form of gp140.
The above experiments show that the introduction of the helix-destabilizing residue Gly or Pro at position 559 increased the tendency of SOS gp140 to form trimers. Hence, this region of gp140 may not be helical in the prefusion configuration of the protein. In the influenza virus hemagglutinin (HA2) protein, a loop-to-helix transition induced by exposure to low pH is essential for the formation of the fusion-intermediate, extended coiled-coil conformation (8). Various proline substitutions in this particular region of HA2 permit the production of proteolytically cleaved trimers, but these mutants are fusion impaired because they cannot undergo the critical loop-to-helix transition (37, 75). We therefore investigated whether proline substitutions at other residues near position 559 of HIV-1 gp41 could have the same, trimer-stabilizing effect as the I559P substitution. However, although most of the mutants could be efficiently expressed, none of the substitutions stabilized the trimeric form of SOS gp140 in the same way that the I559P substitution did (Table 2).
TABLE 2.
Effect of proline substitutions at residues near position 559
We also tested various combinations of amino acid substitutions. Generally, in all of the double or triple mutants that were evaluated, the presence of either the I559P or the I559G change was essential to obtain the trimer-stabilized phenotype. A few other substitutions (e.g., L566V and T569P) had a marginally stabilizing effect on SOS gp140 trimers (Tables 1 and 2).
The I559G and I559P substitutions that conferred increased stability on SOS gp140 were also made in the context of HIV-1LAI expressing membrane-bound Env to determine whether they were compatible with Env function. Both substitutions completely abolished Env function and virus replication (R. W. Sanders and B. Berkhout, unpublished results). This finding is consistent with previous reports of the deleterious effects of most substitutions in this region of gp41 (13, 20, 21, 74, 95, 96).
Stabilized SOS trimers form noncovalently and are cleaved.
Based on the above results, we focused on the SOSgp140 I559P and I559G variants for further analysis. These proteins are designated SOSIP gp140 and SOSIG gp140, respectively. Although SOSIP gp140 was consistently expressed at higher levels than SOSIG gp140, the presence of a glycine residue at position 559 might confer flexibility to the latter protein. We considered that any such flexibility might prove useful if and when the I559G substitution were combined with other modifications. We also further studied the SOS gp140 L566V and T569P variants to determine whether similar results would be obtained with trimers that had been stabilized, even to only a limited extent, by substitutions at positions other than 559.
To investigate whether the oligomeric species present in the SOSIP gp140, SOSIG gp140, L566V, and T569P preparations were covalently or noncovalently associated, we analyzed them by using denaturing but nonreducing SDS-PAGE (Fig. 2B). gp140UNC was included for comparison (Fig. 2B, first lane). As expected, SOS gp140 migrated predominantly as a 140-kDa species (Fig. 2B, second lane). However, some higher-molecular-weight, SDS-resistant species were also present in both the SOS gp140 and the gp140UNC preparations. The higher-molecular-weight species were only a minor component of the SOS gp140 preparation, but they were the predominant form in the gp140UNC preparation (Fig. 2B, first lane). We believe that the higher-molecular-weight forms of these proteins are predominantly covalently associated dimers and, in some cases, tetramers that could be dimers of dimers (see below).
The SOS gp140 variants were all indistinguishable from wild-type SOS gp140, in that the predominant species after SDS treatment were always 140-kDa monomers (Fig. 2B, third through seventh lanes). Thus, the trimeric forms of SOSIP gp140 and SOSIG gp140 are not created by the aberrant formation of intermolecular disulfide bonds; instead, the protein is associated by noncovalent interactions.
Our goal is to make stable, oligomeric gp140 that is properly processed at the cleavage site between the gp120 and gp41 subunits. We therefore analyzed whether the SOSIP gp140, SOSIG gp140, L566V, and T569P variants were processed appropriately. The proteins were boiled in the presence of SDS and dithiothreitol (DTT), to achieve both denaturation and reduction, before SDS-PAGE analysis (Fig. 2C). Under these conditions, each of the SOS gp140 variants was converted to gp120 and gp41ECTO forms (Fig. 2C and data not shown). Thus, each of the gp140 variants was substantially (>90%) cleaved, in that the 140-kDa band did not survive DTT treatment (Fig. 2C, second through seventh lanes). In contrast, gp140UNC was unaffected by DTT and still migrated as a 140-kDa band, because it possesses a peptide bond between the gp120 and gp41 subunits (Fig. 2C, first lane). Thus, the increased trimer stability of the SOSIP gp140, SOSIG gp140, L566V, and T569P variants is not caused by or associated with any cleavage defect.
Most of the SOS gp140 preparations analyzed by SDS-PAGE contained a small percentage of SDS-resistant oligomers, and these forms of gp140 were relatively abundant in the gp140UNC preparation (Fig. 2B). Since these higher-molecular-weight forms of gp140 were not observed when DTT was used to reduce disulfide bonds (Fig. 2C), they presumably represent protein forms that are linked via aberrant, intermolecular disulfide bonds (83). To determine whether the higher-molecular-weight proteins were dimers or trimers, we treated SOSIG gp140 with increasing amounts of SDS and then performed BN-PAGE analysis (Fig. 2D). In the absence of SDS, trimers, dimers, and monomers were present at approximately equal proportions in the SOSIG gp140 preparation (Fig. 2D, second lane). As the SDS concentration was increased, however, the trimer band completely disappeared, whereas the dimer band survived exposure to SDS concentrations even as high as 1% (Fig. 2B, fourth lane, and Fig. 2D). The stronger intensity of the dimer band on the Western blot is probably attributable to increased reactivity of the detecting MAb once gp140 has been denatured with SDS. The more pronounced increase in the intensity of the monomer band suggests that trimers dissociate to three monomers, rather than to a dimer and a monomer (see below). Overall, we conclude that SOSIG gp140 trimers are formed by noncovalent, SDS-sensitive bonds but that dimers are associated via aberrant, intersubunit disulfide bonds. Similar results were obtained with the SOSIP gp140 preparation (data not shown, but see Fig. 4). Wild-type SOS gp140 could not, of course, be tested in this way, as its trimeric form was too unstable.
