Abstract
Cellular receptors for the Fc domain of immunoglobulin G (IgG) (FcγRs) comprise a family of surface receptors on immune cells connecting humoral and cellular immune responses. Several herpesviruses induce FcγR activities in infected cells. Here we identify two distinct human cytomegalovirus (HCMV)-encoded vFcγR glycoproteins of 34 and 68 kDa. A panel of HCMV strains exhibited a slight molecular microheterogeneity between Fcγ-binding proteins, suggesting their viral origin. To locate the responsible genes within the HCMV genome, a large set of targeted HCMV deletion mutants was constructed. The mutant analysis allowed the identification of a spliced UL119-UL118 mRNA to encode vFcγR gp68 and TRL11/IRL11 to encode vFcγR gp34. Both vFcγRs are surface resident type I transmembrane glycoproteins. Significant relatedness of sequences in the extracellular chain of gpUL119-118 and gpTRL11 with particular immunoglobulin supergene family domains present in FcγR I and FcγRs II/III, respectively, indicates a different ancestry and function of gpUL119-118 and gpTRL11. The HCMV-encoded vFcγRs highlight an impressive diversification and redundancy of FcγR structures.
Human cytomegalovirus (HCMV) constitutes a prototype member of the β-subgroup of the herpesvirus family. Herpesviruses are equipped with a large array of gene products which interfere with adaptive and innate immune responses (26, 33, 64). In addition, specific host genes implicated in immune responses are subjected to viral control for exploitation of their function. Subversion of the immune functions is thought to allow the virus to increase the available time window for replication and spread. After the resolution of acute infection, herpesviruses establish lifelong infections characterized by alternate stages of virus productivity and latency. Although producing up to 200 potentially antigenic proteins during the sequential immediate-early (IE), early (E), and late (L) phases of gene expression, HCMV frequently initiates recurrent replication from latency, and transmission to a new host is achieved even in the face of repeatedly boostered antiviral immune responses (45).
The Fc portion of immunoglobulins (Ig) plays a central role during immune responses and forms a well-characterized structure (29). Multiple components of the immune system interact with Fc via distinct binding sites, which include complement, the neonatal Fc receptor transferring maternal IgG to the fetus, and cellular immune receptors (cellular FcRs). FcRs belong to the immunoglobulin supergene family (IgSF) and constitute a group of cell-surface molecules that are expressed on most cells of the immune system. By linking cell-mediated and humoral immune responses, FcRs play a key role in host defenses against pathogens (57). Three classes of cellular FcRs (FcγR) for IgG are known which exhibit homologies in their extracellular IgG-binding chain forming IgSF domains. FcγRI has a high level of affinity for monomeric IgG, whereas FcγRII and FcγRIII exhibit lower levels of affinity for monomeric IgG but bind IgG-containing immune complexes with high avidity. The cross-linking of FcγRI-bound antibodies by multivalent antigens, or the binding of preformed immune complexes with FcγRII or FcγRIII, results in clustering of the FcγR and triggers a variety of effector mechanisms, such as antibody-dependent cellular cytotoxicity (ADCC), phagocytosis, the release of cytokines, enhanced antigen presentation, and the regulation of Ig production (1, 57).
Microbial Ig binding proteins, such as protein A from Staphylococcus aureus and streptococcal protein G, mask the bacteria through immobilized Ig on their surface and modify opsonization, phagocytosis, and complement consumption (38). Coronaviruses (52) and certain herpesviruses, such as herpes simplex virus type I (HSV-1) (3, 53), HSV-2 (10), varicella zoster virus (51), and murine cytomegalovirus (MCMV) (63), produce molecules with FcγR activity. The HSV-encoded vFcγR is formed by a protein complex composed of two type I transmembrane glycoproteins, gE and gI, which are part of the virion structure (32). gE was identified as the IgG-binding protein of HSV-1 but requires association with gI to enhance IgG binding activity (31). This prevents antiviral IgG from neutralizing free virus (16) and engaging in ADCC against HSV-infected cells (18), a mechanism explained by bipolar bridging of specific IgG (20). Other functions of the gE-gI heterodimer are associated with direct cell-to-cell spread of the virus (14, 15). In MCMV-infected cells a vFcγR is encoded by the early gene m138, which has no homolog in the HCMV AD169 sequence (63). The deletion of m138 did not impair viral replication in vitro but resulted in a drastically attenuated phenotype in vivo (11).
Over the last 25 years, several laboratories have reported on Fc binding activities in HCMV-infected cells (21, 22, 36, 58). Biochemical characterization of Fc-binding proteins (FcBPs) in HCMV-infected fibroblasts led to conflicting results, and a viral protein mediating this effect has not yet been identified (48, 62, 67). Alternatively, induction of a cFcγR could account for this effect, although antibodies specific for known cFcγRs did not stain HCMV-infected cells (43).
Here we report on the identification of two HCMV AD169-encoded glycoproteins, designated gpUL119-118 and gpTRL11, which share constitutive capabilities of FcγR, i.e., Fc-dependent binding of nonimmune human IgG and migration along the secretory pathway to the cell surface. To search for the viral genes coding for vFcγRs, we pursued a genomic approach and constructed a panel of HCMV AD169 deletion mutants lacking gene regions predicted to encode glycoproteins. This led to the identification of the viral gene UL119-118 coding for gp68 and confirmed that TRL11/IRL11 expresses gp34 as reported recently (41). While the overall sequence homology of vFcγRs with cFcγRs is moderate, their structural composition, the formation of IgSF-like domains, and the positional conservation of amino acid residues within the extraluminal part of the molecules are consistently preserved in both of the CMV-encoded homologs. On the other hand, the virally encoded homologs indicate an unforeseen plasticity of FcγR structures.
MATERIALS AND METHODS
Cells.
MRC5 fetal human lung fibroblasts (passages 3 to 15) and CV1 monkey kidney cells were grown in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum, penicillin, streptomycin, and 2 mM glutamine.
Viruses, infection conditions, and virus titration.
Virus stocks were prepared by propagating HCMV strains AD169, Toledo, Towne, clinical isolates, and the mutant ts9 (46) in MRC5 cells as described previously (27). Infectious supernatants were harvested when 100% of cells showed cytopathic effects. Virus titers were determined by standard plaque assay. HCMV infection was enhanced by centrifugation at 800 × g for 30 min. In all experiments MRC5 cells were infected with HCMV at a multiplicity of infection of 3 to 5. Selective expression of HCMV IE gene products was achieved as described previously (27). Briefly, infection of cells was performed in the presence of cycloheximide (CHX) (50 μg/ml), which was replaced 4 h later by actinomycin D (ActD) (5 μg/ml). Late phase gene expression was prevented by the use of phosphonoacetic acid (PAA) (250 μg/ml), which arrests CMV-infected cells in the E phase.
Reagents and antibodies.
The following antibodies were used: Fc and F(ab)2 fragments of human IgG (huIgG) (Biotrend, Köln, Germany), complete huIgG (Biotrend), fluorescein isothiocyanate-conjugated Fc fragment of huIgG (huIgG Fc-FITC) (Biotrend), anti-human influenza virus hemagglutinin (HA) epitope-specific rat monoclonal antibody (MAb) 3F10 (Roche, Mannheim, Germany), and anti-FLAG M2 (Sigma, Munich, Germany). Human β2-microglobulin (β2m), huIgG, and Fc and F(ab)2 fragments of huIgG were coupled to cyanogen bromide-activated Sepharose 6MB (Amersham-Pharmacia, Freiburg, Germany) according to the manufacturer's instructions.
Metabolic labeling and precipitation of FcBPs.
Precipitation of proteins from lysates of metabolically labeled cells was performed essentially as described previously (27). In brief, MRC5 cells were infected with HCMV for various time periods. Cells were starved in cysteine- and methionine-free medium prior to labeling with [35S]methionine and [35S]cysteine (1,200 Ci/mmol) (Amersham-Pharmacia) at a concentration of 500 μCi/ml. After 1 h, cells were washed and lysed in lysis buffer (140 mM NaCl, 20 mM Tris [pH 7.6], 5 mM MgCl2, 1 mM phenylmethylsulfonylfluoride, 0.1 mM leupeptin, 1 μM pepstatin A) with 1% NP-40 (Sigma). The incorporation of 35S into proteins was quantified in all experiments by liquid scintillation counting. After removal of cell nuclei by centrifugation, lysates were incubated with huIgG Fc and protein A-Sepharose (Amersham Pharmacia). Digestion of proteins with 2 mU of Endo H (Roche) per sample was performed at 37°C overnight according to the instructions of the manufacturer. Proteins were eluted with sample buffer and analyzed by sodium dodecyl sulfate (SDS)-11.5 to 15% polyacrylamide gel electrophoresis (PAGE). Dried gels were exposed to Kodak BioMaxMR films at −70°C for 1 to 5 days.
Flow cytometry.
To analyze vFcγR surface expression, CV1 cells were infected with vaccinia virus recombinants at a multiplicity of infection (MOI) of 3 and incubated overnight. After washing, cells were incubated for 30 min at 4°C with 10 μg of huIgGFc-FITC/ml before being harvested with cell dissociation solution (Sigma). A total of 104 cells was analyzed for each histogram using a FACScan (Becton Dickinson, Heidelberg, Germany).
