Abstract
Most current evidence suggests that the infection of resting CD4+ T cells by human immunodeficiency virus type 1 (HIV-1) is not productive due to partial or complete blocks in the viral life cycle at steps prior to integration of the viral genome into the host cell chromosome. However, stimulation of an infected resting T cell by antigen, cytokines, or microenvironmental factors can overcome these blocks and allow for the production of progeny virions. In this study, we sought to understand the structure and fate of the virus in unstimulated resting CD4+ T cells. Using a novel linker-mediated PCR assay designed to detect and characterize linear unintegrated forms of the HIV-1 genome, we demonstrate that reverse transcription can proceed to completion following the infection of resting T cells, generating the substrate for the retroviral integration reaction. However, reverse transcription in resting T cells is far slower than in activated T cells, requiring 2 to 3 days to complete. The delay in completing reverse transcription may make the viral DNA genome more susceptible to competing decay processes. To explore the relationship between the formation of the linear viral genome and the stability of the preintegration state, we employed a recombinant HIV-1 virus expressing the enhanced green fluorescent protein to measure the rate at which HIV-1 decays in the preintegration state. Our results demonstrate that the preintegration state is labile and decays rapidly (half-life = 1 day) following the entry of HIV-1 into a resting T cell, with significant decay occurring during the slow process of reverse transcription.
The persistence of human immunodeficiency virus type 1 (HIV-1) in patients treated for long periods of time with powerful combinations of antiretroviral drugs highlights a need for a better understanding of viral reservoirs (reviewed in reference 31). To date, the most extensively characterized long-lived reservoir for HIV-1 in infected individuals is a reservoir composed of latently infected resting CD4+ T lymphocytes carrying integrated provirus (3, 5). In treated patients who have stable suppression of viremia to below the limits of detection (plasma HIV-1 RNA levels below 50 copies per ml), longitudinal analysis of the frequency of resting CD4+ T cells carrying replication-competent provirus demonstrates that this compartment is highly stable (half-life of 6 to 44 months) and has the potential to reinitiate disseminated viremia in infected individuals following the cessation of therapy (6, 12, 13, 29, 41, 47). Therefore, this stable reservoir represents a serious obstacle to HIV-1 eradication with current therapeutic regimens.
HIV-1 latency is best understood in the context of normal CD4+ T-cell physiology. CD4+ T lymphocytes are heterogeneous with respect to a number of parameters including state of differentiation and activation. Following maturation in the thymus, naive CD4+ T cells traffic in a quiescent state through the blood, lymphatics, and peripheral lymphoid tissues, awaiting encounter with cells presenting antigens capable of being recognized by their antigen-specific T-cell receptors (reviewed in reference 16). Exposure to an antigen results in a dramatic increase in the metabolic state of the T cell, up-regulation of large sets of genes involved in immunologic effector function, and entry into the cell cycle (7). Following the clearance of antigens resulting from a successful immune response, the majority of activated T cells die by programmed cell death. However, a small fraction of the activated, antigen-specific T cells survive and return to a resting state. These memory T cells are capable of activation upon reencountering the relevant antigen and comprise the CD4+ component of the memory response (reviewed in reference 8).
A primary target for productive HIV-1 infection is the activated CD4+ T lymphocyte (23). Soon after entry of HIV-1 into an activated T cell, the viral RNA genome is reverse transcribed into DNA, imported into the nucleus, and integrated into a host cell chromosome. These processes allow for the production of progeny viruses, which often results in the death of the infected cell. In contrast, most studies suggest that the interaction of HIV-1 with resting CD4+ T cells does not result in a productive infection. Pioneering studies by Zack et al. (42, 43) and Bukrinsky et al. (2) demonstrated an apparent block in HIV-1 replication in resting CD4+ T cells prior to the integration of the viral genome into the host cell chromosome. Zack et al. suggested that this block reflects an inability to complete the reverse transcription reaction (42, 43). This may be due to insufficient levels of nucleotide precursors (20) and/or a requirement to enter the cell cycle and transit through the G1b phase (19). An alternative hypothesis concerning the block in HIV-1 replication is that a resting T cell does not contain sufficient stores of ATP required for the translocation of the very large preintegration complex (PIC) into the nucleus of the nondividing lymphocyte (39). While these blocks normally prevent the production of progeny virions following the infection of CD4+ resting T cells, infected resting cells can be induced to release virus upon stimulation (2, 43). Therefore, HIV-1 residing in resting CD4+ T cells is in a state of preintegration latency, awaiting stimulation and a transition to productive infection.
Since the majority of CD4+ T lymphocytes in the body are in a resting state, understanding the fate of viral genomes in resting T cells is an important issue in the study of the dynamics of HIV-1 infection in vivo. Although in vitro studies cited above suggest that reverse transcription does not go to completion in resting cells, other studies have demonstrated that resting CD4+ T cells isolated from the blood of HIV-1-infected individuals contain completely reverse transcribed viral DNA. The majority of this DNA is unintegrated (2, 3). Interpretation of these studies is complicated by the fact that, in vivo, it is not possible to establish the activation state of the cell at the time of virus entry and reverse transcription. Additionally, the microenvironment in which the cell is exposed to virus can influence the degree to which HIV-1 can perform the reverse transcription and integration reactions. For example, exposure of resting CD4+ T cells to cytokines and chemokines has been shown to partially activate resting T lymphocytes to a degree sufficient to allow not only for complete reverse transcription but also for integration and virus gene expression, events that are not generally observed in resting T cells in the absence of an activating stimulus (40). Resting T cells trafficking through peripheral tissues in vivo may be exposed to stimulatory cytokines or chemokines that allow for viral gene expression, as demonstrated by in situ RNA hybridization studies (48). Recent work by Eckstein and colleagues supports these observations by demonstrating that naive T cells isolated from tonsils have the capacity to produce virus in the absence of antigenic stimulation, perhaps due to the complex microenvironment of the histocultures (10). Taken together, these studies demonstrate that subtle changes in the activation state of a resting T cell may influence the outcome of HIV-1 infection. Further studies are required to understand at a molecular level the fate of the HIV-1 in resting cells.
In patients with untreated HIV-1 infection, most of the HIV-1 DNA present in resting CD4+ T cells isolated from the blood is unintegrated, suggesting that, in vivo, virus in the preintegration state is common (2, 3). A recent longitudinal study examined the frequency of resting CD4+ T cells capable of producing virus following in vitro stimulation in patients treated soon after exposure to HIV-1 (1). Most of the patients enrolled in this study had high levels of virus in the plasma prior to the initiation of highly active antiretroviral therapy. The amount of virus that could be rescued from resting CD4+ T cells by activation decayed in a biphasic fashion, suggesting differential stability of virus in the preintegration and postintegration states.
In this study, we sought to understand the structure and fate of the virus in the preintegration state of latency in unstimulated resting CD4+ T cells. Our results demonstrate that the preintegration state is labile due to a rapid decay of functional PICs prior to the completion of reverse transcription.
MATERIALS AND METHODS
Isolation of highly purified CD4+ resting T cells.
Highly purified resting CD4+ T cells were isolated from peripheral blood mononuclear cells (PBMC) of HIV-1-negative donors as described previously (3, 13). Briefly, PBMC were obtained by centrifugation on Ficoll-Hypaque gradients. Adherent cells were removed during overnight culture in RPMI 1640 supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine (minimal medium [MM]). Purification of resting CD4+ T cells was accomplished in two steps. An initial enrichment involved staining cells with monoclonal antibodies against early, intermediate, and late markers of T-cell activation (CD69, CD25, and HLA-DR) as well as unwanted cell lineages (CD8, CD19, CD16, and CD14), followed by removal of cells decorated with antibodies by using magnetic beads coated with goat antibodies specific for murine immunoglobulin G. The fractionated cells were then stained with a phycoerythrin-conjugated anti-CD4 antibody and a fluorescein isothiocyanate-labeled anti-HLA-DR antibody and sorted on an Elite cell sorter. The resulting population of highly purified CD4+ resting cells was typically contaminated by less than 1% of cells expressing activation marker HLA-DR.