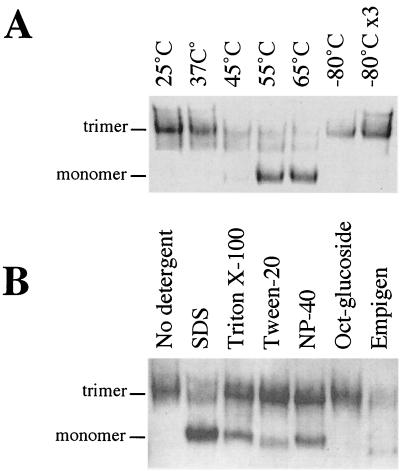
FIG. 4.
Stability of SOSIP gp140 trimers. Fractions 6 and 7 from a gel filtration profile similar to that shown in Fig. 3 were pooled as a source of SOSIP gp140 trimers. The trimers were incubated for 1 h at the temperatures indicated (A) or exposed to a 0.1% concentration of various detergents for 1 h at 25°C (B). The proteins were then analyzed by BN-PAGE and Western blotting. −80°C ×3, three freeze-thaw cycles at −80°C; NP-40, Nonidet P-40; Oct-glucoside, n-octyl β-d-glucopyranoside.
Fractionation of oligomeric gp140 species by gel filtration.
The BN-PAGE analyses showed that amino acid substitutions, in particular those at position 559, can stabilize SOS gp140 trimers. To corroborate this finding by an independent technique, unpurified gp140 secreted from Env-transfected 293T cells was studied by analytical gel filtration chromatography with a Superdex 200 column. Proteins of known molecular masses provided reference standards. These were catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa). However, it should be noted that fully glycosylated gp120 and gp140 molecules are nonglobular in shape, so gel filtration cannot precisely determine their absolute molecular masses (14, 15, 83).
The eluate fractions were collected and then analyzed by SDS-PAGE and Western blotting to identify the migration positions of various Env forms (Fig. 3). Because the input Env was unpurified, unrelated proteins are also present in the eluate, so the fractions in which Env forms eluted could not be determined by nonspecific methods. SOS gp140 was found predominantly in fractions 13, 14, and 15, corresponding to an apparent molecular mass similar to that of catalase (232 kDa) (Fig. 3A, top panel). Thus, the average apparent molecular mass of the eluted SOS gp140 monomer was ∼240 kDa, a value consistent with the value of ∼220 kDa that was reported previously, also by gel filtration (83). The small amount of gp120 present in this preparation of SOS gp140 had a slightly lower apparent molecular mass of ∼220 kDa (Fig. 3A). In most preparations of SOS gp140, a small quantity of covalently linked oligomers, probably dimers, was also seen, centered around fractions 9 and 10 (data not shown). These results confirm the previous report that SOS gp140 is usually predominantly a monomeric protein (83) (Fig. 1A).
In contrast, SOSIP gp140 eluted over a broad range in fractions 1 to 16, indicating that both oligomeric and monomeric species are present (Fig. 3A, bottom panel). A small amount of covalently linked oligomers was also observed (Fig. 2B, third lane), just as similar oligomers were usually present in SOS gp140 preparations (Fig. 2B, second lane). To resolve the different oligomeric species, the same SOSIP gp140 gel filtration fractions were then analyzed by BN-PAGE. This analysis showed that trimeric, dimeric, and monomeric proteins had been clearly resolved on the Superdex 200 column; the trimers were predominantly in fractions 4 to 9, the dimers were in fractions 7 to 11, and the monomers were in fractions 11 to 15 (Fig. 3B). Similar results were obtained with an SOS gp140 triple mutant containing the I559P, L566V, and T569P mutations (data not shown).
The apparent molecular mass of the SOSIP gp140 monomer corresponds to what was observed with wild-type SOS gp140 (∼240 kDa; see above). The retention of dimers, centered around fraction 9, corresponds to an average apparent molecular mass of ∼410 kDa, whereas the trimer (peak fraction 6) has an average apparent molecular mass of ∼520 kDa. It is notable that the trimers do not elute in a position consistent with their expected size of three times the size of a gp140 monomer. They elute at a position corresponding to a molecular mass of ∼520 kDa, as opposed to the “expected” ∼660 to 720 kDa (i.e., 3 × ∼220 to 240 kDa). The same is true, to a lesser extent, for the dimers, which elute at ∼410 kDa, compared to the “expected” ∼440 to 480 kDa (i.e., 2 × ∼220 to 240 kDa). The explanation for these results is probably that the trimers, and perhaps also the dimers, are folded into a conformation which is more compact than that of gp140 monomers. Electron microscopy studies may be able to confirm this suggestion. Alternatively, the nature and extent of glycosylation of the different oligomeric forms of gp140 may vary, because glycosylation sites on the trimer could be less accessible to modifying enzymes than the same sites on the monomer. Overall, given the limitations of gel filtration for estimating the molecular masses of nonglobular proteins (14, 15, 83), other techniques will need to be used (e.g., mass spectrometry) to establish the absolute molecular masses of the various purified oligomeric species of SOS gp140.