Plasmids.
p7.5kUL119-118FLAG was generated in two steps using PCR amplification products. First, UL119 was amplified from the HCMV bacterial artificial chromosome (BAC) plasmid pHB5 (4) using the primer pair az-UL119-1 (5′GACTTAGATCTACATGTGTTCCGTACTGGCG-3′) and az-UL119-5 (5′GAGATGAAGCTTACCTTCATTATCGGG-3′). The PCR fragment was digested by BglII and HindIII (indicated by italics) and cloned into the BamHI and HindIII sites of vector p7.5k (28), resulting in plasmid p7.5kUL119. A second PCR fragment was generated using primer az-UL119-6 (5′GGAAAGCTTCATCTCTCCTACAACGCTACT-3′) and az-UL119F1 (5′AAGAATTCTACTTGTCGTCGTCGTCCTTGTAGTCAGCAGCCCACTGCTTGAAGTAGGGCACCG-3′) encoding a FLAG epitope from the underlined sequence. The PCR product was treated with HindIII and EcoRI and cloned into the respective sites of p7.5kUL119, resulting in p7.5kUL119-118FLAG. The IRL/TRL11 open reading frame (ORF) was amplified from pHB5 using the primers az-irl11-1 (5′GCTTAGGGATCCATGCAGACCTACAGCACCCC-3′) and az-irl11-F1 (5′GCTTAAGATATCCTACTTGTCGTCGTCGTCCTTGTAGTCCTGTAAATCCCCGTCCACCG-3′). Primer az-irl-11-1 contains a BamHI restriction site (shown in italics), and az-irl11-F1 contains the coding sequence for the FLAG epitope and an EcoRV site (underlined). The PCR fragment was cloned into p7.5k via the BamHI and EcoRV sites. Correct insertion of HCMV coding sequences and FLAG epitopes was confirmed by sequencing.
BAC mutagenesis.
Recombinant CMV genomes were generated in Escherichia coli using a recently established new method that relies on homologous recombination of a linear PCR fragment with the CMV BAC plasmid (65). For construction of mutant BAC plasmids containing extended deletions, PCR products were used that were generated with primers which contained homologies of 60 bp upstream or downstream of the positions of the deleted ORFs. The 3′ ends of the primers had the sequence 5′CGATTTATTCAACAAAGCCACG-3′ in the case of the upstream primers and 5′GCCAGTGTTACAACCAATTAACC-3′ in the case of the downstream primers. The primer pairs were used to amplify the Knr gene from plasmid pACYC177 (New England Biolabs, Frankfurt/Main, Germany). The resulting PCR products contained the Knr gene flanked by homologies of 60 bp upstream and downstream of the respective target sequences. After purification, this PCR fragment was inserted into pHB5 by homologous recombination in E. coli which was mediated by the recombination plasmid pBADαβγ (69) leading to simultaneous deletion of the respective target genes. Correct mutagenesis was confirmed by restriction pattern analysis and by PCR analysis using an internal primer pair within the deleted sequences. Double mutants were constructed using primer pairs containing 60-bp homologous sequences as described above, followed by the sequences 5′CCAGTGAATTCGAGCTCGGTAC-3′ for upstream primers and 5′GACCATGATTACGCCAAGCTCC-3′ for downstream primers and the plasmid pCP16 (9) encoding a tetracycline resistance gene as the template. The resulting PCR fragment was inserted into pHB5-ΔIRL as described above. Plasmid pSLFRTKn, which was used as the template for PCR-based BAC mutagenesis, was generated as follows: the kanamycin resistance gene and its promoter (Knr) from the plasmid pACYC177 (New England Biolabs) were amplified by PCR using the contiguous primer pair EcoRI-FRT-Kn-3 (5′CACCGAATTCGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCGCCAGTGTTACAACCAATTAACC-3′) and EcoRI-FRT-Kn-5 (5′CCGAATTCGACTACAAGGACGACGACGACAAGTAAGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCCGATTTATTCAA-3′). Besides the priming region specific for the Knr gene in pACYC177, both primers contain a minimal 34-bp FRT site in the same orientation (underlined sequences) and an EcoRI site. The PCR fragment was inserted into the EcoRI site of plasmid pSL301 (Invitrogen, Karlsruhe, Germany), generating plasmid pSLFRTKn, which thus contains the Knr gene flanked by minimal FRT sites. Mutated BAC plasmids were transfected into MRC5 fibroblasts, and DNA of reconstituted virus mutants was analyzed in parallel with BAC plasmid DNA by Southern blotting and PCR to prove the correct deletions of DNA sequences. For construction of an HCMV genome with a 3′-terminal addition of the HA-tag sequence to the UL118 ORF, a PCR fragment was generated using the plasmid pSLFRTKn as template DNA and the contiguous primers 5′-UL118-HA (5′-GTCAGCGAAATAAAGACAACACAGCAGCCACTCCTCTCGTCTCGGGCCCGTCGTGGAATGCCTTCGAATTC-3′) and 3′-UL118-HA (5′-CCCGTTGAGGAAAAGAAACACCCGGTGCCCTACTTCAAGCAGTGGTACCCATACGATGTTCCAGATTACGCGTAGACAAGGACGACGACGACAAGTAA-3 (the HA sequence is underlined). The resulting PCR fragments contained the Knr gene (flanked by minimal FRT sites), next to Knr the HA sequence plus a stop codon, and finally, on both ends, homologies of about 50 bp upstream and downstream to the native stop codon of the UL118 ORF within pHB5. After purification and DpnI digestion (65), this PCR fragment was inserted into pHB5 by homologous recombination in E. coli, which led to simultaneous deletion of the native UL118 stop codon and to generation of the HCMV BAC plasmid pUL118Kn-HA with a HA fusion to UL118. The Knr downstream of the tagged gene was excised from pUL118-HA via the flanking FRT sites by Flp-mediated recombination as described previously (65), generating the HCMV BAC plasmid pUL118-HA. Correct mutagenesis was confirmed by restriction pattern analysis and sequencing of the 3′-terminal part of the tagged gene.
Generation of HCMV recombinants.
Recombinant HCMV was reconstituted from mutant HCMV BAC plasmids as described previously (4).
Viral nucleic acid isolation and analysis.
DNA from cells infected with recombinant HCMV was isolated and analyzed as described previously (4). For analysis of the UL119-115 transcription unit, total RNA was purified 120 h postinfection (p.i.) from cells infected with HB5 or HB5-ΔUL118-120 by using the RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. RNA was reverse transcribed using Superscript reverse transcriptase (Gibco BRL, Karlsruhe, Germany) and a random hexanucleotide primer mix (Roche) for 1 h at 45°C. First strands from this reaction were used as the template for PCRs using primer az-UL119-1 (5′GACTTAGATCTACATGTGTTCCGTACTGGCG-3′) containing a BglII restriction site as the forward primer and primers az-UL118-2 (5′-GGCATTGCATGCTACCACTGCTTGAAGTA-3′), az-UL117-1 (5′-TCCAAGCATGCTCAT GAGGTGGGCAGGCG-3′), and UL116-1 (5′-GTTCCGCATGCTCAAGTCTGCGGCACGAT-3′), respectively, all containing an SphI site as backward primers. PCR products were subcloned via the BglII und SphI sites into either the pUC19 or the p7.5K131 vector and subjected to sequence analysis.
Generation of vaccinia virus recombinants.
Vaccinia virus recombinants were constructed as described previously (28). Briefly, p7.5K131 constructs were used to generate vaccinia virus recombinants after homologous recombination with the vaccinia virus strain Copenhagen. The recombinant vaccinia viruses expressing the gene of interest were isolated by selection with BrdU using tk-143 cells.
Nucleotide sequence accession number.
The GenBank accession number for the UL119-118 nucleotide sequence reported in this article is AY065993.
RESULTS
Identification and characterization of FcBPs in HCMV-infected cells.
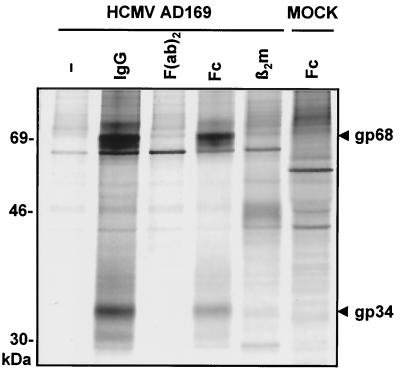
Detection of FcBP activity within HCMV virions (62), on the cell surface (2, 21, 43), and in the cytoplasm following infection (22, 36) has been reported by a number of laboratories. Both single and multiple biochemical activities associated with different molecular weights have been proposed to mediate Fc-dependent IgG binding, but a specific polypeptide responsible for this effect has not been demonstrated unequivocally. To identify proteins with IgG and Fc-dependent binding properties, Sepharose beads were covalently coupled with complete huIgG, Fc fragments, and F(ab)2 fragments thereof and a control protein, β2m. HCMV AD169-infected cells were pulse labeled with [35S]methionine at 72 h p.i., and lysates were incubated with Sepharose beads. Elution and separation of precipitated proteins by SDS-PAGE revealed distinct proteins with an approximate molecular mass of 68 and 34 kDa which did not bind to F(ab)2 fragments or β2m (Fig. 1). While the Fcγ binding proteins did not bind human IgA or human IgM, they strongly bound huIgG of the subclasses IgG1, IgG2, IgG3, and IgG4 (Table 1). However, the FcBPs clearly differed in their specificity for IgG of different mammal species (Table 1).