Infection of highly purified resting T lymphocytes.
High-titer HIV-1 IIIb stocks (108 50% tissue culture infectious doses/ml) were obtained commercially (Advanced Biotechnology Inc.) and treated with DNase I (Boehringer Mannheim) in the presence of 5 mM MgCl2 for 45 min at 37°C. Cells were infected for 3 h with HIV-1 in a small volume of MM (100 to 400 μl) and washed three times in 40 ml of phosphate-buffered saline (PBS; pH 7.2) supplemented with 2% fetal calf serum, 0.1% glucose, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 12 mM HEPES, pH 7.2 (wash medium [WM]). At various times, cells were harvested, washed once in WM, and lysed for DNA isolation (Gentra). Lysates were stored at −20°C until analysis. To account for intravirion reverse transcription products (44, 45, 46) or contaminating DNA in the viral stocks, an aliquot of infected cells was washed three times in 40 ml of WM immediately following application of the virus (referred to as time zero) and lysed for DNA isolation.
Detection of intermediates of reverse transcription.
Intermediates of reverse transcription were detected by a modification of a previously described PCR method (42, 43). The initiation of reverse transcription was detected with primers M667 and AA55 (long terminal repeat [LTR]). The translocation of the positive-strand strong-stop molecule that occurs near the end of the reverse transcription reaction was detected with primers M667 and M661 (LTR-gag). A mass of 200 ng of DNA was loaded into each PCR mixture, which contained 1 μM (each) primer, 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, and either 2.5 U of Expand High Fidelity Taq polymerase (Boehringer Mannheim) or 2.5 U of Platinum Taq (Life Technologies) in 1× proprietary buffer. A 3-min hot start at 94°C was performed, followed by cycling between 95°C for 30 s and 65°C for 30 s for 25 cycles. Products of the reaction were subjected to 2% agarose electrophoresis followed by alkaline transfer to nitrocellulose and hybridization with an end-labeled oligonucleotide probe (5′GTGCTTCAAGTAGTGTGTGCCC3′). To control for the relative number of cells in each lysate, β-globin was amplified and detected by Southern hybridization as described previously (4).
Detection of linear forms of HIV-1 DNA by LM-PCR.
The biologically relevant product of the retroviral reverse transcription reaction is a linear molecule with blunt phosphorylated ends. To specifically detect this linear form of the HIV-1 genome, a linker-mediated PCR (LM-PCR) approach was employed. This methodology involves the addition of a double-stranded linker onto the termini of linear viral genomes by using T4 DNA ligase, followed by detection of the junction of the linker and viral genome by PCR (2).
This procedure was optimized by using a previously described plasmid, pLTR4202, that contains the two HIV-1 LTRs orientated in a head-to-head fashion (3). There are several novel restriction endonuclease recognition sites at the junction of the two LTRs in this plasmid. Restriction enzyme digestion allows for the generation of a linear molecule flanked by LTRs that closely approximates the linear form of HIV-1. Restriction of pLTR4202 with StuI generates a linear molecule with blunt ends, while cleavage with AccI produces a linear molecule with a 2-bp overhang. The latter is similar in structure to the ends of the HIV-1 genome produced following processing of the reverse-transcribed viral cDNA by the viral integrase.
Linkers were assembled by annealing two partially complementary unphosphorylated oligonucleotides (45 μM each) together in the presence of 1× T4 DNA ligase buffer (Life Technologies). Annealing was performed in a PCR thermocycler programmed to incubate for 5 min at 99°C and slowly cool to room temperature in increments of 5°C with 5 min at each increment. To detect blunt forms of the viral genome, oligonucleotide 25t (5′GCGGTGACCCGGGAGATCTGAATTC3′) was annealed to oligonucleotide 11b (5′GAATTCAGATC3′). A linker capable of detecting linear forms of the viral genome that had been processed by HIV-1 integrase was composed of oligonucleotides 25t and 11-GTb (5′GTGAATTCAGATC3′). This linker complements the two-base overhang generated by removal of the 3′ GT dinucleotide from the 3′ end of each strand of viral DNA by integrase. This linker was capable of detecting both blunt and processed forms of the viral genome (see Results). To detect pLTR4202 following cleavage with AccI, a linker composed of oligonucleotides 25t and 11-GCb (5′GCGAATTCAGATC3′) was used.
To detect the linear form of HIV-1 DNA in infected cells, a ligation reaction mixture was assembled in a 30-μl volume containing 200 ng of genomic DNA, 4.5 μM double-stranded linker, and 10 U of T4 DNA ligase. This ligation reaction mixture was incubated for 12 to 16 h at room temperature. A nested-PCR strategy was employed to detect the addition of linker onto the U3 terminus of the viral genome. The linker ligation reaction mixture was diluted 1:5 in water and loaded into a PCR mixture assembled as described above. The first PCR used as primers oligonucleotide 25t and outer LPCR-L (nucleotides 446 to 460 of HIV-1 LAI: 5′TAACCAGAGAGACCCAGTACAGGC3′) and was run for 20 cycles of 95°C for 30 s and 65°C for 30 s. The products of this reaction were diluted 1:20 and loaded into a second PCR mixture containing oligonucleotide 25t and LPCR-L (nucleotides 130 to 154 of HIV-1 LAI: 5′ TGGTACTAGCTTGAAGCACCATCCA3′). This nested reaction involved 25 cycles of 95°C for 30 s and 65°C for 30 s. Reaction products were run on 2% agarose gels followed by alkaline transfer to nitrocellulose and hybridization with an end-labeled oligonucleotide probe in the LTR (nucleotides 59 to 81 of HIV-1 LAI: 5′CACACACAAGGCTACTTCCCTGA3′).
Characterization of the molecular structure of linear forms of HIV-1 DNA.
To detect the three most terminal nucleotides of the U3 terminus of HIV-1, which represent one-half of a ScaI restriction endonuclease recognition site, we designed a screen using a modified linker ligation reaction employing a linker containing the other half of the ScaI site. This linker was assembled as described above from oligonucleotide 25 SCAt (5′GCGGTGACCCGGGAGATCTGAATTCAGT3′) and oligonucleotide 11 SCAb (5′ACTGAATTCAGATC3′). LM-PCR was performed as described above, and the products were cloned with a topoisomerase I-mediated cloning system (TOPO-TA). Clones were restricted with ScaI, run on a 1% agarose gel, and subjected to Southern blotting with a probe annealing in the LTR (5′CACACACAAGGCTACTTCCCTGA3′). Sequencing the PCR insert was performed to validate the approach by determining the identities of nucleotides adjacent to the attached linker.
Analysis of linear forms of HIV-1 DNA processed by the viral integrase.
To detect linear forms of HIV-1 DNA that had been processed by integrase, a modification of the LM-PCR approach in which the two-base AC overhang resulting from 3′ processing is removed prior to the attachment of the linker was employed. Removal of these nucleotides was accomplished by treating viral and genomic DNA with mung bean nuclease (MBN), which is highly specific for single-stranded DNA and which has no activity on double-stranded DNA. After the processed form of the viral genome is blunted, the terminal residues of the linear form of HIV-1 are TGG, which constitute one-half of an NcoI restriction endonuclease recognition sequence. The use of a linker containing the other half of the NcoI recognition site allowed the terminal residues of linear DNA to be assayed rapidly by LM-PCR as described above.