Clearly, SOSIP gp140 preparations are not pure trimers prior to fractionation by gel filtration (Fig. 2A and 3); monomers, dimers, and tetramers are also present. We estimate that typically 40% of an SOSIP gp140 preparation elutes in the trimer fraction. We assume that the dimers and tetramers are aberrant forms of Env that are generated by much the same, albeit unknown, processes that also cause them to be present in gp140UNC preparations (Fig. 2) (28-30, 33, 83, 101, 102). The small amount of monomers present in SOSIP gp140 preparations probably arises because the effect of the trimer-stabilizing I559P substitution is imperfect. We are presently determining how to purify SOSIP gp140 trimers from other non-Env forms that elute from gel filtration columns at similar positions.
Stability of SOSIP gp140 trimers.
The Superdex 200 column fractions corresponding to the trimer peak of the SOS gp140 variant I559P (fractions 6 and 7) were pooled for analysis of their stability (Fig. 4). The trimers were stable when incubated for 1 h at 25 and 37°C (Fig. 4A, first and second lanes), but some monomers became visible after 1 h at 45°C (third lane) and almost all of the protein was in the monomeric form after heating for 1 h at 55 or 65°C (fourth and fifth lanes). Three freeze-thaw cycles at −80°C did not convert the trimers into monomers (Fig. 4A, seventh lane). We next incubated the fractionated trimers with various detergents for 1 h at 25°C (Fig. 4B). The trimers dissociated into monomers upon incubation with 0.1% SDS, an anionic detergent (Fig. 4B, second lane), but they were at least partially resistant to the same concentration of the nonionic or zwitterionic detergents Triton X-100, Tween 20, Nonidet P-40, n-octyl β-d-glucopyranoside, and Empigen (third through seventh lanes). We also observed that SOSIP gp140 trimers did not dissociate into monomers in the presence of NaCl concentrations of up to 1.0 M or after exposure to mild acid (pH 4.0) (data not shown). The effects of various adjuvants and of long-term storage at various temperatures on the trimeric state of purified SOSIP gp140 trimers are currently under investigation.
Dimers were present at only very low levels in heat- or detergent-treated SOSIP gp140 trimers. This result suggests that the assembly units of the trimers are three equivalent monomers rather than a monomer and a dimer.
Antigenic structure analysis of stabilized SOS gp140.
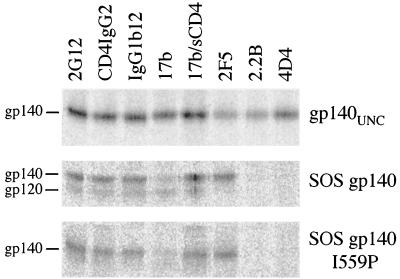
Since our goal is to make cleaved, stable gp140 trimers that mimic as closely as possible the antigenic structure of virion-associated Env, we studied the reactivity of unpurified SOSIP gp140 with a panel of MAbs and CD4-based reagents. SOS gp140 and gp140UNC were also studied for comparison (Fig. 5). Both wild-type SOS gp140 and SOSIP gp140 were immunoprecipitated by the CD4 IgG2 molecule, indicating that the CD4BS was intact on the gp120 subunits of both proteins (Fig. 5, second lane). Neutralizing MAb b12 to a CD4BS-associated epitope also bound to both proteins efficiently, as did neutralizing MAb 2G12 to a mannose-dependent gp120 epitope (80, 93) (Fig. 5, first and third lanes). Furthermore, soluble CD4 induction of the 17b epitope was highly efficient in both SOS gp140 and SOSIP gp140 (Fig. 5, fourth and fifth lanes). This epitope overlaps the CD4-inducible coreceptor binding site on gp120 (77, 91, 97, 98). Thus, the I559P substitution in gp41ECTO does not affect the ability of the gp120 subunits of SOS gp140 to bind the CD4 receptor and then to undergo receptor-mediated conformational changes. SOSIP gp140 appears indistinguishable from wild-type SOS gp140 in this regard.
FIG. 5.
Antigenic structure analysis of gp140. Radioimmunoprecipitation assays were performed with gp140UNC, SOS gp140, and SOSIP gp140 (SOS gp140 I559P) and various neutralizing and nonneutralizing MAbs as previously described (5). sCD4, soluble CD4.
There are three predominant epitope clusters in gp41ECTO. One cluster is recognized by neutralizing MAbs 2F5, 4E10, and z13 and is located close to the C terminus of gp41ECTO (68, 88, 106) (Fig. 1B). This region of gp41ECTO is well exposed on SOS gp140 and SOSIP gp140, as indicated by their efficient binding of 2F5 (Fig. 5, sixth lane) and 4E10 (data not shown). The cluster I and cluster II gp41ECTO epitopes are highly immunogenic. However, antibodies to these regions of gp41 are nonneutralizing because their epitopes are occluded in the native, prefusion form of the Env complex, either by interactions between gp41ECTO moieties or because of the presence of the gp120 subunits (5, 42, 59, 65, 71, 82, 90). MAbs to cluster I (2.2B) and cluster II (4D4) epitopes interact with gp140UNC efficiently but do not bind to SOS gp140 or SOSIP gp140 (Fig. 5, seventh and eighth lanes). Similar results were obtained with other MAbs to these epitope clusters (data not shown). This pattern of results is consistent with previous reports on the antigenic structure of SOS gp140 (5, 79, 83). It is possible that the introduction of the cysteine substitution at position 605 in SOS gp140 directly perturbs the nearby epitopes for some or all of the cluster I MAbs. However, this cannot be the case for the cluster II MAb epitopes, since these are located in the C-terminal helical region, approximately 40 residues from the Cys-605 substitution (Fig. 1B).
The SOS gp140 variants I559G, L566V, and T569P were also tested for reactivity with the above MAbs and CD4-based reagents. Each of them behaved in a manner similar to that of SOS gp140 and SOSIP gp140 (data not shown). Further studies on the receptor and MAb binding properties of purified SOSIP gp140 trimers will be described elsewhere in a comparison with other forms of gp140 (M. Vesanen, R. W. Sanders, and J. P. Moore, unpublished results). These experiments, together with other planned structural analyses, should determine whether the gp41ECTO moieties of SOSIP gp140 adopt a native configuration or whether the helix-destabilizing effect of the I559P substitution creates a nonnative gp41ECTO conformation that is not recognized by some anti-gp41 MAbs.