FIG. 1.
Precipitation of FcBPs from HCMV AD169-infected cells. Cells were labeled with [35S]methionine at 72 h p.i. for 1 h. The precipitation from lysates was done with Sepharose covalently coupled with complete nonimmune huIgG, Fc fragments of huIgG, F(ab)2 fragments of huIgG, and β2m. Proteins were separated by SDS-11.5 to 15% PAGE. FcBPs gp34 and gp68 are indicated by arrowheads.
TABLE 1.
Ig binding specifity of the Fc-binding proteins gp34 and gp68a
| Species | Ig subtype | Ig binding by:
|
|
|---|---|---|---|
| gp34 | gp68 | ||
| Human | IgG1 | ++ | ++ |
| Human | IgG2 | ++ | ++ |
| Human | IgG3 | ++ | ++ |
| Human | IgG4 | ++ | ++ |
| Human | IgM | − | − |
| Human | IgA | − | − |
| Rabbit | IgG | ++ | − |
| Mouse | IgG2b | − | − |
| Rat | IgG1 | + | − |
| Bovine | IgG | − | − |
| Goat | IgG | − | − |
MRC5 cells were infected at an MOI of 3 for 72 h with HCMV AD169 before cells were metabolically labeled with [35S]methionine and proteins from lysates were precipitated with indicated antibodies and protein A Sepharose. Precipitated proteins were separated by SDS-11.5 to 15% PAGE. ++ indicates strong binding; + indicates weak binding; − indicates no binding.
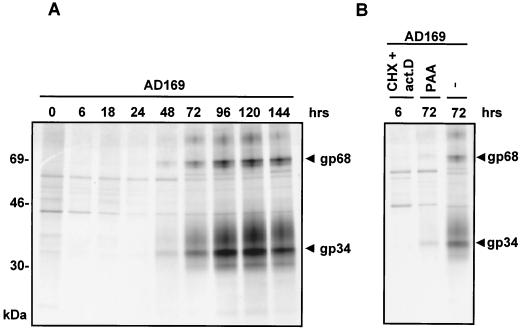
Next, synthesis of FcBPs was assessed at different stages of the HCMV replication cycle. Synthesis of the 34- and 68-kDa protein was readily detected by 48 h p.i. and reached a maximum at 96 h p.i. (Fig. 2A). At this time point a further prominent band of 31 kDa occurred (Fig. 2A). Under conditions of selective and enhanced IE gene expression using CHX and ActD, no FcBPs were recovered (Fig. 2B). In the presence of the late phase inhibitor PAA, the synthesis of all FcBPs was significantly decreased, indicating an E-L pattern of gene expression. As demonstrated in Fig. 2A, a proportion of FcBPs acquired an electrophoretic mobility of 34 to 41 kDa or 95 to 105 kDa, compatible with posttranslational modification of N-linked glycans in the trans-Golgi network. To assess the transport and maturation status of FcBPs, the precipitated material was digested with Endo H, which cleaves high-mannose N-linked glycans that have not been processed by medial-Golgi enzymes to complex glycans. Endo H treatment of precipitated FcBPs from AD169-infected cells resulted in a shift of the 31- and 34-kDa proteins to a single band of 24 kDa, while the 34- to 41-kDa band had acquired Endo H resistance (Fig. 3, compare lanes 9 and 10). Likewise, the glycans of the 95- to 105-kDa protein were not removed by Endo H, while the 68-kDa protein was reduced to 33 kDa, indicating extensive N-linked glycosylation (Fig. 3, lanes 9 and 10). To further understand the precursor-product relationship of FcBPs and to determine their half-lives, pulse-chase labeling of fibroblasts infected with AD169 for 72 h was performed. All gp34 molecules were rapidly processed to a higher-molecular-mass form of 34 to 41 kDa, which completely disappeared within 4 h of chase, while gp68 was processed to a polypeptide of 95 to 105 kDa, which also showed a significant loss within 4 h of chase (R. Atalay and H. Hengel, unpublished data). In agreement with recently published work (41), we concluded that in AD169-infected cells two distinct and independently expressed short-lived Fcγ binding glycoproteins of 34 and 68 kDa were synthesized under E and L phase conditions.
FIG. 2.
Expression kinetics of FcBPs during the HCMV replication cycle. (A) MRC5 cells were infected with HCMV strain AD169 at an MOI of 3 for the times indicated. Cell lysates were precipitated with Fc fragments of huIgG, and precipitates were separated by 11.5-to-15%-gradient SDS-PAGE. FcBPs gp34 and gp68 are indicated by arrowheads. (B) Selective and enhanced synthesis of HCMV IE proteins was used by infecting cells in the presence of CHX, which was replaced by ActD (act.D), as described in Materials and Methods. PAA was used as an inhibitor of late phase gene expression.
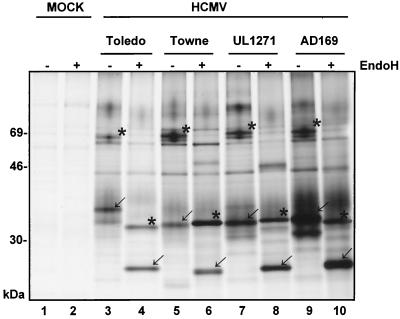
FIG. 3.
Expression of FcBPs in HCMV strains and clinical isolates. MRC5 cells were infected at an MOI of 3 for 72 h with HCMV laboratory strains and the clinical isolate UL1271. Cells were metabolically labeled with [35S]methionine, and proteins from lysates were precipitated with Fc fragments and protein A Sepharose. Before separation of proteins by SDS-11.5 to 15% PAGE, an aliquot of each precipitate was deglycosylated with Endo H. FcBPs and their deglycosylated products are indicated (gp68 and p33 by asterisks; gp34 and p24 by arrows).
Expression of FcBPs in HCMV strains and clinical isolates.
The Fcγ binding activity has been detected on the cell surfaces of HCMV-infected fibroblasts, but MAbs specific for FcγRI, FcγRII, and FcγRIII did not stain infected cells (R. Atalay and H. Hengel, unpublished data) (43). This suggested that either viral proteins account for the Fcγ binding activity observed or HCMV induces other members of the diverse families of cellular FcR molecules, two of which have been reported very recently (12, 25, 59). A number of HCMV proteins have been demonstrated to exhibit subtle electrophoretic differences between HCMV strains. To compare FcBPs between HCMV laboratory strains and clinical isolates, fibroblasts were infected with a panel of viruses and metabolically labeled proteins were subjected to Fc-dependent precipitation. Before separation of proteins by SDS-PAGE, an aliquot of precipitated material was deglycosylated with Endo H. Comparative analysis revealed that proteins similar to gp34 and their deglycosylated product p24 of AD169 were present in all HCMV strains and isolates tested (Fig. 3). The same result was observed for gp68. However, subtle differences in the electrophoretic mobilities were observed, e.g., the gp34 FcBP of AD169 migrated in HCMV Toledo-infected cells at 37 kDa, compatible with the presence of a further site for N-linked glycosylation. Likewise, the deglycosylated product of gp68 in cells infected with the clinical isolate UL1271 migrated slightly more slowly compared to p33 in AD169-infected cells. Thus, the biochemical data suggest that the expression of FcBPs is conserved between HCMV strains. The observed microheterogeneity between FcBPs supported the notion that the factors are encoded by HCMV itself rather than being due to HCMV-induced cellular gene expression.
Construction and analysis of FcBP expression in AD169 deletion mutants.