A mass of 2 μg of genomic DNA isolated from infected cells was treated with 36 U of MBN (GIBCO) for 15 min at 37°C. The reaction was stopped with two extractions of an equal volume of phenol-chloroform-isoamyl alcohol. Next, the remaining organic solvents were extracted with ether. The resulting DNA was loaded into a ligation reaction mixture containing a linker composed of oligonucleotide 25 NCOt (5′GCGGTGACCCGGGAGATCTGAATTCCCA3′) and oligonucleotide 11-NCOb (5′TGGGAATTCAGATC3′), followed by a nested LM-PCR as described above. The products of this reaction were cloned into the TOPO-TA vector (Invitrogen), and the resulting clones were restricted with NcoI and assayed by Southern blotting. To validate the assay, a fraction of the positive clones were sequenced to determine the identity of the nucleotides at the junction of the linker and the viral genome.
Construction of an HIV-1 reporter vector.
To detect the productive infection of cells by HIV-1, we constructed a reporter virus that encodes the enhanced green fluorescent protein (EGFP) in place of HIV-1 env. Manufacture of this vector involved replacing the KpnI-NheI env fragment of the HIV-1 NL4-3 provirus with a PCR fragment containing the EGFP coding sequence. This fragment was amplified from a pEGFP-N1 plasmid template (Clontech) with primers containing KpnI and NheI sites (EGFP-KpnI: 5′-ATTGGGTACCTAAGGCCTATCCACCGGTCGCCACCATG-3′; EGFP-NheI: 5′-GTCCGTGCTAGCTTACAGCTCGTCCTTGTACAGCTCGTCCATGCC-3′). The reverse primer introduces an in frame endoplasmic reticulum (ER) retention sequence (KDEL) and a stop codon at the end of the EGFP coding sequence. To insert the EGFP coding sequence into pNL4-3, a three-way ligation involving the KpnI-NheI-restricted PCR product, a EcoRI-KpnI fragment of pNL4-3, and the EcoRI- and NheI-restricted pNL4-3 backbone was set up. The correct insertion of EGFP into NL4-3 was verified by sequencing. In the resulting construct, pNL4-3-GFP, the EGFP coding region is inserted in frame downstream of an env sequence encoding a small N-terminal fragment containing the signal peptide. The hybrid protein is directed into the ER by the env-encoded signal peptide and retained in the ER by the KDEL sequence.
Reporter virus production and infection.
Infectious virions were produced from the pNL4-3-GFP vector as described previously (33). Thirty million 293T cells were transfected with 40 μg of pNL4-3-GFP and 20 μg of pVSV-G in a 150T flask by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Five hours later, the medium was replaced with fresh MM. Supernatant containing vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped NL4-3-GFP virions were harvested 48 h posttransfection and concentrated with ultracentrifugation (25,000 rpm in a Beckman SW28 rotor at 4°C for 145 min). Viral stocks were titered by infection of Jurkat T cells with serial dilutions of the stock, with analysis of EGFP expression 2 days later by flow cytometry. Typical titers ranged from 3 × 107 to 10 × 107 infectious units per ml. Primary CD4+ T cells were infected at various multiplicities of infection in 0.5 ml of MM for 2 h at 37°C during centrifugation as described previously (28). After each infection, cells were washed and cultured at 2 × 106 cells per ml in MM until the time of analysis.
Activation of resting CD4+ T cells.
Purified resting CD4+ T cells were activated with the mitogen phytohemagglutanin (PHA; Wellcome Diagnostics) and a 10-fold excess of irradiated allogeneic PBMC in 12-well plates. To clearly distinguish the PBMC feeders from infected resting T cells during fluorescence-activated cell sorter (FACS) analysis, feeders were labeled with excitable dye PKH26 prior to addition to infected-resting-T-cell cultures. Staining was performed by incubation of irradiated PBMC with PKH26 (Sigma) at 25°C for 5 min according to the manufacturer's instructions. Labeled cells were washed three times in WM and mixed with HIV-1-infected resting CD4+ T cells in the presence of 0.5 μg of PHA/ml in six-well plates. PHA was removed the next day, and the culture was maintained in MM until analysis was performed. To monitor cell division after stimulation, CD4+ T cells (5 × 107/ml) were incubated with carboxyfluorescein diacetate succinimydyl ester (5 μm) in PBS for 3 min at room temperature. An equal volume of fetal calf serum was added to stop the labeling reaction. After being washed three times with WM, CD4+ T cells were cultured for various periods of time before activation, and the change in CFSE fluorescence intensity in the cells was monitored by FACS analysis.
Analysis of resting CD4+ T cells infected with NL4-3-GFP.
To determine the rate with which virus in the preintegration state decays, the percentage of reporter virus-infected resting T cells expressing EGFP was determined by FACS following activation. Infected cells were fixed in PBS in presence of 1% paraformaldehyde for 30 min on ice. FACS analysis was performed on a FACScan using Cell Quest software (Becton Dickinson).
RESULTS
Detecting the linear form of HIV-1 DNA by LM-PCR.
The biologically relevant product of the reverse transcription reaction is a full-length linear form of HIV-1 DNA that is recognized by the viral integrase and that is inserted into the host cell chromosome (reviewed in reference 14). The blunt, phosphorylated ends of this molecule distinguish linear HIV-1 DNA from all other forms of the viral genome in the cell. To detect and characterize linear HIV-1 DNA in infected cells, we performed LM-PCR as shown in Fig. 1. First, a blunt linker is attached to the phosphorylated termini of the viral cDNA with T4 ligase. Next, the junction of the linker and the viral genome was detected by a PCR employing a sense primer in the linker itself and an antisense primer in the R region of the 5′ HIV-1 LTR.
FIG. 1.
Detection of the linear form of HIV-1 DNA using LM-PCR. The final product of the reverse transcription reaction is a linear genome with blunt ends. To detect linear products of the reverse transcription reaction, genomic DNA was isolated from infected cells and loaded into a ligation reaction mixture containing a vast molar excess of an asymmetric blunt linker. The ligation of linker onto the end of the genome was detected by PCR with an HIV-1-specific primer and one complementary to the linker. In some experiments, a heminested PCR approach was employed. Production of an HIV-1-specific amplicon was universally T4 ligase dependent.
To study the specificity of the linker ligation process, control experiments were performed using a linearized plasmid that mimics the linear HIV-1 genome. HIV-1 infection results in the production of at least two different forms of linear viral DNA. Reverse transcription produces a linear viral genome with blunt ends. The processing of this blunt form by the viral integrase results in the removal of two bases from the 3′ strand at each end of the linear genome. To evaluate the abilities of different types of linkers to discriminate between these two forms of the viral genome, control experiments were performed using linear plasmid DNA with either blunt ends or ends resembling those of processed forms of the HIV-1 genome. Plasmid pLTR4202 was restricted with StuI or AccI to generate a linear molecule with blunt or asymmetric ends (two-base 5′ overhangs), respectively. These linear molecules were then used as targets in linker ligation reactions employing either a blunt linker or an asymmetric linker with a 2-bp overhang that complements the asymmetric termini of the target (Fig. 2A). These control studies demonstrated that the ligation of a blunt linker onto blunt linear target molecules occurs efficiently but not ligation onto targets with a two-base 5′ overhang, such as the one generated by HIV-1 integrase (Fig. 2B, compare lanes 1 and 3). Surprisingly, an asymmetric linker with a two-base 3′ overhang had the capacity to detect blunt and asymmetric targets with equal efficiencies (Fig. 2B, compare lanes 2 and 4). This result may reflect the fact that the overhanging nucleotides of the linker, which is present in large excess, are not involved in the chemistry of ligation to the phosphate present on the 5′ end of the genome. Therefore, asymmetric linkers have the capacity to detect both the blunt and integrase-processed forms of the viral genome and may provide a generalized strategy for the detection of linear forms of HIV-1 in infected cells.
FIG. 2.
Specificity of LM-PCR for molecules with blunt ends. (A) The specificity of LM-PCR for blunt targets representing completed reverse transcripts (top) or asymmetric targets representing the processed form of the HIV-1 genome (bottom) was evaluated with both blunt linkers and linkers with complementary overhangs. (B) Ten thousand copies of a linear molecule with blunt (StuI restricted) or asymmetric ends (AccI restricted) were loaded into a LM-PCR mixture. Ligation of the linker onto the terminus of the plasmid was evaluated with a 28-cycle PCR employing oligonucleotide 25t and LPCR-L. Numbers refer to the reactions shown in panel A.