Destabilization of the six-helix bundle form of gp41.
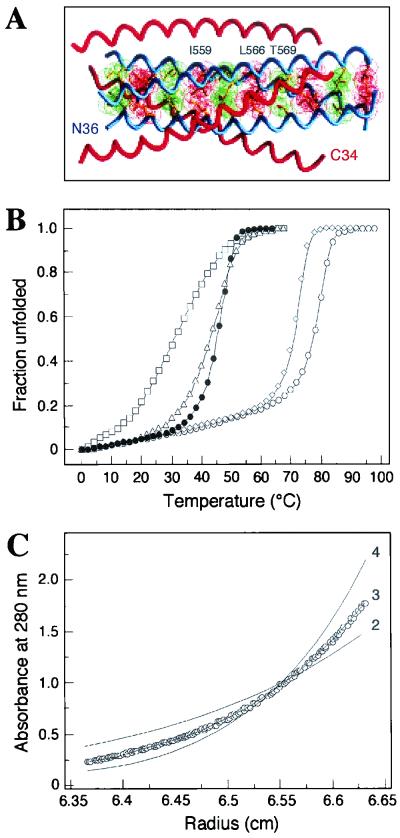
Our mutagenesis results indicate that SOS gp140 trimers can be stabilized by the I559G, I559P and, to some extent, L566V and T569P substitutions in the N-terminal heptad repeat region of gp41ECTO. Given that hydrophobic interactions are a dominant factor in the stabilization of the gp41 core (40, 51), it would appear that these amino acid substitutions destabilize the six-helix bundle structure. To directly test this hypothesis, we determined the effects of each of the four amino acid substitutions on the overall structure and stability of the JR-FL gp41ECTO core. To do this, we constructed a recombinant peptide model of the soluble gp41 core. This model peptide, designated N36(L6)C34, consists of the N36 and C34 peptides connected via a short peptide linker that replaces the disulfide-bonded loop region of gp41ECTO (Fig. 6A ) (51). The N36 peptide consists of residues 546 to 581 and the C34 peptide consists of residues 628 to 661 of the JR-FL gp41 sequence. Each of the above four amino acid changes was introduced into the N36(L6)C34 peptide. Sedimentation equilibrium analysis showed that the molecular weights of the N36(L6)C34 variants, except for the I559P mutant, were all within 10% those calculated for an ideal trimer, with no systematic deviation of the residuals (data not shown).
FIG. 6.
HIV-1 gp41ECTO core structure and mutant peptides. (A) The N36-C34 crystal structure is shown with one N36 helix and one C34 helix labeled at the amino termini. Three C34 helices (red) pack against the N36 trimeric coiled coil (blue). The van der Waal surfaces of residues at positions a (red) and d (green) are superimposed on the helix backbone of the N36 coiled coil. Amino acids substituted in this study are indicated above the a and d layers. The diagram was prepared by using the program GRASP (64). (B) Thermal melting transition curves of the N36(L6)C34 (open circles), I559G (closed circles), I559P (open squares), L566V (open diamonds), and T569P (open triangles) peptides were determined by CD spectroscopy at 222 nm and at a peptide concentration of 10 μM in PBS. The increase in the fraction of unfolded molecules is shown as a function of temperature. All melts were reversible. Superimposable folding and unfolding curves were observed, and >90% of the signal was regained upon cooling. (C) Equilibrium sedimentation analysis of the T569P peptide.Representative data for this peptide were collected at 20°C, 20,000 rpm, and a peptide concentration of ∼30 μM in PBS. The data fit best to a trimer model (curve 3). Curves for a dimer (curve 2) and a tetramer (curve 4) are depicted for comparison. Analyses of residual differences from curve 3 did not reveal a systematic error.
CD measurements indicated that the N36(L6)C34 wild-type peptide and the I559G, L566V, and T569P variant peptides are each >95% alpha helical at 4°C, whereas the I559P variant peptide is apparently only ∼75% alpha helical (Table 3). Under these conditions, the Tm of the I559G, I559P, L566V, and T569P peptides are 46, 34, 72, and 44°C, respectively; the Tm for the wild-type peptide is 78°C (Table 3). The pre- and posttransitional slopes and the steepness of the main transition are very similar for the N36(L6)C34, I559G, and L566V peptides (Fig. 6B). In contrast, the I559P and T569P peptides display broad thermal unfolding transitions (Fig. 6B). Sedimentation equilibrium experiments indicate that the N36(L6)C34 peptide and the I559G, L566V, and T569P variant peptides each sediment as discrete trimers over a 10-fold range of peptide concentrations (10 to 100 μM) (Table 3 and Fig. 6C). The I559P peptide is also trimeric in solution at concentrations of less than 15 μM but exhibits a systematic deviation from the trimer molecular weight at 10 to 100 μM, indicating that it is prone to aggregation. Taken together, these results indicate that the Ile-559-to-Gly and Thr-569-to-Pro substitutions each lead to an appreciable destabilization of the six-helix bundle structure but do not affect its overall fold. In contrast, the Ile-559-to-Pro substitution essentially disrupts the six-helix bundle structure. Moreover, the Leu-566-to-Val change is associated with a small, unfavorable, residual destabilization of the six-helix bundle structure.
TABLE 3.