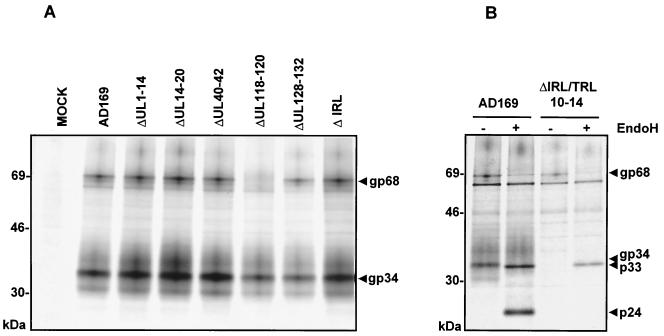
Based on the size of deglycosylated proteins and the estimated number of N-linked glycosylation sites, multiple ORFs predicted for the AD169 genome could represent possible gene candidates. In addition to the gene products predicted from the sequence, several AD169-encoded glycoproteins have been identified (37, 47). Based on this rationale, a genetic approach was followed to identify genes coding for FcBPs. A panel of recombinant HCMV mutants with targeted deletions in gene regions predicted to encode glycoproteins was constructed (Table 2). Altogether, 52 predicted ORFs encompassing 39,795 bp were deleted by homologous recombination of the HCMV AD169-derived infectious BAC pHB5 in E. coli (4). Analysis of replication kinetics demonstrated efficient growth of mutant viruses in human fibroblasts (unpublished data). Deletion mutants were screened for a loss of FcBP expression by Fc precipitation of lysates and SDS-PAGE. When fibroblasts were infected with a mutant lacking the genes UL118-120, a complete loss of gp68 expression was observed, while synthesis of the gp34 FcBP was maintained (Fig. 4A). This finding located the genomic region required for gp68 expression to involve the ORFs UL118-120 of the unique long component of the AD169 genome. Cells infected with the deletion mutants listed in Table 2 expressed gp34 FcBP, indicating that the responsible gene does not reside within one of these gene regions. The AD169 IRL (internal repeat, long segment) gene region represents a duplication of the TRL (terminal repeat, long segment) ORFs TRL1 through TRL14 (8) and includes several predicted glycoprotein genes. Therefore, a further deletion of the TRL10-14 ORFs was introduced into the deletion mutant ΔIRL, resulting in the deletion mutant ΔTRL10-14/IRL lacking 16,278 bp. When this mutant was analyzed, a loss of gp34 synthesis was noted (Fig. 4B), pointing to the ORF TRL11/IRL11 as a likely candidate gene which was predicted to encode a polypeptide with properties similar to that of the gp34 FcBP. In summary, the analysis of HCMV deletion mutants confirmed the expression of two independent FcBPs, gp34 and gp68, by HCMV AD169. The gene encoding gp68 was located within the region UL118-120, and the gene encoding gp34 was located within TRL10-14. Moreover, for the first time it was formally demonstrated that the genomic regions encompassing UL1-14, UL14-20, UL40-42, UL118-120, UL128-132, IRL, and TRL10-14 are dispensable for HCMV replication in vitro.
TABLE 2.
Expression of FcγR homologs by HCMV ts9- and pHB5-derived deletion mutantsa
| Mutant | Deletion | Expression of:
|
|
|---|---|---|---|
| gp34 | gp68 | ||
| ts9b | ΔUS1-15 | + | + |
| HB5-ΔUL1-14 | 11660-21440c | + | + |
| HB5-ΔUL14-20 | 20883-26433 | + | + |
| HB5-ΔUL40-42 | 53401-55537 | + | + |
| HB5-ΔUL118-120 | 168430-169579 | + | − |
| HB5-ΔUL128-132 | 175840-178619 | + | + |
| HB5-ΔIRL | 179150-192329 | + | + |
| HB5-ΔTRL10-14/IRL | 8051-11150 and 179150-192329 | − | + |
MRC5 cells were infected at an MOI of 3 for 72 h with the HCMV mutant indicated. Cells were metabolically labeled with [35S]methionine, and proteins from lysates were precipitated with Fc fragments and protein A Sepharose and separated by SDS11.5 to 15% PAGE. + indicates presence of the FcBP; − indicates absence of the FcBP.
See reference 46.
Ranges represent nucleotide positions.
FIG. 4.
Analysis of FcBPs in HCMV pHB5-derived deletions mutants. (A) MRC5 fibroblasts were infected with HCMV deletion mutants for 72 h, labeled with [35S]methionine, and lysed. Lysates were precipitated with Fc fragments and protein A Sepharose before analysis by SDS-11.5 to 15% PAGE. (B) MRC5 cells were infected with ΔTRL10-14/IRL for 72 h, labeled with [35S]methionine, and lysed. Precipitation of lysates was performed with Fc fragments and protein A Sepharose. Precipitates were digested with Endo H before analysis by SDS-11.5 to 15% PAGE.
Identification of the UL119-118 transcription unit to encode the vFcγR gp68.
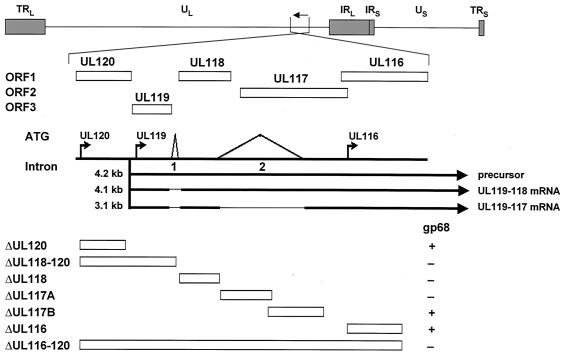
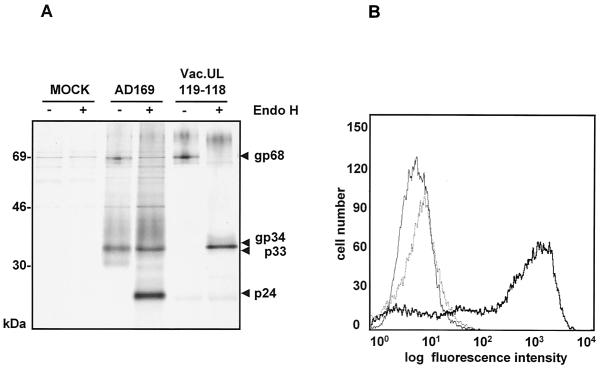
The UL115-119 transcription unit represents a genomic region of high complexity, giving rise to a 4.2-kb RNA precursor undergoing multiple differential splicing events (39). Based on the predicted molecular weight and the number of N-linked glycosylation sites, neither the ORFs UL120, UL119, and UL118, the expression of which is affected in the ΔUL118-120 mutant, nor any of the three hypothetical proteins derived from the UL115-119 transcription unit is compatible with the observed properties of FcBP gp68. To narrow the gene sequences responsible for expression of gp68, a further set of pHB5-derived deletion plasmids was constructed, and reconstituted HCMV mutants were analyzed (Fig. 5). As expected, the mutant ΔUL116-UL120 did not express gp68. Mutants with deletions of UL118 and the 5′ part of UL117 had lost FcBP gp68 synthesis. In contrast, expression was maintained in ΔUL120 and in deletion mutants lacking the 3′ part of UL117 and the coding sequence of UL116 (Fig. 5). These data indicated that the ORFs UL119 and UL118 were required to express gp68. Hence, we postulated that the UL119-115 transcription unit could give rise to a further, as yet unidentified spliced transcript of 4.1 kb (Fig. 5). To test this hypothesis, cDNAs of spliced transcripts derived from the common 4.2-kb mRNA precursor encompassing intron 1 and intron 2 were amplified by reverse transcription-PCR. As a control, amplification of cDNA derived from RNA of ΔUL118-120-infected cells was negative. Sequence analysis of two cloned cDNAs derived from AD169-infected cells identified fairly similar exon-intron boundaries as predicted by Leatham et al., i.e., intron 1 spanning from bp 423 to 511 and intron 2 spanning from bp 932 to 1989 downstream from the UL119 start ATG. As presumed, two cDNA populations were found, including one which connected the UL119 and UL118 ORFs by excision of intron 1, thus coding for a predicted glycoprotein of 33 kDa compatible with FcBP gp68. In the other cDNA introns 1 and 2 were excised, corresponding to the transcript described previously (39). To test the Fc-binding function of both gene products, recombinant vaccinia viruses containing both genes were constructed, vacUL119-118 and vacUL119-117. Precipitation of proteins from the lysate of cells infected with vacUL119-118 using Fc fragments and Protein A Sepharose revealed the presence of a FcBP of 68 kDa with N-linked glycans identical to the FcBP gp68 precipitated from a lysate of AD169-infected cells (Fig. 6A). In contrast, no FcBP was detected in vaccUL119-117-infected cells (A. Zimmermann, R. Atalay, and H. Hengel, unpublished data). As demonstrated by cytofluorometric analysis, Fc fragments strongly decorated the cell surface of vacUL119-118-infected cells, confirming cell surface exposition of the vFcγR gp68 (Fig. 6B). To confirm that the C terminus of vFcγR gp68 is encoded within the UL118 ORF in the context of the HCMV genome, an influenza virus HA epitope tag was inserted into the pHB5 BAC sequence just in front of the native stop codon of the UL118 ORF. After reconstitution of the UL118-HA-tagged virus, biochemical analysis revealed correct features of the HA-bearing vFcγR gp68 (R. Atalay, M. Wagner, and H., Hengel, unpublished data). In conclusion, the data identified a new spliced UL119-118 transcript from the UL119-115 transcription unit of HCMV to encode the vFcγR gp68, designated gpUL119-118.
FIG. 5.
Identification of the transcription unit coding for FcBP gp68 within the HCMV UL120-UL116 gene region. The prototype arrangement of the viral genome consisting of long (UL) and short (US) unique components bounded by terminal repeats (TR) and inverted internal repeats (IR) as well as the position and the direction of transcription of the UL120-UL116 genes are indicated at the top panel. Below, open boxes designate the UL120-UL116 ORFs. The region contains three possible start ATGs indicated by arrows and two possible intron sequences. The mRNAs derived from the 4.2-kb mRNA precursor transcribed from the UL119 promoter are shown as lines interrupted by thin lines indicating splicing events. The positions of deletion mutants within the UL120-UL116 genomic region is given by boxes at the bottom. +, presence of vFcγR gp68 synthesis in cells infected with HCMV deletion mutants; −, absence of vFcγR gp68 synthesis.