Analysis of the progress of the reverse transcription reaction in highly purified resting T cells infected with HIV-1.
Pioneering studies by Zack and colleagues first characterized the reverse transcription reaction in quiescent CD4+ T cells infected with HIV-1 (42, 43). These initial studies demonstrated that, compared to reverse transcription in activated T cells, reverse transcription in resting cells is inefficient and fails to proceed to completion during the first day of infection. These observations suggested that the failure of HIV-1 to replicate in resting CD4+ T cells might be due to a block in the reverse transcription reaction. To extend these observations, we studied the progress of reverse transcription in resting T cells through the detection of the linear, double-stranded DNA products using LM-PCR. Highly purified populations of resting CD4+ T cells were obtained as previously described (13) and were infected at a multiplicity of infection of 1.0 with HIV-1 IIIb. At various times, total cellular DNA was isolated, and a linker ligation reaction was performed with an asymmetric linker in the presence or absence of T4 DNA ligase. The ligation of the linker onto the U3 terminus of the viral genome was detected by nested PCR. Analysis of the U3 end of the linear form of HIV DNA was selected because previous studies demonstrated that the U3 end is formed after the U5 end and thus represents the final step in the reverse transcription reaction (25).
Using this approach we demonstrated that linear double-stranded DNA products of appropriate size are produced by reverse transcription in resting CD4+ T cells, but with kinetics that are markedly slower than those observed in a transformed T-cell line (MT-2) infected in parallel (Fig. 3). While reverse transcription in T-cell lines appears to proceed to completion within hours of infection, our studies demonstrated that, in resting T cells, linear forms of the viral genome accumulated over the course of several days. This increase in linear HIV-1 DNA in resting T cells was not the result of a low level of spreading infection because identical results were obtained when a high concentration (60 μM) of HIV-1 protease inhibitor indinavir was added 3 h postinfection (data not shown). Previous studies by Zack et al. demonstrate the inability of the virus to complete the reverse transcription reaction in resting cells in the first 24 h postinfection (42, 43). Our analysis detected significant amounts of apparently complete reverse transcripts 3 days postinfection but not during the first day of infection, consistent with the idea that reverse transcription in resting cells is a slow process but one that is eventually capable of proceeding to completion, at least in some resting CD4+ T cells.
FIG. 3.
Reverse transcription proceeds with protracted kinetics in highly purified resting T cells but can yield double-stranded HIV-1 DNA molecules of the appropriate length. Highly purified resting T cells were obtained from HIV-1-negative donors by cell sorting. Control MT-2 cells and resting CD4+ T cells were infected with HIV-1 IIIb at a multiplicity of infection of 1. At the indicated times, DNA was isolated and nested LM-PCR was performed to detect the U3 end of the linear HIV-1 genome. The LM-PCR involved an initial reaction of 15 cycles with outer LPCR-L and oligonucleotide 25t. Reaction products were diluted 1:4 in water, and a second 25-cycle PCR was performed using oligonucleotide 25t and LPCR-L. Reaction products were run on a 2% gel and Southern blotted with a U3-specific probe.
Characterization of the linear form of HIV-1 DNA in resting T cells.
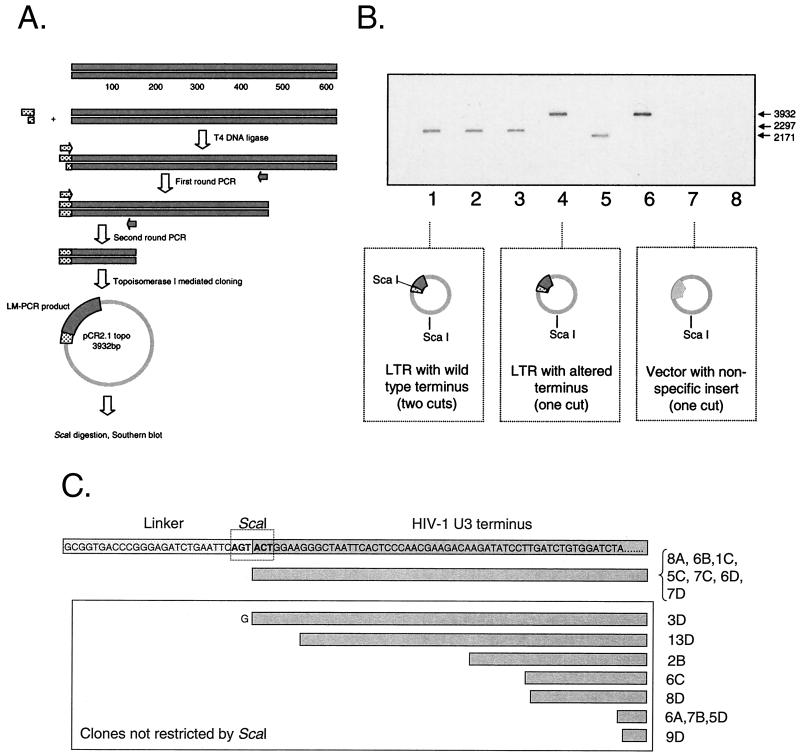
To demonstrate that the linear HIV-1 DNA molecules detected using LM-PCR are complete and are suitable substrates for the integrase enzyme, we further characterized the structure of linear reverse transcription products in resting CD4+ T cells. HIV-1 integrase binds the ends of the viral cDNA through the recognition of the most terminal five or six nucleotides of the linear viral genome (11). We therefore characterized the terminal structure of linear HIV-1 molecules found in infected resting CD4+ T cells in order to evaluate their capacity to serve as substrates for the viral integrase. The terminal nucleotides of the U3 end of HIV-1, ACTGG, are highly conserved. The three most terminal bases represent one-half of a ScaI restriction site. To determine the structure of the ends of linear forms of HIV-1 DNA, a blunt linker encoding the other half of a ScaI site was used in the linker ligation strategy described above. Addition of this linker to full-length, blunt, linear forms of the viral genome creates a novel ScaI site at the junction of the linker and the end of the viral genome (Fig. 4A).
FIG. 4.
Characterization of complete reverse transcripts cloned from resting T lymphocytes. (A) LM-PCR assay for complete reverse transcripts. The ligation of an asymmetric linker containing one-half of a ScaI site onto the terminus of a full-length HIV-1 genome creates a novel ScaI site, allowing rapid screening of cloned LM-PCR products. (B) Restriction analysis of cloned LM-PCR products obtained from resting CD4+ T cells. Highly purified resting CD4+ T cells were infected with HIV-1 IIIB. Three days after infection, cells were lysed and DNA was isolated for LM-PCR. Cloned LM-PCR products were restricted with ScaI and analyzed by Southern blotting using an LTR-specific probe. The analysis allowed the identification of blunt linear molecules with either the predicted U3 end (lanes 1, 2, 3, and 5) or clones bearing alterations that result in a change in the sequence of the most-terminal U3 nucleotides (lanes 4 and 6). Because the insert was cloned in either a forward or reverse orientation, inserts of two different sizes were predicted for clones containing the novel ScaI site (lanes 1, 2, 3, and 5). Some clones did not contain an insert that contained a sequence from the HIV-1 LTR and were not characterized further (lanes 7 and 8). (C) Sequence analysis of LM-PCR clones obtained from infected resting CD4+ T cells.