Biophysical characterization of JR-FL gp41 core mutantsa
| Mutant | θ222 (degrees/cm2/dmol) | Tm (°C) | Mobs/Mcalc |
|---|---|---|---|
| N36(L6)C34 | 32,500 | 78 | 3.1 |
| I559G | 32,300 | 46 | 2.9 |
| I559P | 25,600 | 34 | —b |
| L566V | 32,100 | 72 | 3.1 |
| T569P | 29,400 | 44 | 3.0 |
All CD scans and melts were performed with 10 μM peptide solutions in PBS (pH 7.0). Tm was estimated from the thermal dependence of θ222. Sedimentation equilibrium results are reported as a ratio of the experimentally determined molecular weight to the calculated molecular weight for a monomer (Mobs/Mcalc).
—, Aggregated, as determined by sedimentation equilibrium analysis.
Overall, these experiments confirm that the I559P, I559G, L566V, and T569P substitutions do, in fact, destabilize the six-helix bundle, postfusion conformation of gp41ECTO. Indeed, the I559P change appears sufficient to completely prevent the formation of the postfusion state by destabilizing the N-terminal helix. The results obtained by using model peptides are therefore consistent with what was observed when the corresponding amino acid substitutions were introduced into SOS gp140. Moreover, both sets of results support the underlying hypothesis that destabilizing or otherwise preventing the formation of the six-helix bundle form of gp41ECTO helps maintain gp140 in its native, trimeric, prefusion configuration.
DISCUSSION
We describe here the generation and characterization of soluble, cleaved HIV-1 Env trimers. In these gp140 molecules, the gp120-gp41 interactions are stabilized by an intermolecular disulfide bond, and the gp41-gp41 interactions are stabilized by specific amino acid substitutions in the N-terminal heptad repeat region of gp41ECTO, most notably at position 559. The need for this study arose when we observed that SOS gp140 was unstable, in that it dissociated into gp140 monomers and could not be purified in trimeric form (83). The fragility of the SOS gp140 trimer is created by the proteolytic cleavage event that eliminates the peptide bond between gp120 and gp41ECTO as the gp140 precursor is processed to maturity. Thus, gp140UNC forms stable oligomers, whether or not the disulfide bond that characterizes SOS gp140 is also present (2, 83, 87, 101, 102, 105). Since we want to make cleaved, stable Env trimers for structural and immunogenicity studies, we needed to find a way to overcome the instability of the gp41-gp41 interactions in the prefusion form of gp140.
Our efforts were guided by the hypothesis that destabilization of the postfusion state of gp41 might lead to stabilization of the native, trimeric SOS gp140 complex. The native HIV-1 Env complex is metastable and undergoes a transition to the postfusion, six-helix bundle structure after activation by receptor binding, probably losing gp120 in the process (17, 24, 25, 31, 61, 99). The gp120 moiety of SOS gp140 can bind CD4 and undergo conformational changes that expose the coreceptor binding site. We do not know whether any additional conformational changes are initiated in the gp41ECTO moiety of SOS gp140 upon CD4 and/or coreceptor binding. Any such changes cannot be completed. However, whether or not SOS gp140 has bound to CD4, the gp41-gp41 interactions that are responsible for its trimerization are unstable, and the protein readily becomes a monomer (83).
Our hypothesis for stabilizing the trimeric form of SOS gp140 was based on an increasing understanding of the delicate balance between the metastable, prefusion state of gp140 and its stable, postfusion, six-helix bundle configuration. The balance involves not only the trimeric interactions between gp41ECTO moieties but also the association between gp120 and gp41ECTO. Several lines of evidence support our experimental approach. SIVmac gp140 seems to be a more stable trimeric protein than HIV-1 gp140 (15, 19). In contrast, the postfusion state of SIVmac Env is less stable than that of HIV-1 Env (50); it may or may not be relevant that SIVmac Env contains a valine and not an isoleucine residue at position 559. Furthermore, SIVmac Env with an unusually strong gp120-gp41 association has a destabilized, postfusion, six-helix bundle conformation, unlike the parental virus from which it evolved (47, 50). Also consistent with our hypothesis are studies with an HIV-1LAI variant engineered to contain SOS substitutions and so to have gp120 covalently linked to gp41. This virus is minimally infectious but evolves in cultures to a more infectious form via reversions at a region of gp41ECTO that corresponds to the trimer interface of the postfusion form (Sanders and Berkhout, unpublished). These changes in gp41ECTO may influence the interactions both between gp120 and gp41 and between the gp41 moieties.
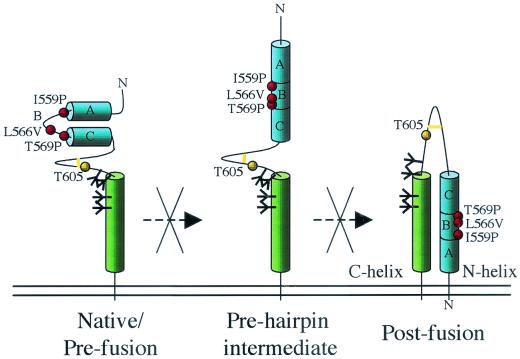
We believe that the substitutions that we have introduced at position 559 to make SOSIP gp140 or SOSIG gp140 block one or more of the conformational transitions in gp41ECTO (39). A model of how the substitutions might act is presented in Fig. 7. We argue that preventing these transitions stabilizes SOS gp140 as a trimer. However, we do not yet know which of the several conformational transitions that gp41ECTO undergoes during fusion is impaired by the changes at position 559. Two possibilities are that the transition from the elusive native state to the prehairpin intermediate is affected or that the subsequent transition from the prehairpin intermediate to the six-helix bundle structure is prevented (Fig. 7). The observation that the N36(L6)C34 I559P peptide is only ∼75% helical argues in favor of the first possibility—interference with the formation of the N-terminal helix—although additional studies will be required to confirm this notion. Peptide-based studies of the gp41 N-terminal helix have shown that the first part of this helix, including the region around position 559, is more flexible than the last part (18). There is evidence that the prehairpin intermediate may exist as an equilibrium between a monomeric state and a trimeric coiled coil and that disrupting the homotrimeric coiled coil is an efficient way to block fusion (4, 11, 72). Preventing the formation of the inherently unstable prehairpin intermediate state may therefore stabilize the Env complex in its native conformation.