FIG. 6.
Expression of vFcγR gp68 by vacUL119-118. CV1 cells were infected at an MOI of 3 for 14 h. (A) After labeling with [35S]methionine was done, precipitation of proteins was performed with Fc fragments and protein A Sepharose. Precipitates were digested with Endo H and analyzed by SDS-11.5 to 15% PAGE. (B) Cells were stained with huIgGFc-FITC, harvested using cell dissociation solution, and analyzed by flow cytometry. Thin line, CV1 cells mock infected; dotted line, wild-type vaccinia-infected cells, bold line, cells infected with vacUL119-118.
The HCMV ORF TRL11 encodes vFcγR gp34.
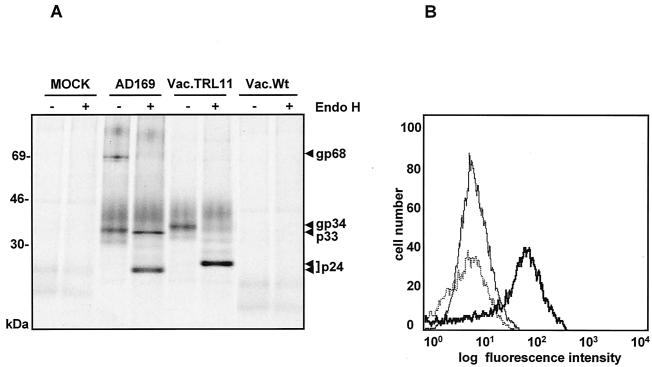
The loss of FcBP gp34 expression in ΔTRL10-14/IRL-infected cells located the responsible gene to the TRL10-14 ORFs. Among those ORFs, the product of TRL11 closely matches the biochemical properties of gp34 in terms of molecular size and number of glycosylation sites. Therefore, the TRL11 sequence was amplified from pHB5 DNA by PCR and cloned into the p7.5K vector, followed by the construction of a vaccinia virus recombinant expressing the TRL11-FLAG sequence. Proteins with FcBP properties were precipitated with Fc fragments from vacTRL11-infected cells, which migrated slightly more slowly than gp34 of HCMV AD169 (Fig. 7A). In addition, immunoprecipitation with an anti-FLAG MAb retrieved a glycoprotein of 34 kDa from the lysate of vacTRL11-infected cells (unpublished data). Endo H digestion of gp34 resulted in a single deglycosylated protein with a mass of 24 kDa, as seen in AD169-infected cells (Fig. 7A). Cytofluorometric analysis of cells infected with vacTRL11 exhibited greater Fcγ-binding activity on the cell surface than control cells (Fig. 7B). In agreement with recently published information (41), our data prove that the TRL11 ORF and its identical copy, IRL11, encode the gp34 vFcγR, designated gpTRL11.
FIG. 7.
Expression of vFcγR gp34 by vacTRL11. CV1 cells were infected at an MOI of 3 for 14 h. (A) After labeling with [35S]methionine was done, precipitation of proteins was performed with Fc fragments and protein A Sepharose. Precipitates were digested with Endo H digestion before analysis by SDS-11.5 to 15% PAGE. (B) Cells were stained with huIgGFc-FITC, harvested using cell dissociation solution, and analyzed by flow cytometry. Thin line, CV1 cells mock infected; dotted line, wild-type vaccinia-infected cells; bold line, cells infected with vacTRL11.
Sequence analysis of the HCMV UL119-118- and TRL11/IRL11-encoded vFcγRs and alignment with cFcγR sequences.
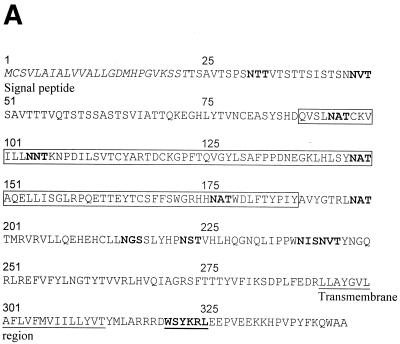
The spliced UL119-118 gene encodes a predicted 347-amino-acid (aa) type Ia transmembrane protein with 12 potential N-glycosylation sites. The predicted signal sequence comprises aa 1 to 25 and the transmembrane region aa 294 to 314 (Fig. 8A ). A potential intracytoplasmic immunoreceptor tyrosine-based-like inhibition motif (consensus sequence I/V/L/SxYxxL) has been noted (57). Likewise, the TRL11 sequence is predicted to encode a 234-aa type Ia transmembrane glycoprotein with three potential N-linked glycosylation sites, a transmembrane sequence of 21 aa, and a cytoplasmic region of 31 aa (Fig. 8B). The cytoplasmic tail contains a dileucine consensus motif (DXXXLL, where X is unknown) indicating a potential function in intracellular targeting of the protein to the endocytic route (40).
FIG. 8.
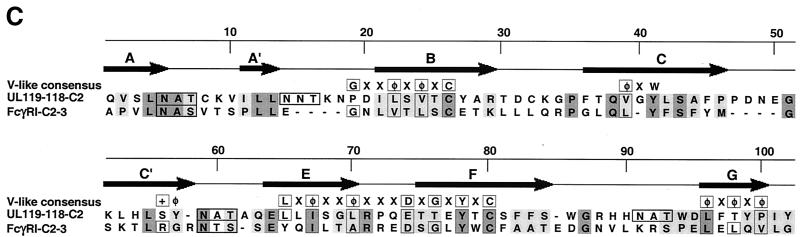
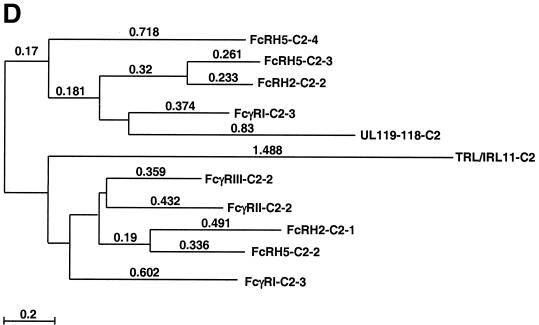
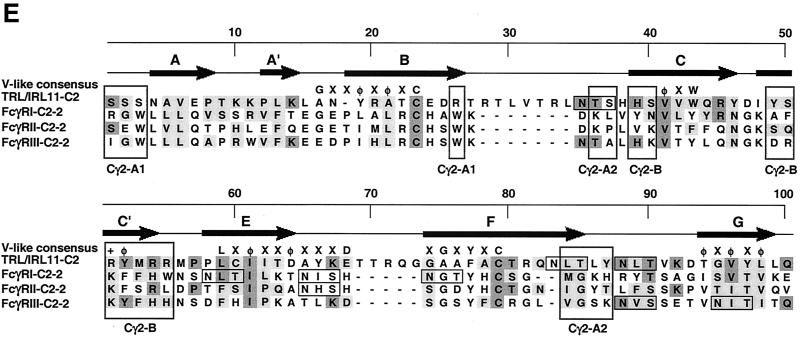
Sequence analysis and alignment of gpUL119-118 and gpTRL11/IRL11. The deduced amino acid sequences of the UL119-118 ORF (A) and the TRL11/IRL11 ORF (B) are shown. The putative signal peptides are shown in italics, the putative transmembrane regions are underlined, and N-glycosylation sites are given in bold. The boxes indicate the putative IgSF domains of gpUL119-118 and gpTRL11. Putative cytoplasmic signaling motifs are in bold and underlined, i.e., a potential tyrosine-based motif in gpUL119-118 and an internalization motif for gpTRL11. Sequence alignments were performed by the program MacVector 7.02r using the ClustalW method with a PAM350 similarity matrix. For detailed analysis, the gpUL119-118 IgSF-like domain (aa 91 to 190) was aligned with the third domain of human FcγRI (aa 191 to 282; GenBank accession number X14356). (C) Amino acid residues are represented by the one-letter code, with dashes indicating gaps in the alignments. Identical residues are shaded in dark grey; residues showing similarity, defined as acidic (D or E), basic (H, K, or R), hydrophobic (A, F, I, L, M, P, V, or W) or polar (C, G, N, Q, S, T, or Y) are shaded in light grey. The N-glycosylation sites are boxed in black. The V-like consensus sequence (56) is shown above the alignment (+ represents a basic residue; φ represents a hydrophobic residue). Residues fitting the consensus are boxed in dark grey. The arrows above the sequence indicate the putative β-sheets deduced from the crystal structure of FcγRII (60). (D) A phylogenetic tree was generated by alignment of the IgSF-like domains of gpUL119-118 (aa 91 to 190) and gpTRL (aa 24 to 122) with the Fc-binding second domains of human FcγRI (aa 102 to 187; GenBank accession no. X14356), FcγRII (aa 121 to 207; accession no. M31932), and FcγRIII (aa 106 to 192; accession no. X16863), the third domain of human FcγRI (aa 191 to 282), and different domains of two human cellular Fc receptor homologs (12), FcRH2 (GenBank accession no. AY043465) domains 1 (aa 17 to 103) and 2 (aa 110 to 198), as well as FcRH5 (accession no. AF397453) domains 2 (aa 102 to 187), 3 (aa 193 to 282), and 4 (aa 288 to 377). Alignment of the TRL11 IgSF-like domain (aa 24 to 122) with the Fc-binding second domains of human FcγRI (aa 102 to 187; GenBank accession number X14356), FcγRII (aa 121 to 207; accession number M31932), and FcγRIII (aa 106 to 192; accession number X16863) is shown (E). The arrows above the sequence indicate the β-sheet structure of FcγRII/III based on crystal structure analysis (60, 68). The boxes indicate the contact regions for the two Fc-chains of huIgG (Cγ2-A and Cγ2-B) as determined by crystal structure analysis (61).