To characterize the linear viral genome formed following infection of quiescent lymphocytes, DNA was isolated from resting CD4+ T cells 3 or 4 days after infection with HIV-1 IIIb. LM-PCR was performed on the U3 terminus with the linker described above, and the products were cloned. Clones were selected at random for restriction by ScaI and analysis by Southern blotting using a probe specific for the HIV-1 LTR. This approach allowed us to discriminate between full-length linear viral genomes and those that do not contain the highly conserved terminal residues. Our analysis included 38 clones derived from three different resting T-cell infections. Twenty-seven of these clones were derived from full-length linear viral genomes, as evidenced by the presence of a novel ScaI site at the junction of the linker and the viral genome (Fig. 4B). Sequence analysis of several ScaI-restricted clones (eight of eight) confirmed that they contained the entire U3 LTR fused directly to the linker (Fig. 4C). These results definitively demonstrate that full-length linear HIV-1 DNA molecules containing sequences recognized by HIV-1 integrase are generated in infected resting CD4+ T cells.
Interestingly, sequencing identified linear forms of the viral genome that were blunt (by virtue of their recognition by a blunt linker) but that lacked the highly conserved terminal residues. Eleven clones contained a portion of the U3 LTR but not the diagnostic ScaI site at the linker-LTR junction. The majority of these clones (9 of 11) had deletions of various lengths of the terminal nucleotides of the linear viral genome (Fig. 4C). Published analysis of the activity of HIV-1 integrase on different linear substrates allows us to predict that viral genomes bearing such deletions are not competent to integrate into the host cell chromosome (11). Interestingly, similar deletions were not observed during screens of a large number of clones derived from infections of activated CD4+ T cells (0 of 25) (data not shown). One clone from resting cells (3D) contained an additional guanine nucleotide at the junction of the linker and viral genome. The origin of this additional G is likely inappropriate cleavage of the 3′ polypurine tract by RNase H during reverse transcription. Similar additions of polypurine tract or primer binding site sequences at the junction of the two LTRs found in two-LTR circles have been reported (21, 37). Finally, one clone (4C) had a single point mutation at the terminus of the viral genome, abolishing ScaI recognition and cleavage. This mutation may have arisen during PCR amplification, and its biological relevance cannot be established.
A fraction of the linear viral DNA in infected resting T cells has been processed by HIV-1 integrase.
The experiments presented above demonstrate the presence of full-length forms of the linear viral genome in resting T cells infected with HIV-1. Next, we asked whether HIV-1-infected resting CD4+ T cells contained linear viral genomes that were associated with functional PICs. Following reverse transcription, the ends of the linear viral genome are bound tightly by the integrase protein. Integrase then catalyzes the removal of two nucleotides from the 3′ strand of each end. To determine whether integrase bound the end of the viral genome in resting T lymphocytes, a further modified linker ligation assay was employed to specifically detect linear molecules with the 2-bp 5′ overhang resulting from integrase 3′ processing.
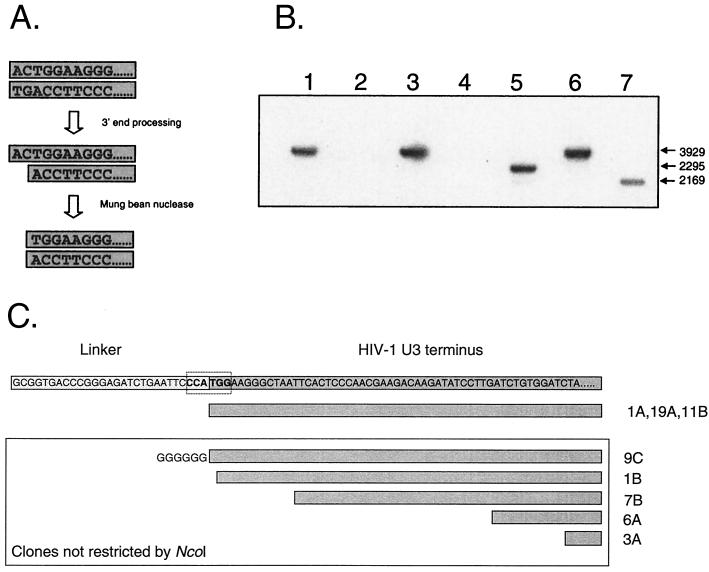
An initial attempt to detect molecules that had been processed by integrase involved using asymmetric linkers that complement the 2-bp GT overhang on the viral DNA. However, as shown in Fig. 2, these linkers have no power to distinguish between blunt and asymmetric forms of the HIV-1 linear genome. Instead, we employed a single-strand-specific nuclease to remove the 2-bp 5′ overhang generated by integrase processing, generating a blunt end that is missing 2 bp at the end of the LTR. MBN does not cleave double-stranded DNA. Removal of the overhanging two bases with MBN exposes one-half of an NcoI site at the U3 end of the viral genome (Fig. 5A). Addition of a linker complementing this NcoI half site, followed by LM-PCR, allows for the rapid identification of linear genomes that have been processed by the viral integrase.
FIG. 5.
Infected resting CD4+ T cells contain linear HIV-1 DNA molecules that have 3′ processing by integrase. (A) LM-PCR assay for complete reverse transcripts that have undergone 3′ processing by integrase. As an initial step in the integration reaction, integrase removes the terminal 2 nucleotides from the 3′ end of each strand of the full-length, linear HIV-1 DNA. The product of this reaction can be detected by removing the 5′ overhang with single-strand-specific MBN, generating a blunt-ended molecule whose terminal nucleotides form one-half of an NcoI site. The linker used in these experiments creates a novel NcoI site when ligated to the linear genome of HIV-1 that had been processed by integrase and cleaved by MBN. LM-PCR clones were restricted with NcoI, run on a 1% gel, and probed by Southern blotting with an LTR-specific probe. The vector used in these studies has an NcoI site; therefore, clones with inserts derived from linear DNA that has been processed by integrase will be cleaved twice in a manner analogous to that for the ScaI screen described in the legend for Fig. 4. (B) LM-PCR analysis of HIV-1 DNA from infected resting CD4+ T cells. Highly purified resting CD4+ T cells were infected with HIV-1 IIIB. Three days after infection, cells were lysed and DNA was isolated for LM-PCR. Restriction analysis of cloned LM-PCR products identified cloned inserts that were cleaved by NcoI, indicating prior processing by integrase (lanes 5 and 7) and inserts that were not cleaved (lanes 1, 3, and 6). Because the insert was cloned in either a forward or reverse orientation, inserts of two different sizes were predicted for clones containing the novel NcoI site. Some clones did not contain an HIV-1-derived insert (lanes 2 and 4). (C) Sequence analysis of LM-PCR clones obtained from MBN-treated DNA isolated from infected resting T lymphocytes.
The ends of linear DNA molecules may contain some single-stranded character, and thus, to validate this approach, it was necessary to demonstrate that MBN treatment of the viral DNA did not artifactually cleave two bases off the terminus of the genome. To do this, a control virus (HIV-1 D64N) deficient in all integrase catalytic function was used to infect MT-2 cells. This virus, bearing a D-to-N mutation in the catalytic site, should not generate any clones containing the novel NcoI site formed at the junction of the linker and the viral genome. LM-PCR was performed on genomic DNA isolated from infected cultures and analyzed as described above by using the blunt-NcoI site-containing linker. None of the seven clones identified in our screen could be restricted with NcoI, suggesting that MBN treatment does not result in the nonspecific removal of two bases from the end of the HIV-1 genome (data not shown).
To determine if any of the linear HIV-1 DNA in infected resting T cells had been processed by the viral integrase, we isolated DNA from resting T cells infected with HIV-1 IIIb 3 days previously. Following treatment with MBN, LM-PCR was performed as described in Materials and Methods. The products of this reaction were cloned and analyzed by NcoI digestion and Southern blotting with an LTR-specific probe. A total of 17 clones derived from linear forms of HIV-1 were identified in this screen. Six of these clones contained an NcoI site at the junction of the linker and the viral genome, suggesting that they had been processed by HIV-1 integrase prior to treatment with MBN (Fig. 5B and C). A fraction (6 of 11) of the remaining clones were studied by sequencing. These clones were characterized by deletions of various numbers of nucleotides from the terminus of the linear genome, similar to the truncations described above. Taken together, these results suggest that reverse transcription proceeds to completion in infected resting T lymphocytes and that the resulting DNA product is associated with a functional PIC.