FIG. 7.
Model of gp41ECTO and its transitions during fusion. (Left panel) Hypothetical, native prefusion configuration of gp41 (39). (Middle panel) Prehairpin intermediate form. (Right panel) Postfusion state. In the prefusion configuration, the N-terminal helix is not present, and the region around positions 559 to 569 is not helical. The I559P and related substitutions are proposed to disrupt either the formation of the N-terminal helix in the prehairpin intermediate form or the formation of the six-helix bundle. As a result, modified SOS gp140 is maintained in the prefusion configuration. The position of the T605C substitution that creates SOS gp140 is also specified, as is the adjacent intermolecular disulfide bond (yellow bar). The positions of N-linked glycans are represented as black branching structures. Only two helices from one gp41 molecule are shown, for clarity. The letters A, B, and C are added to assist with orientation of the helices.
Stabilizing the prefusion, trimeric structure of a fusogenic viral glycoprotein by destabilizing or disrupting its N-terminal helix via a proline substitution is not without precedent. In the influenza virus HA2 glycoprotein, a stretch of 22 amino acids is not helical in the prefusion form. However, upon exposure to low pH to trigger fusion, this region of HA2 undergoes a loop-to-helix transition to form the fusion-active configuration of the protein (8). Proline substitutions here in HA2 allow the expression of properly processed but fusion-incompetent proteins (37, 75). Similarly, a proline substitution at position 559 in HIV-1 gp41 is known to abolish the fusogenicity of an otherwise infectious virus (20, 21).
The SOSIP gp140, SOSIG gp140, L566V, and T569P variants do not suffer from any proteolytic cleavage defects, in contrast to the cleavage defects that are caused by amino acid substitutions at the same positions in the context of wild-type gp160 (13, 20, 21, 74). An explanation for the apparent discrepancy may be provided by the earlier observation that the presence of the SOS intersubunit disulfide bond can rescue some cleavage defects in gp140 (79). Moreover, the SOS gp140 variants are expressed in the presence of cotransfected furin, so the increased concentration of this enzyme may compensate for any partial reduction in cleavage efficiency. The use of basic substitutions within the gp140 cleavage site to increase the efficiency with which SOS gp140 variants are processed by endogenous and exogenous furin proteases has been described elsewhere (6). The utility of these cleavage site changes in the context of SOSIP gp140 is now being evaluated.
Several unpurified and purified gp140UNC preparations from both HIV-1 and SIVmac have been described elsewhere (2, 15, 19, 28, 86, 87, 105). There is general agreement that SIVmac gp140UNC is predominantly trimeric (15, 19). In contrast, the oligomeric state of HIV-1 gp140UNC varies from study to study. After a purification procedure that included an earlier size exclusion chromatography step, HIV-1 strain ADA gp140UNC was eluted homogeneously from size exclusion columns with a molecular weight that indicated that it was a trimer (105). The production of oligomeric US4 gp140UNC that might be trimeric has also been reported (86). The presence of both dimers and tetramers in uncleaved versions of both membrane-bound gp160 and soluble gp140 molecules has been observed in several studies (27-30, 33, 101-103). Our own experience, with the BN-PAGE assay, is that a mixture of dimers, trimers, and tetramers is present in unpurified and purified HIV-1 strain JR-FL gp140UNC preparations, with dimers being the most abundant species (83). Moreover, the dimeric and tetrameric forms of HIV-1 gp140UNC and gp160 are probably oligomerized by aberrant intermolecular disulfide bonds (Fig. 2) (67, 83). It is unlikely that oligomeric gp140 of this type will fully mimic the native conformation of Env. Indeed, we have found that unpurified gp140UNC has an antigenic structure different from that of unpurified SOS gp140 monomers and SOSIP gp140 trimers. Thus, nonneutralizing antibody epitopes in both the gp120 and the gp41ECTO moieties are exposed to a much greater extent in gp140UNC than in SOS gp140 (Fig. 5) (5, 79, 83). We are now comparing purified, cleaved and uncleaved trimers as well as purified, uncleaved dimers and tetramers to assess their antigenic structures, their migration in BN-PAGE analysis after various treatments, and their receptor binding properties. These and other planned structural studies should help determine whether SOSIP gp140 truly has properties that mimic those of the native Env trimer or whether the I559P or related substitutions have converted the gp41ECTO moieties into a nonnative, irrelevant conformation. It is possible that the gp41ECTO component of SOSIP gp140 is in a fusion-incompetent configuration without any adverse effect on the native conformation of the gp120 moieties.
The trimer-stabilizing substitutions that we have described here should simplify the production of cleaved Env trimers, which may be useful in vaccine design and for structural studies. Whether cleaved, stabilized trimeric Env forms such as SOSIP gp140 will turn out to be better or worse immunogens than other forms of Env (e.g., gp140UNC) can be determined only empirically. It may be necessary to further modify the structure of SOSIP gp140 to increase its immunogenicity, for example, by removing the variable loops to increase the exposure of underlying conserved epitopes or by reducing the extent to which the protein is glycosylated (2, 7, 23, 53, 62, 76, 104). Proteins with such changes are now being constructed for future structural and immunogenicity studies.
Acknowledgments
We are grateful to James Robinson, George Lewis, Hermann Katinger, and Dennis Burton for the gifts of MAbs used in these studies. We thank Steven Harrison for providing SIV gp140UNC trimers for the calibration of gel filtration columns.