The common Fcγ binding property and plasma membrane expression link HCMV-encoded vFcγRs with cFcγRs, i.e., FcγRI, which is the high-affinity receptor for monomeric IgG, and FcγRII and FcγRIII, which are low-affinity receptors binding aggregated IgG (57). The extracellular chains of FcγRII and FcγRIII consist of two IgSF-like domains arranged in tandem, while that of FcγRI comprises three IgSF-like domains. IgSF domains are characterized by a structural motif termed the immunoglobulin fold (66). This feature is a specialized β-barrel of polypeptide strands, forming antiparallel β-pleated sheets. Each Ig domain is composed of two β-pleated sheets, one containing four β strands, the other consisting of at least three β strands. Loops of variable length connect the different strands. Stability is provided by a disulfide bond near the domain's core, covalently linking the two sheet layers. Ig domains are divided into several sets according to the sequence and folding topology. IgSF domains of cFcγRs share features of the C2 set and I set of Ig domains and are structurally related to killer cell Ig-like receptors (13). The binding of Fc is mediated by the membrane-proximal second IgSF domain of cFcγRs (61), while the membrane-proximal third domain of FcγRI is required to achieve high-affinity binding of IgG (30). To identify potential common features in sequence and structure, the predicted amino acid sequence of gpUL119-118 and gpTRL11 was used to identify sequences homologous to human cFcγRs. The sequences of vFcγRs and cFcγRs, aligned by the program MacVector, revealed significant homology between the extracellular domains of gpUL119-118 and gpTRL11 molecules, respectively, and cFcγRs. Specifically, a single IgSF-like domain each present in both vFcγRs was aligned with IgSF domains of cFcγRs. The potential IgSF-like domains of the vFcγRs fulfill the criteria for IgSF domains (66) in that domain-sized sequences are present which include a conserved disulfide bond, they share similarity to Ig-related domains, and they are likely to share key structural features of the Ig-fold. The IgSF-like domain identified in gpUL119-118 ranging from aa 91 to 190 is in good agreement with the V-like domain consensus sequence (56), including a conserved disulfide bond between β strand B and F, while sequences of the loop residues varied considerably (Fig. 8C). The gpUL119-118 IgSF domain showed its highest degree of homology to the third domain of FcγRI (20% aa identity, 31% aa similarity) (Fig. 8D), including conserved N-glycosylation sites. This degree of relatedness is in a similar range as found for homologous cFcγR domains (e.g., second domains of FcγRII and FcγRIII, which have 39% identity and 14% similarity) and heterologous cFcγR domains (e.g., the third domain of FcγRI and the second domain of FcγRII, which share 15% identity and 17% similarity). In contrast, sequence relatedness of gpUL119-118 with the second domain of FcγRII/III and particularly with the IgSF-like domain of gpTRL11 is clearly at a lower level (12% identity, 18% similarity; 7% identity, 17% similarity, respectively).
Alignment of the gpTRL11 IgSF-like domain (aa 24 to 122) revealed the second domain of FcγRIII as the closest cellular homolog (12% identity, 27% similarity) (Fig. 8D), including one conserved N-glycosylation site. The three-dimensional crystal structures of the extracellular part of FcγRIII (68) as well as of the IgG-FcγRIII complex (61) have been determined. Alignment of the gpTRL11 IgSF-like domain with the respective second domains of cFcγRs is shown in Fig. 8E. Sequence comparison of gpTRL11 residing at equivalent positions with those forming the three main contact areas between FcγRIII and Fc revealed a good agreement concerning the interface with the second Fc chain (C2γ-B) within the C/C′ region (Fig. 8E). In contrast, residues mediating contact to the other Fc-chain (C2γ-A) are either not conserved (C2γ-A1) in gpTRL11 or allow the addition of a new N-linked glycan (C2γ-A2). Taken together, alignment of the gpUL119-118 as well as of the gpTRL11 vFcγR amino acid sequences with cFcγR domains predicts the presence of a single IgSF-like domain in the extracellular regions of each viral FcγR homolog.
DISCUSSION
Here, we report on the identification and expression of HCMV genes which code for type I transmembrane glycoproteins with IgG Fc binding properties. Taking advantage of a panel of HCMV strains that allowed us to demonstrate biochemical microheterogeneity between FcBPs, the hypothesis that HCMV-induced cellular FcγR genes may account for the observed FcBPs became implausible. In a systematic approach, deletion of a total of almost 40-kbp encompassing 52 predicted ORFs, including 36 ORFs coding for bona fide nonessential glycoproteins, was performed to locate the responsible gene regions within the 229,351-bp AD169 genome. This approach appeared promising, since the genes m138 of MCMV and US7/US8 of HSV coding for vFcγR molecules are dispensable for virus replication in vitro (11, 42, 50). Indeed, this assumption was confirmed in the cases of both of the HCMV-encoded vFcγRs. Moreover, we extend the list of predicted HCMV ORFs not required for replication in vitro by 52. Altogether three early genes of HCMV AD169 were demonstrated to code for two vFcγR molecules with distinct biochemical properties. The ORF IRL11 and its identical copy, TRL11, encode the vFcγR gp34, confirming recently published work (41). The spliced gene UL119-118 codes for the vFcγR gp68. Expression of the viral genes during the HCMV replication cycle occurs simultaneously yet independently. Isolated expression of their products proved that each molecule has intrinsic Fc binding capabilities. Both FcBPs readily reach the cell surface, thus constituting genuine FcγRs.
Simultaneous synthesis of two vFcγRs is a property unique to HCMV among the herpesviruses. At a first glance, this feature adds another example to the equipment with redundant glycoproteins that HCMV employs to modulate major histocompatibility complex (MHC) molecules (26) or the multitude of G coupled receptor homologs within the HCMV genome (8). According to rough structural, functional, and genetic criteria, such as binding specificities for IgG of different species and their sequence relatedness with cFcγRs, gpTRL11 and gpUL119-118 can be readily discriminated. Many viruses, particularly herpesviruses, have captured and modified cellular genes to decoy the host immune response. HCMV encodes homologs of MHC class I (8), chemokine receptors (8), chemokines (54), and interleukin-10 (37). Both vFcγRs appear phylogenetically congruent with host FcγRs and are predicted to form IgSF-like domains. Specifically, gpUL119-118 relates most closely to the third domain of FcγRI, while the gpTRL11 is reminiscent of the second domain of FcγRII/III. Facilitated by the available ultrastructural information on cFcγRs (60, 61, 68), this study identifies for the first time the presence of structural homologs of cellular FcγRs within a cytomegaloviral genome. Detailed sequence alignment of the MCMV m138 gene reveals significant homology with mouse FcγRs and predicts the preservation of three FcγR-derived putative IgSF domains (A. Zimmermann and H. Hengel, unpublished data), further highlighting the impressive diversification and redundancy of FcγR structures. It is tempting to speculate that the presence of vFcγR homologs may indeed constitute a typical feature of CMV genomes.
Distinct IgSF-like domains within vFcγRs reflect genealogical differences of the inserted genes and independent integration into the relatively stable HCMV DNA genome. Differential conservation and variation of vFcγRs during millions of years of HCMV coevolution with its host is likely to reflect adaptation to and selection for specific molecular purposes. While the conserved hydrophobic core residues of β sheets promote folding of IgSF domains, the loop residues are free to vary considerably and may serve as a substrate for selection in phylogeny. New state-of-the-art algorithms (34) (available from the internet at http://bioinf.cs.ucl.ac.uk/psiform.html) providing largely correct structural information of already crystallized FcγRII and FcγRIII suggest from the amino acid sequence of gpUL119-118 a composition of seven β sheets for the putative IgSF-like domain in gpUL119-118 resembling the architecture of IgSF domains of cFcγRs (Zimmermann and Hengel, unpublished). β sheets within the IgSF-like domain of gpTRL11 predicted by this algorithm are supposed to be interrupted by an α-helical conformation. Interestingly, the Fc binding contact interface of the neonatal Fc receptor includes a helical domain as determined by crystal structure (5, 6). Given such a scenario, gpUL119-118 might have been selected for the maintenance of the IgSF domain during evolution, while a different selection pressure allowed the disruption of the IgSF fold of the gpTRL11 molecule while retaining its Fc binding capability.