Measuring the rate with which HIV-1 decays in the preintegration state.
Although reverse transcription can proceed slowly to completion in resting CD4+ T cells, the infection of resting CD4+ T cells generally does not progress to the stage of viral gene expression. Previous studies have demonstrated a labile form of latent infection in resting CD4+ T cells, generally referred to as preintegration latency (1, 12, 43). To explore the relationship between the stability of the virus in the preintegration state and the kinetics of reverse transcription in resting CD4+ T cells, we infected purified resting CD4+ T cells with a recombinant VSV-G-pseudotyped HIV-1 expressing the green fluorescent protein (GFP) (Fig. 6A). At various times following infection, a fraction of the infected resting cells were activated with mitogen in order to render the cells fully permissive for viral replication. By flow cytometry, the fraction of infected resting T cells from which viral gene expression could be rescued following cellular activation was determined in order to quantitate decay processes occurring during early stages of the infection of resting cells. These experiments were done with a VSV-G-pseudotyped recombinant HIV-1 so that higher numbers of infected cells could be generated and so that rescue of virus gene expression could be precisely quantitated at the single-cell level. Experiments in which resting cells infected with HIV-1 were cultured and then activated gave similar results; however, quantitation required a cumbersome and less precise limiting-dilution virus culture assay.
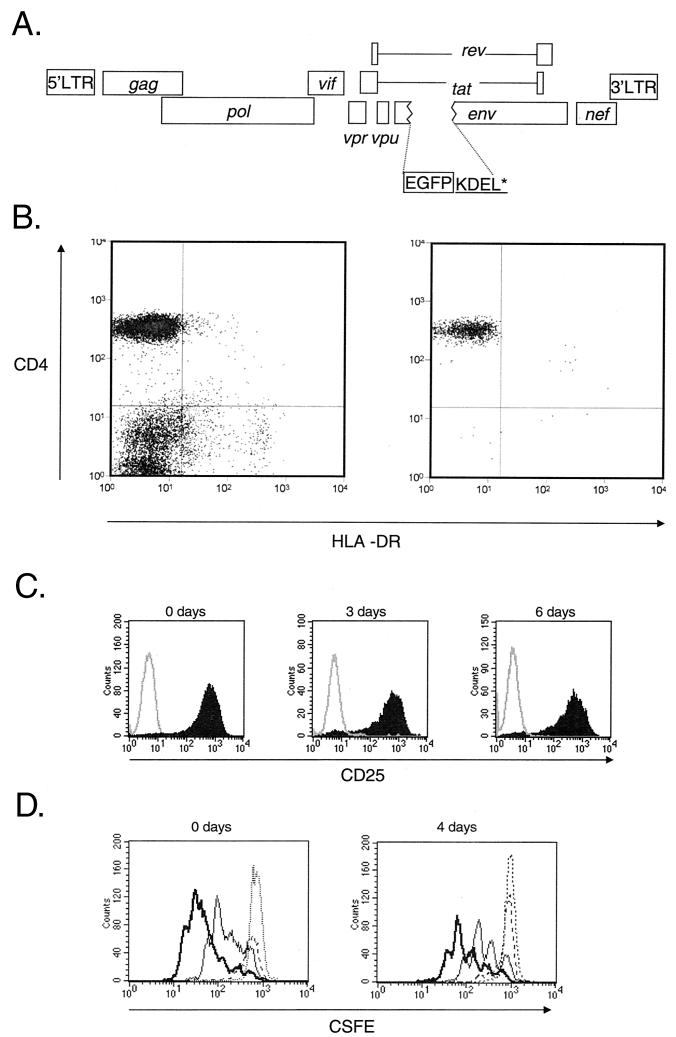
FIG. 6.
Analysis of the decay of the preintegration state of HIV-1 latency. (A) Map of reporter virus NL4-3-GFP used to monitor the fate of HIV-1 in infected resting CD4+ T cells. NL4-3-GFP contains the EGFP coding sequence inserted in frame in the env gene, followed by codons for the KDEL sequence to retain GFP in the ER and a stop codon. Highly purified resting CD4+ T cells were infected with VSV-G-pseudotyped NL4-3-GFP. Following infection, the cells were extensively washed and cultured for various times before being subjected to activation with mitogen PHA and irradiated allogeneic PBMC. Expression of GFP by the infected cells was monitored by flow cytometry. (B) Purity of resting CD4+ T-cell preparations. Resting CD4+ T cells were isolated from the peripheral blood of HIV-negative donors by magnetic bead depletion and cell sorting. Cells stained with antibodies to CD4 and activation marker HLA-DR before (left) and after (right) magnetic bead depletion and cell sorting were analyzed by flow cytometry. (C) Efficiency of in vitro activation of CD4+ T cells. Uninfected resting CD4+ T cells were activated with PHA and irradiated feeders immediately after isolation (0 days) or after 3 or 6 days of culture in MM. Cells were stained with an anti-CD25 antibody 72 h after activation. Solid histogram, CD25 staining; open histogram, isotype control. (D) Proliferative responses of resting CD4+ T cells following activation with PHA and irradiated allogeneic PBMC. Resting CD4+ T cells were stained by CFSE and activated immediately after isolation (left) and after 4 days of culture (right). CFSE intensity was measured at 24 (dotted line), 48 (broken line), 72 (thin line), and 96 h (thick line) after activation.
Purified resting CD4+ T cells were obtained by a previously described (13, 30) procedure that routinely produced cell preparations with <1% contaminating activated cells (Fig. 6B). The purified resting cells were infected with VSV-G-pseudotyped NL4-3-GFP and then washed and maintained in MM. At the indicated times, a fraction of the infected culture was removed and activated with PHA and a 10-fold excess of irradiated allogeneic PBMC labeled with red fluorescent dye PKH26. Cells were harvested 48 to 72 h after activation and fixed in 2% paraformaldehyde for analysis by flow cytometry. The percentage of resting T cells in which cellular activation induced the expression of viral genes was calculated by determining the percentage of PKH26-negative cells expressing EGFP.
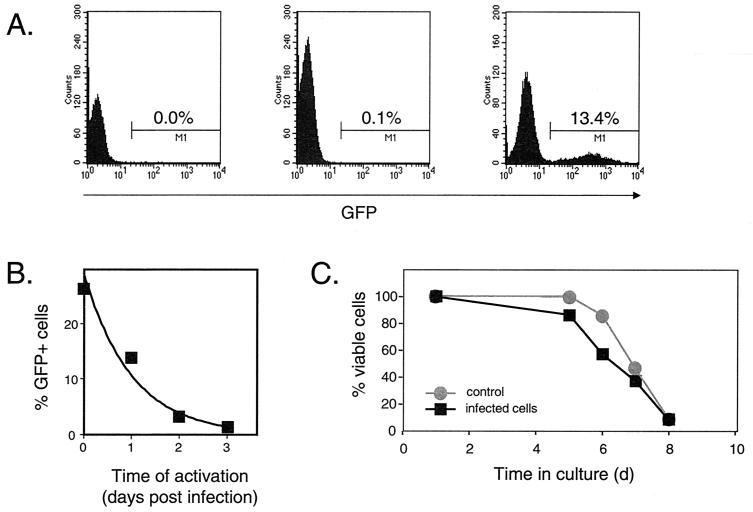
In control experiments, we established that this activation procedure results in activation of 100% of the input resting CD4+ T cells under conditions that closely mimic physiological activation with antigen and antigen-presenting cells. Uniform activation was demonstrated by up-regulation of CD25, the alpha chain of the interleukin-2 receptor, on 100% of the cells by 72 h after activation (Fig. 6C). Uniform activation was observed whether the resting cells were activated immediately after infection (day 0) or after 3 or 6 days in culture (Fig. 6C). These activation conditions induce the vast majority of the resting cells to enter the cell cycle and proliferate, as demonstrated by the dilution of dye CFSC (Fig. 6D). Using this approach, we examined the fate of the virus following infection of resting CD4+ T cells by varying the time between infection and activation. In this system, expression of viral genes is heavily dependent on cellular activation, as few GFP-positive cells are observed unless the infected resting-cell culture is activated (Fig. 7A). However, if resting cells are activated, GFP-expressing cells are readily detectable 3 days later. By varying the time between infection and activation, we observed a remarkably rapid decay of rescuable viral gene expression during the first 3 days after infection of resting CD4+ T cells (Fig. 7B; Table 1). Longitudinal analysis of six separate experiments demonstrated that virus in the preintegration state decays with a half-life of 0.9 ± 0.2 days during this period (Fig. 7B and Table 1). These results suggest that, during the time that reverse transcription is proceeding to completion, irreversible degradative processes prevent virus rescue in a significant fraction of the recently infected resting CD4+ T cells (>85%). As shown in Fig. 6C and D, the loss of rescuable virus does not result from any decrease in the susceptibility of the resting cells to activation. Furthermore, the viability of the resting-cell cultures, in the presence or absence of infection, does not begin to decline until day 5 (Fig. 7C).