This work was supported by NIH grants AI39420, AI42382, AI45463, and AI49764. J.P.M. is a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at the Weill Medical College, Cornell University, gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M. C. Gauduin, R. A. Koup, J. S. McDougal, and P. J. Maddon. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 11:533-539. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, H. B., S. P. Tucker, S. R. Kar, S. A. McPherson, D. T. McPherson, J. W. Dubay, J. Lebowitz, R. W. Compans, and E. Hunter. 1995. Oligomerization of the hydrophobic heptad repeat of gp41. J. Virol. 69:2745-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bewley, C. A., J. M. Louis, R. Ghirlando, and G. M. Clore. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. 277:14238-14245. [DOI] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S.-L. Hu, W. C. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolmstedt, A., J. Hinkula, E. Rowcliffe, M. Biller, B. Wahren, and S. Olofsson. 2001. Enhanced immunogenicity of a human immunodeficiency virus type 1 env DNA vaccine by manipulating N-glycosylation signals. Effects of elimination of the V3 N306 glycan. Vaccine 20:397-405. [DOI] [PubMed] [Google Scholar]
- 8.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., and P. W. Parren. 2000. Vaccines and the induction of functional antibodies: time to look beyond the molecules of natural infection? Nat. Med. 6:123-125. [DOI] [PubMed] [Google Scholar]
- 10.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caffrey, M., J. Kaufman, S. Stahl, P. Wingfield, A. M. Gronenborn, and G. M. Clore. 1999. Monomer-trimer equilibrium of the ectodomain of SIV gp41: insight into the mechanism of peptide inhibition of HIV infection. Protein Sci. 8:1904-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantor, C., and P. Schimmel. 1980. Biophysical chemistry. Part III. W. H. Freeman & Company, New York, N.Y.
- 13.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Center, R. J., P. L. Earl, J. Lebowitz, P. Schuck, and B. Moss. 2000. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J. Virol. 74:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Center, R. J., P. Schuck, R. D. Leapman, L. O. Arthur, P. L. Earl, B. Moss, and J. Lebowitz. 2001. Oligomeric structure of virion-associated and soluble forms of the simian immunodeficiency virus envelope protein in the prefusion activated conformation. Proc. Natl. Acad. Sci. USA 98:14877-14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 17.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 18.Chang, D. K., V. D. Trivedi, S. F. Cheng, and S. Francis. 2001. The leucine zipper motif of the envelope glycoprotein ectodomain of human immunodeficiency virus type 1 contains conformationally flexible regions as revealed by NMR and circular dichroism studies in different media. J. Pept Res. 57:234-239. [DOI] [PubMed] [Google Scholar]
- 19.Chen, B., G. Zhou, M. Kim, Y. Chishti, R. E. Hussey, B. Ely, J. J. Skehel, E. L. Reinherz, S. C. Harrison, and D. C. Wiley. 2000. Expression, purification, and characterization of gp160e, the soluble, trimeric ectodomain of the simian immunodeficiency virus envelope glycoprotein, gp160. J. Biol. Chem. 275:34946-34953. [DOI] [PubMed] [Google Scholar]
- 20.Chen, S. S. 1994. Functional role of the zipper motif region of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 68:2002-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, S. S., S. F. Lee, H. J. Hao, and C. K. Chuang. 1998. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J. Virol. 72:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, Y. H., J. T. Yang, and K. H. Chau. 1974. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry 13:3350-3359. [DOI] [PubMed] [Google Scholar]
- 23.Cherpelis, S., I. Shrivastava, A. Gettie, X. Jin, D. D. Ho, S. W. Barnett, and L. Stamatatos. 2001. DNA vaccination with the human immunodeficiency virus type 1 SF162DeltaV2 envelope elicits immune responses that offer partial protection from simian/human immunodeficiency virus infection to CD8+ T-cell-depleted rhesus macaques. J. Virol. 75:1547-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 25.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Earl, P. L., C. C. Broder, R. W. Doms, and B. Moss. 1997. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J. Virol. 71:2674-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earl, P. L., and B. Moss. 1993. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res. Hum. Retrovir. 9:589-594. [DOI] [PubMed] [Google Scholar]
- 30.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 32.Edelhoch, H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6:1948-1954. [DOI] [PubMed] [Google Scholar]
- 33.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fouts, T. R., A. Trkola, M. S. Fung, and J. P. Moore. 1998. Interactions of polyclonal and monoclonal anti-glycoprotein 120 antibodies with oligomeric glycoprotein 120-glycoprotein 41 complexes of a primary HIV type 1 isolate: relationship to neutralization. AIDS Res. Hum. Retrovir. 14:591-597. [DOI] [PubMed] [Google Scholar]
- 35.Gallo, S. A., A. Puri, and R. Blumenthal. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40:12231-12236. [DOI] [PubMed] [Google Scholar]
- 36.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 74:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruenke, J. A., R. T. Armstrong, W. W. Newcomb, J. C. Brown, and J. M. White. 2002. New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J. Virol. 76:4456-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harbury, P. B., P. S. Kim, and T. Alber. 1994. Crystal structure of an isoleucine-zipper trimer. Nature 371:80-83. [DOI] [PubMed] [Google Scholar]
- 39.Hunter, E. 1997. gp41, a multifunctional protein involved in HIV entry and pathogenesis, p. III-55-III-73. In B. Korber, B. Hahn, B. Foley, J. W. Mellors, T. Leitner, G. Myers, F. McCutchan, and C. L. Kuiken (ed.), Human retroviruses and AIDS 1997. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 40.Jelesarov, I., and M. Lu. 2001. Thermodynamics of trimer-of-hairpins formation by the SIV gp41 envelope protein. J. Mol. Biol. 307:637-656. [DOI] [PubMed] [Google Scholar]
- 41.Ji, H., C. Bracken, and M. Lu. 2000. Buried polar interactions and conformational stability in the simian immunodeficiency virus (SIV) gp41 core. Biochemistry 39:676-685. [DOI] [PubMed] [Google Scholar]
- 42.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson, M. L., J. J. Correia, D. A. Yphantis, and H. R. Halvorson. 1981. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 36:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 45.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold. Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 46.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in a complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, M. Marsh, and J. A. Hoxie. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69:5217-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, England.