The topological composition of vFcγRs differs from the extracellular part of cFcγRs. FcγRI possesses an extra IgSF domain compared with the lower-affinity receptors FcγRII and FcγRIII, which shows less similarity with the other two domains. The third domain of FcγR is responsible for high-affinity Fc binding. Analysis of mutant and chimeric FcγRs showed that removal of the third domain of FcγRI abrogates high-affinity binding to monomeric IgG, although domains 1 and 2 were sufficient to retain a weak affinity for IgG (30). It is, therefore, remarkable that single IgSF-like domains of the HCMV-encoded FcγRs are sufficient to mediate strong Fc-dependent binding of IgG. Moreover, the relative position of IgSF domains in both vFcγR molecules of HCMV differs in that they are located in the N′ terminal part of the molecule. The presence of a single IgSF-domain near the N′ terminus is a structural feature resembling the recently discovered Fcα/μR (59). Assessment of binding affinities of IgG subclasses, domain swap experiments, and determination of the disulfide bonding pattern and crystal structures are required to investigate the impact of the Ig-SF-like domains for the interaction of gpTRL11 and gpUL119-118 with Fc.
At present, the biological role of the HCMV-encoded vFcγRs is still unclear. A paradigm for the function of virus-encoded FcγRs is represented by HSV gE/gI binding both IgG aggregates and monomers (7, 17). Alignment of the extracellular HSV gE residues required for IgG binding (aa 322 to 359) reveals some homology with the second domain of cFcγRs (17). Binding the Fc part of immune IgG that recognizes its target antigen on the surface of infected cells or viral particles, HSV gE/gI initiates a process called antibody bipolar bridging (20). As a consequence, the vFcγR blocks antiviral activities mediated by the Fc domain of IgG, e.g., complement fixation and activation as well as ADCC (18). In vivo experiments using a mouse model of HSV infection support the notion that this mechanism may provide significant protection to the virus against antibody-mediated immunity and thus gives a replicative advantage (49). The function of the MCMV m138-encoded vFcγR is less clear. As expected for its hypothetical role in immune evasion, an MCMV mutant lacking m138 expression showed drastically reduced replication in vivo (11). However, attenuation of the mutant was also seen in antibody-deficient μMT− mice, which was interpreted to mean that this phenotype is not mainly dependent on the Ig binding property. This conclusion could be premature for two reasons. First, deletion of m138 may affect the transcription of flanking genes, since several ORFs of this gene region share common mRNA transcription units (63). Neighboring genes of this region have been implicated in macrophage tropism and thereby MCMV replication in vivo (23, 24). Second, replication of the Δm138 mutant was assessed only during the primary phase of MCMV infection. In this situation, antibodies do not contribute to viral control (35). The data do not rule out that the m138-encoded vFcγR still has a role during recurrent phases of infection when antibodies efficiently limit virus dissemination and transmission (35, 55).
Besides interacting with immune IgG, vFcγRs could serve further purposes. Given the fact that CMV targets cells constitutively expressing cFcγRs, like monocytes, macrophages, and dendritic cells for latent and productive infection, vFcγRs could act as an agonist or competitive antagonist for cFcγRs. The cFcγR-Ig interaction may be a promising target for the downregulation of immune responses or the control over the activation state of myelomonocytic cells. Notably, HCMV-encoded FcγRs share not only structural features and the Fc-binding capability with cFcγRs, but also the propensity to enter the endocytic pathway. The restricted half-life for both gpTRL11 and gpUL119-118 is due to a rapid degradation in endolysosomal compartments (R. Atalay and H. Hengel, unpublished data). Considering the abundant synthesis and fast endocytosis of gpTRL11 and gpUL119-118 at late times of infection, vFcγRs could serve as an efficient transport device, disposing antiviral antibodies from the cell surface to the endolysosome for destruction. Hypothetically, this pathway could be exploited by antibody-coated viral particles to enter CMV-infected cells. Certain viruses, e.g., human immunodeficiency virus (HIV), are able to initiate gene expression and replication after entry via endosomal compartments (19). Indeed, HCMV-induced Fc binding molecules have been proposed to confer HIV susceptibility in an antibody-dependent manner to human fibroblasts which lack the CD4 receptor (44). This could open up a CD4-negative compartment for HIV replication where HIV may benefit from the multitude of stealth mechanisms by which HCMV avoids immune attack.
Acknowledgments
R.A. and A.Z. contributed equally to this work.
We are grateful to Katja Wichmann and Bettina Bauer for expert technical assistance, Henrike Reinhard for constructing vaccinia virus recombinants, and Thomas Mertens, University of Ulm, for generously providing clinical isolates.
This study was supported by grants from the Deutsche Forschungsgemeinschaft through SFB 421 project A8 and SFB 455 projects A6, A2, and A7.
REFERENCES
- 1.Amigorena, S., and C. Bonnerot. 1999. Fc receptor signaling and trafficking: a connection for antigen processing. Immunol. Rev. 172:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Antonsson, A., and P. J. Johansson. 2001. Binding of human and animal immunoglobulins to the IgG Fc receptor induced by human cytomegalovirus. J. Gen. Virol. 82:1137-1145. [DOI] [PubMed] [Google Scholar]
- 3.Baucke, R. B., and P. G. Spear. 1979. Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J. Virol. 32:779-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burmeister, W. P., L. N. Gastinel, N. E. Simister, M. L. Blum, and P. J. Bjorkman. 1994. Crystal structure at 2.2 A resolution of the MHC-related neonatal Fc receptor. Nature 372:336-343. [DOI] [PubMed] [Google Scholar]
- 6.Burmeister, W. P., A. H. Huber, and P. J. Bjorkman. 1994. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 372:379-383. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, T. L., I. You, I. M. Joseph, P. J. Bjorkman, S. L. Morrison, and M. Raghavan. 1999. Characterization of the interaction between the herpes simplex virus type I Fc receptor and immunoglobulin G. J. Biol. Chem. 274:6911-6919. [DOI] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Costa, J., C. Yee, Y. Nakamura, and A. Rabson. 1978. Characteristics of the Fc receptor induced by herpes simplex virus. Intervirology 10:32-39. [DOI] [PubMed] [Google Scholar]
- 11.Crnkovic-Mertens, I., M. Messerle, I. Milotic, U. Szepan, N. Kucic, A. Krmpotic, S. Jonjic, and U. H. Koszinowski. 1998. Virus attenuation after deletion of the cytomegalovirus Fc receptor gene is not due to antibody control. J. Virol. 72:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, R. S., Y. H. Wang, H. Kubagawa, and M. D. Cooper. 2001. Identification of a family of Fc receptor homologs with preferential B cell expression. Proc. Natl. Acad. Sci. USA 98:9772-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis, G., Jr., H. Kubagawa, and M. D. Cooper. 2000. Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc. Natl. Acad. Sci. USA 97:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowler, K. W., and R. W. Veltri. 1984. In vitro neutralization of HSV-2: inhibition by binding of normal IgG and purified Fc to virion Fc receptor (FcR). J. Med. Virol. 13:251-259. [DOI] [PubMed] [Google Scholar]
- 17.Dubin, G., S. Basu, D. L. Mallory, M. Basu, R. Tal-Singer, and H. M. Friedman. 1994. Characterization of domains of herpes simplex virus type 1 glycoprotein E involved in Fc binding activity for immunoglobulin G aggregates. J. Virol. 68:2478-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubin, G., E. Socolof, I. Frank, and H. M. Friedman. 1991. Herpes simplex virus type 1 Fc receptor protects infected cells from antibody-dependent cellular cytotoxicity. J. Virol. 65:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fackler, O. T., and B. M. Peterlin. 2000. Endocytic entry of HIV-1. Curr. Biol. 10:1005-1008. [DOI] [PubMed] [Google Scholar]
- 20.Frank, I., and H. M. Friedman. 1989. A novel function of the herpes simplex virus type 1 Fc receptor: participation in bipolar bridging of antiviral immunoglobulin G. J. Virol. 63:4479-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frey, J., and B. Einsfelder. 1984. Induction of surface IgG receptors in cytomegalovirus-infected human fibroblasts. Eur. J. Biochem. 138:213-216. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa, T., E. Hornberger, S. Sakuma, and S. A. Plotkin. 1975. Demonstration of immunoglobulin G receptors induced by human cytomegalovirus. J. Clin. Microbiol. 2:332-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 75:6292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson, L. K., J. S. Slater, Z. Karabekian, H. W. T. Virgin, C. A. Biron, M. C. Ruzek, N. van Rooijen, R. P. Ciavarra, R. M. Stenberg, and A. E. Campbell. 1999. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J. Virol 73:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzivassiliou, G., I. Miller, J. Takizawa, N. Palanisamy, P. H. Rao, S. Iida, S. Tagawa, M. Taniwaki, J. Russo, A. Neri, G. Cattoretti, R. Clynes, C. Mendelsohn, R. S. Chaganti, and R. Dalla-Favera. 2001. IRTA1 and IRTA2, novel immunoglobulin superfamily receptors expressed in B cells and involved in chromosome 1q21 abnormalities in B cell malignancy. Immunity 14:277-289. [DOI] [PubMed] [Google Scholar]