FIG. 7.
Analysis of preintegration latency. (A) Dependence of viral gene expression on cellular activation. Resting CD4+ T cells were left uninfected (left) or were infected with pseudotyped NL4-3-GFP virus at a multiplicity of infection (MOI) of 1 (center and right). Four days after infection, GFP expression was analyzed by flow cytometry. In the absence of an activating stimulus (center), the percentage of cells with green fluorescence 4 days after infection was very low (0.1%), only slightly higher than the background observed in uninfected cells (left). However, if the resting cells were activated with PHA and irradiated allogeneic PBMC on day 1 postinfection, a significant portion of infected CD4+ T cells expressed GFP on day 4 postinfection (right). (B) Decay of HIV-1 in the preintegration state. Resting CD4+ T cells were infected with pseudotyped NL4-3-GFP at a MOI of 7, washed, and maintained in RPMI 1640 supplied with 10% fetal calf serum (FCS) for the indicated times before activation with PHA and allogeneic PBMC. The decay of virus in the preintegration state was measured as a decrease in the percentage of GFP-positive cells determined by FACS analysis 48 to 72 h after activation. The experiment shown is representative of six independent experiments, which are summarized in Table 1. (C) Viability of resting CD4+ T cells infected with pseudotyped NL4-3-GFP. Highly purified resting CD4+ T lymphocytes were isolated and infected with pseudotyped NL4-3-GFP at a MOI of 2. Both HIV-1-infected cells and noninfected resting T cells were maintained in RPMI 1640 supplemented in 10% FCS. Cell viability was measured over time by exclusion of vital dye trypan blue.
TABLE 1.
Half-life measurements for preintegration latency in infected resting CD4+ T cellsa
| Expt no. | MOIb | Acid treatmentc | Decay rated (days−1) | Half-lifee (days) | R2 of regression |
|---|---|---|---|---|---|
| 1 | 2 | No | 0.59 | 1.2 | 0.99 |
| 2 | 5 | No | 0.78 | 0.89 | 0.86 |
| 3 | 7 | No | 1.0 | 0.69 | 0.98 |
| 4 | 10 | Yes | 0.82 | 0.84 | 0.88 |
| 5 | 10 | Yes | 0.78 | 0.89 | 0.98 |
| 6 | 20 | Yes | 0.96 | 0.72 | 0.89 |
Highly purified resting CD4+ T cells were infected with NL4-1-GFP and incubated for various times up to 3 days before activation and analysis of induced GFP expression.
MOI, multiplicity of infection.
In some experiments, infected cells were washed briefly at an acidic pH to promote direct fusion of the virus with the plasma membrane.
Decay rates were determined by modeling the decay data from the first 3 days postinfection as an exponential process.
The average half-life for the six experiments was 0.9 ± 0.2 days.
HIV-1 infection is mediated by the binding of gp120 to CD4 and a chemokine receptor on the surface of a cell (reviewed in reference 9). In contrast, the VSV-G virions employed in these experiments enter the infected cell by endocytosis. The efficacy of the endocytosis of VSV-G-pseudotyped virions into resting T cells has not been measured directly. To exclude the possibility that the rapid decay of virus in the preintegration state observed in our experiments was due to the decay of bound extracellular virions, the resting CD4+ T cells were infected with VSV-G-pseudotyped virus as described above and washed briefly in an acidic pH to promote direct fusion of the virus with the plasma membrane. We demonstrated that the rapid decay of virus in the preintegration state could be observed even under conditions where direct fusion of attached viruses is stimulated by low pH (Table 1). Taken together, our results suggest that the decrease in the number of infected resting CD4+ T cells that can be induced to express viral genes by mitogenic stimulation is most likely due to decay processes operating on PICs after virus entry and during the time period required for the completion of reverse transcription.
To determine what fraction of the virus rescued from the preintegration state came from cells containing completely reverse-transcribed DNA, infected resting T cells were treated with the reverse transcriptase inhibitor lamivudine (3TC) (10 μM) 4 h prior to activation with PHA and irradiated feeders. Under these conditions, only cells containing complete reverse transcripts can go on to express viral genes following stimulation. We found that if cells were treated with 3TC and activated in the first 3 days after infection, the rescue of virus from the preintegration state was blocked (Fig. 8). In contrast, if resting cells are infected and then cultured for 3 days or longer before treatment with 3TC and activation, viral gene expression becomes insensitive to 3TC. However, the total number of cells with rescuable viral gene expression is much lower, reflecting the decay processes occurring during the time that it takes for reverse transcription to progress to completion in resting CD4+ T cells. Figure 8B shows the relative contribution of cells with complete reverse transcripts to the total number of cells containing rescuable forms of the virus. This analysis demonstrates that, in most resting CD4+ T cells, the completion of reverse transcription requires approximately 3 days. The experiment shown in Fig. 8 is representative of four experiments done with resting cells from different donors, all showing the same phenomenon. Thus in resting CD4+ T cells, irreversible decay processes occurring during the time that it takes to complete reverse transcription prevent the rescue of viral gene expression in most recently infected resting CD4+ T cells, even after the cells are rendered competent for viral replication by activating stimuli.
FIG. 8.
Contribution of completely reverse transcribed DNA. (A) Decay of HIV-1 in the preintegration state was measured as described for Fig. 7 in the presence (triangles) or absence (circles) of reverse transcription inhibitor 3TC at 10 μM. Resting CD4+ T cells were infected at a multiplicity of infection of 10 and then incubated for the indicated times before the addition of 3TC and activation with PHA and allogeneic feeder cells. 3TC was added 4 h prior to activation. (B) Relative contribution of completely reverse transcribed DNA to the rescue of virus from the preintegration state. The relative contribution was determined by expressing the number of cells with inducible GFP expression in the presence of 3TC as a percentage of the cells with induced GFP expression in the absence of 3TC.
DISCUSSION
Understanding the state of preintegration latency in HIV-1 infection is important for several reasons. At steady state in vivo, only a minority of CD4+ T lymphocytes in the blood and peripheral lymphoid tissues are in an activated state. Therefore, it is likely that HIV-1 most frequently encounters resting CD4+ T cells, cells that are incapable of sustaining productive infection due to the replication blocks discussed above. In untreated patients, the state of preintegration latency is quantitatively dominant over postintegration latency; at any given time, the fraction of infected resting cells in the preintegration state of latency is larger than the fraction containing integrated HIV-1 DNA (2, 3). This must be considered in the interpretation of all DNA PCR studies carried out by standard methods that are incapable of distinguishing between the preintegration and postintegration states. As shown here, this preintegration state is extremely labile due to irreversible decay processes that occur during the prolonged process of reverse transcription in resting cells. Appreciating the distinctions between pre- and postintegration latency may allow the development of novel clinical assays capable of monitoring the recent infection of cells by HIV-1. Thus although the dynamics of this preintegration form of latency are not directly relevant to the stable postintegration form of HIV-1 latency that represents a major barrier to eradication of the infection with antiretroviral drugs (3, 5, 6, 12, 13, 41), a more complete understanding of the structure of the viral genome in resting T cells may allow for new insights into HIV-1 pathogenesis and viral dynamics.