- 49.Liu, J., W. Shu, M. B. Fagan, J. H. Nunberg, and M. Lu. 2001. Structural and functional analysis of the HIV gp41 core containing an Ile573 to Thr substitution: implications for membrane fusion. Biochemistry 40:2797-2807. [DOI] [PubMed] [Google Scholar]
- 50.Liu, J., S. Wang, J. A. Hoxie, C. C. LaBranche, and M. Lu. 2002. Mutations that destabilize the gp41core are determinants for stabilizing the Simian Immunodeficiency Virus-CPmac envelope glycoprotein complex. J. Biol. Chem. 277:12891-12900. [DOI] [PubMed] [Google Scholar]
- 51.Lu, M., H. Ji, and S. Shen. 1999. Subdomain folding and biological activity of the core structure from human immunodeficiency virus type 1 gp41: implications for viral membrane fusion. J. Virol. 73:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Nunberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 75:11146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu, S., R. Wyatt, J. F. Richmond, F. Mustafa, S. Wang, J. Weng, D. C. Montefiori, J. Sodroski, and H. L. Robinson. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res. Hum. Retrovir. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 54.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCune, J. M., L. B. Rabin, M. B. Feinberg, M. Lieberman, J. C. Kosek, G. R. Reyes, and I. L. Weissman. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55-67. [DOI] [PubMed] [Google Scholar]
- 56.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore, J. P., and J. Binley. 1998. HIV. Envelope's letters boxed into shape. Nature 393:630-631. [DOI] [PubMed] [Google Scholar]
- 58.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 59.Moore, J. P., P. W. H. I. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell. Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 62.Mori, K., Y. Yasutomi, S. Ohgimoto, T. Nakasone, S. Takamura, T. Shioda, and Y. Nagai. 2001. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J. Virol. 75:4023-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469:121-132. [DOI] [PubMed] [Google Scholar]
- 64.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 65.Nyambi, P. N., M. K. Gorny, L. Bastiani, G. van der Groen, C. Williams, and S. Zolla-Pazner. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olofsson, S., and J. E. Hansen. 1998. Host cell glycosylation of viral glycoproteins—a battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 30:435-440. [DOI] [PubMed] [Google Scholar]
- 67.Owens, R. J., and R. W. Compans. 1990. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology 179:827-833. [DOI] [PubMed] [Google Scholar]
- 68.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schulke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parren, P. W., D. R. Burton, and Q. J. Sattentau. 1997. HIV-1 antibody—debris or virion? Nat. Med. 3:366-367. [DOI] [PubMed] [Google Scholar]
- 70.Parren, P. W., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 72.Peisajovich, S. G., and Y. Shai. 2001. SIV gp41 binds to membranes both in the monomeric and trimeric states: consequences for the neuropathology and inhibition of HIV infection. J. Mol. Biol. 311:249-254. [DOI] [PubMed] [Google Scholar]
- 73.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 74.Poumbourios, P., K. A. Wilson, R. J. Center, W. El Ahmar, and B. E. Kemp. 1997. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic alpha-helical/leucine zipper-like sequence. J. Virol. 71:2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiao, H., S. L. Pelletier, L. Hoffman, J. Hacker, R. T. Armstrong, and J. M. White. 1998. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J. Cell Biol. 141:1335-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 77.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 78.Robinson, W. E., Jr., T. Kawamura, M. K. Gorny, D. lake, J. Y. Xu, Y. Matsumoto, T. Sugano, Y. Masuho, W. M. Mitchell, E. Hersh, and S. Zolla-Pazner. 1990. Human monoclonal antibodies to the human immunodeficiency virus type 1 (HIV-1) transmembrane glycoprotein gp41 enhance HIV-1 infection in vitro. Proc. Natl. Acad. Sci. USA 87:3185-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 83.Schuelke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, D. J. Anselma, A. R. Villa, P. W. H. I. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shu, W., H. Ji, and M. Lu. 1999. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry 38:5378-5385. [DOI] [PubMed] [Google Scholar]
- 85.Shugars, D. C., C. T. Wild, T. K. Greenwell, and T. J. Matthews. 1996. Biophysical characterization of recombinant proteins expressing the leucine zipper-like domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 70:2982-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Srivastava, I. K., L. Stamatatos, H. Legg, E. Kan, S. Coates, L. Leung, A. Fong, M. Wininger, A. R. Tipton, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2002. Purification and characterization of oligomeric glycoprotein from a primary R5 subtype B human immunodeficiency virus for vaccine applications. J. Virol. 76:2835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stamatatos, L., M. Lim, and C. Cheng-Mayer. 2000. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 16:981-994. [DOI] [PubMed] [Google Scholar]
- 88.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 89.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taniguchi, Y., S. Zolla-Pazner, Y. Xu, X. Zhang, S. Takeda, and T. Hattori. 2000. Human monoclonal antibody 98-6 reacts with the fusogenic form of gp41. Virology 273:333-340. [DOI] [PubMed] [Google Scholar]
- 91.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 93.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 95.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weng, Y., Z. Yang, and C. D. Weiss. 2000. Structure-function studies of the self-assembly domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 74:5368-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 98.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 100.Xu, J. Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65:4832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang, C. W., Y. Chishti, R. E. Hussey, and E. L. Reinherz. 2001. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J. Biol. Chem. 276:39577-39585. [DOI] [PubMed] [Google Scholar]
- 106.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]