- 26.Hengel, H., W. Brune, and U. H. Koszinowski. 1998. Immune evasion by cytomegalovirus—survival strategies of a highly adapted opportunist. Trends Microbiol. 6:190-197. [DOI] [PubMed] [Google Scholar]
- 27.Hengel, H., C. Esslinger, J. Pool, E. Goulmy, and U. H. Koszinowski. 1995. Cytokines restore MHC class I complex formation and control antigen presentation in human cytomegalovirus-infected cells. J. Gen. Virol. 76:2987-2997. [DOI] [PubMed] [Google Scholar]
- 28.Hengel, H., J. O. Koopmann, T. Flohr, W. Muranyi, E. Goulmy, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623-632. [DOI] [PubMed] [Google Scholar]
- 29.Huber, R., J. Deisenhofer, P. M. Colman, M. Matsushima, and W. Palm. 1976. Crystallographic structure studies of an IgG molecule and an Fc fragment. Nature 264:415-420. [DOI] [PubMed] [Google Scholar]
- 30.Hulett, M. D., N. Osman, I. F. McKenzie, and P. M. Hogarth. 1991. Chimeric Fc receptors identify functional domains of the murine high affinity receptor for IgG. J. Immunol. 147:1863-1868. [PubMed] [Google Scholar]
- 31.Johnson, D. C., and V. Feenstra. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J. Virol. 61:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, D. C., M. C. Frame, M. W. Ligas, A. M. Cross, and N. D. Stow. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, D. C., and A. B. Hill. 1998. Herpesvirus evasion of the immune system. Curr. Top. Microbiol. Immunol. 232:149-177. [DOI] [PubMed] [Google Scholar]
- 34.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 35.Jonjic, S., I. Pavic, B. Polic, I. Crnkovic, P. Lucin, and U. H. Koszinowski. 1994. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J. Exp. Med. 179:1713-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller, R., R. Peitchel, J. N. Goldman, and M. Goldman. 1976. An IgG-Fc receptor induced in cytomegalovirus-infected human fibroblasts. J. Immunol. 116:772-777. [PubMed] [Google Scholar]
- 37.Kotenko, S. V., S. Saccani, L. S. Izotova, O. V. Mirochnitchenko, and S. Pestka. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 97:1695-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langone, J. J. 1982. Protein A of Staphylococcus aureus and related immunoglobulin receptors produced by streptococci and pneumonococci. Adv. Immunol. 32:157-252. [PubMed] [Google Scholar]
- 39.Leatham, M. P., P. R. Witte, and M. F. Stinski. 1991. Alternate promoter selection within a human cytomegalovirus immediate-early and early transcription unit (UL119-115) defines true late transcripts containing open reading frames for putative viral glycoproteins. J. Virol. 65:6144-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letourneur, F., and R. D. Klausner. 1992. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell 69:1143-1157. [DOI] [PubMed] [Google Scholar]
- 41.Lilley, B. N., H. L. Ploegh, and R. S. Tirabassi. 2001. Human cytomegalovirus open reading frame TRL11/IRL11 encodes an immunoglobulin G Fc-binding protein. J. Virol. 75:11218-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longnecker, R., S. Chatterjee, R. J. Whitley, and B. Roizman. 1987. Identification of a herpes simplex virus 1 glycoprotein gene within a gene cluster dispensable for growth in cell culture. Proc. Natl. Acad. Sci. USA 84:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacCormac, L. P., and J. E. Grundy. 1996. Human cytomegalovirus induces an Fc gamma receptor (Fc gammaR) in endothelial cells and fibroblasts that is distinct from the human cellular Fc gammaRs. J. Infect. Dis. 174:1151-1161. [DOI] [PubMed] [Google Scholar]
- 44.McKeating, J. A., P. D. Griffiths, and R. A. Weiss. 1990. HIV susceptibility conferred to human fibroblasts by cytomegalovirus-induced Fc receptor. Nature 343:659-661. [DOI] [PubMed] [Google Scholar]
- 45.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 46.Mockenhaupt, T., M. Reschke, E. Bogner, B. Reis, and K. Radsak. 1994. Structural analysis of the US-segment of a viable temperature sensitive human cytomegalovirus mutant. Arch. Virol. 137:161-169. [DOI] [PubMed] [Google Scholar]
- 47.Mullberg, J., M. L. Hsu, C. T. Rauch, M. J. Gerhart, A. Kaykas, and D. Cosman. 1999. The R27080 glycoprotein is abundantly secreted from human cytomegalovirus-infected fibroblasts. J. Gen. Virol. 80:437-440. [DOI] [PubMed] [Google Scholar]
- 48.Murayama, T., S. Natsuume-Sakai, K. Shimokawa, and T. Furukawa. 1986. Fc receptor(s) induced by human cytomegalovirus bind differentially with human immunoglobulin G subclasses. J. Gen. Virol. 67:1475-1478. [DOI] [PubMed] [Google Scholar]
- 49.Nagashunmugam, T., J. Lubinski, L. Wang, L. T. Goldstein, B. S. Weeks, P. Sundaresan, E. H. Kang, G. Dubin, and H. M. Friedman. 1998. In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J. Virol. 72:5351-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neidhardt, H., C. H. Schroder, and H. C. Kaerner. 1987. Herpes simplex virus type 1 glycoprotein E is not indispensable for viral infectivity. J. Virol. 61:600-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogata, M., and S. Shigeta. 1979. Appearance of immunoglobulin G Fc receptor in cultured human cells infected with varicella-zoster virus. Infect. Immun. 26:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oleszak, E. L., and J. L. Leibowitz. 1990. Immunoglobulin Fc binding activity is associated with the mouse hepatitis virus E2 peplomer protein. Virology 176:70-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Para, M. F., R. B. Baucke, and P. G. Spear. 1980. Immunoglobulin G(Fc)-binding receptors on virions of herpes simplex virus type 1 and transfer of these receptors to the cell surface by infection. J. Virol. 34:512-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Penfold, M. E., D. J. Dairaghi, G. M. Duke, N. Saederup, E. S. Mocarski, G. W. Kemble, and T. J. Schall. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA 96:9839-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polic, B., H. Hengel, A. Krmpotic, J. Trgovcich, I. Pavic, P. Luccaronin, S. Jonjic, and U. H. Koszinowski. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 188:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raghavan, M., and P. J. Bjorkman. 1996. Fc receptors and their interactions with immunoglobulins. Annu. Rev. Cell Dev. Biol. 12:181-220. [DOI] [PubMed] [Google Scholar]
- 57.Ravetch, J. V., and S. Bolland. 2001. IgG Fc receptors. Annu. Rev. Immunol. 19:275-290. [DOI] [PubMed] [Google Scholar]
- 58.Sakuma, S., T. Furukawa, and S. A. Plotkin. 1977. The characterization of IgG receptor induced by human cytomegalovirus. Proc. Soc. Exp. Biol. Med. 155:168-172. [DOI] [PubMed] [Google Scholar]
- 59.Shibuya, A., N. Sakamoto, Y. Shimizu, K. Shibuya, M. Osawa, T. Hiroyama, H. J. Eyre, G. R. Sutherland, Y. Endo, T. Fujita, T. Miyabayashi, S. Sakano, T. Tsuji, E. Nakayama, J. H. Phillips, L. L. Lanier, and H. Nakauchi. 2000. Fc alpha/mu receptor mediates endocytosis of IgM-coated microbes. Nat. Immunol. 1:441-446. [DOI] [PubMed] [Google Scholar]
- 60.Sondermann, P., R. Huber, and U. Jacob. 1999. Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 A resolution. EMBO J. 18:1095-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sondermann, P., R. Huber, V. Oosthuizen, and U. Jacob. 2000. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature 406:267-273. [DOI] [PubMed] [Google Scholar]
- 62.Stannard, L. M., and D. R. Hardie. 1991. An Fc receptor for human immunoglobulin G is located within the tegument of human cytomegalovirus. J. Virol. 65:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thale, R., P. Lucin, K. Schneider, M. Eggers, and U. H. Koszinowski. 1994. Identification and expression of a murine cytomegalovirus early gene coding for an Fc receptor. J. Virol. 68:7757-7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 65.Wagner, M., and U. H. Koszinowski. Mutagenesis of viral BAC plasmids with linear DNA fragments (ET recombination). In Z. Shaying (ed.), Methods in molecular biology, vol. 23, Bacterial artificial chromosomes: methods and protocols, in press. Humana Press, Tolowa, N.J.
- 66.Williams, A. F., and A. N. Barclay. 1988. The immunoglobulin superfamily—domains for cell surface recognition. Annu. Rev. Immunol. 6:381-405. [DOI] [PubMed] [Google Scholar]
- 67.Xu, B., T. Murayama, K. Ishida, and T. Furukawa. 1989. Characterization of IgG Fc receptors induced by human cytomegalovirus. J. Gen. Virol. 70:893-900. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, Y., C. C. Boesen, S. Radaev, A. G. Brooks, W. H. Fridman, C. Sautes-Fridman, and P. D. Sun. 2000. Crystal structure of the extracellular domain of a human Fc gamma RIII. Immunity 13:387-395. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]