The molecular state of HIV-1 in infected resting T cells.
A defining characteristic of a retrovirus is its ability to convert a positive-strand genomic RNA into a double-stranded, linear DNA molecule that may then be integrated into the host cell chromosome. The ability of HIV-1 to complete the reverse transcription reaction in resting CD4+ T cells has been examined in several studies. Initial studies of the progress of the reverse transcription reaction in resting T lymphocytes demonstrated that it was inefficient in comparison to reverse transcription in activated T cells (42, 43). This analysis was made possible by the development of assays capable of distinguishing between intermediates formed at the initiation and near the completion of the reverse transcription reaction. By employing this strategy, Zack et al. established that, while reverse transcription was initiated in an infected resting cell soon after virus entry, it did not progress to completion during the first 24 h of infection (42, 43). Subsequent experiments in which the production of reverse transcription intermediates was monitored for longer periods suggested that reverse transcription may in fact eventually proceed to near completion in infected resting lymphocytes (30, 38).
The present study extends and clarifies previous observations by assaying directly for the presence of the biologically relevant final product of the reverse transcription reaction in highly purified resting CD4+ T cells infected with HIV-1. We demonstrate that, in infected resting CD4+ T cells, reverse transcription proceeds slowly to completion to generate a full-length linear DNA molecule that represents a suitable substrate for integration into the host cell chromosome. In addition, we detected linear molecules that had been processed by HIV-1 integrase in a reaction that normally occurs in the context of a functional PIC and that is required for integration into the host cell genome.
The kinetics of reverse transcription are markedly slower in resting cells than in activated T cells, macrophages, or T-cell lines (20, 27, 43). Our results clearly show that 3 days are required for the reaction to be completed in most resting CD4+ T cells. The delayed kinetics may be a result of smaller pools of nucleotides or other cellular factors in resting T cells (22, 24). Interestingly, it has been shown that the addition of exogenous nucleotides increases the rate at which reverse transcription proceeds (20). This supports the notion that the inefficiency of reverse transcription in resting T cells is not necessarily the result of an absolute block but rather a problem of mass action in which insufficient reactants are present.
In addition to full-length linear forms of HIV-1 DNA, we identified in resting CD4+ T cells a significant number of viral genomes incapable of integration due primarily to an apparent truncation or terminal degradation process. Viral genomes bearing terminal deletions were not identified during parallel analysis of linear HIV-1 DNA detected in transformed T-cell lines infected with HIV-1. Previous work has demonstrated that treatment of resting T lymphocytes with exogenous nucleosides increases the rate of the reverse transcription reaction and can allow for its completion (20). Interestingly, the inefficiency with which virus can be rescued from these nucleoside-treated, HIV-1-infected, resting T cells following cellular activation supports the finding that not all reverse transcription products have the capacity to serve as integrase substrates (20). An intriguing possibility is that the formation of these aberrant linear molecules may be a consequence of the slow kinetics of reverse transcription in resting cells, which may expose the linear genome to nucleases for extended periods of time. Therefore, degradation of the linear viral genome may be a reflection of the processes contributing to the labile nature of HIV-1 in the preintegration state. However, the rapid decrease in the amount of virus that can be rescued from the preintegration state during the first few days of infection, prior to the completion of the reverse transcription process, suggests that mechanisms other than degradation of full-length linear DNA also play a role in the lability of preintegration latency.
Previous studies have analyzed two-LTR circles in order to determine the structure of the ends of retroviral DNA. Several studies of infections of cell lines with HIV-1 (15) or with murine (35, 36) or avian retroviruses (17, 18) have demonstrated deletions at the terminus of the linear genome, especially at the U3 end. It was suggested that the relatively high frequency of circles bearing deletions at the circle junction was a function of an increased ability of linear forms bearing deletions to become circular through end-to-end ligation (15). In light of these findings, one possible interpretation of our results is that the truncated linear HIV-1 DNA molecules formed following the infection of an activated cell are the preferential substrate of the two-LTR circle formation process and as a result are underrepresented in the population of linear DNA molecules sampled from activated cells. The detection in resting cells of a significant fraction of the linear viral genomes bearing deletions may be a result of the inability of the viral genome to form circles in these cells (32), presumably due to the inability to access the nucleus of the infected cell.
Several limitations in the application of LM-PCR to the analysis of linear HIV-1 DNA should be pointed out. First, the LM-PCR approach used in this study is highly specific for molecules with blunt ends. Therefore, we cannot rule out the possibility that other forms of linear HIV-1 DNA, which are not blunt, exist in infected cells. Analysis of linear molecules without blunt ends, such as those processed by integrase, required a modified approach. Second, the LM-PCR approach used in these studies has limited quantitative power because the sensitivity of the assay is affected by subtle differences in the number of double-strand breaks in genomic DNA suitable for ligation by the linker. Additionally, the heterogeneity of the linear forms of HIV-1 DNA also makes quantitation difficult, as one assay lacks the capacity to detect all forms of the linear genome. Development of a more quantitative approach may allow for the use of linear forms of HIV-1 DNA in studies of viral dynamics.
The lability of the preintegration state of HIV-1 latency.
The results presented here provide new insight into the short-lived nature of the preintegration form of HIV-1 latency. The ability to rescue HIV-1 from the infected resting CD4+ T cells decayed with a half-life of approximately 1 day. The molecular mechanisms responsible for the decay of the preintegration state are unclear and merit further study. The half-life obtained from these rescue experiments is markedly similar to estimates for the half-life of unintegrated HIV-1 DNA in resting T cells (42). It is important that the majority of the decay observed in these studies occurs during the slow process of reverse transcription in the resting T cell, making it unlikely that degradation of full-length linear viral genomes is a major mechanism by which PICs are inactivated in the cell.
The PIC is a dynamic association of several viral and cellular proteins responsible for the trafficking of the viral genome through the nuclear membranes and integration of the viral genome into a host cell chromosome. Several recent reports suggest a potential role for the proteasome in the degradation of functional PICs. Schwartz and colleagues demonstrate that inhibition of the proteasome results in increased levels of linear and circular forms of proviral DNA, which may reflect the role of the proteasome in the degradation of the PIC prior to integration (34). Additionally, HIV-1 integrase is degraded via the N-end rule pathway, supporting the notion that it is a suitable substrate for the proteasome (26).
The analysis of the resting CD4+ T-cell compartment in patients receiving no therapy or nonsuppressive therapy is complicated by the contribution of recent infection of resting CD4+ T cells by HIV-1, which does not generally result in a stably integrated provirus. Recently, Blankson et al. suggested that the preintegration state decays in vivo with a half-life of 1 week (1). This is significantly longer that the half-life for virus in the preintegration state reported here. The analysis of in vivo decay rates is complicated by the additional entry of virus into the resting-T-cell compartment due to incomplete suppression of viral replication during the first few weeks of therapy. In addition, the decay of virus in the first phase reported by Blankson et al. may reflect the combined effects of a very rapid decay process acting on the PIC during the first few days after infection and a slower decay process that acts on full-length linear HIV-1 genomes once reverse transcription is complete. Current efforts involve evaluating the latter possibility.
The experiments presented here provide additional insight into the fate of the virus following the infection of highly quiescent lymphocytes such as the resting CD4+ T cells found in the circulation. We demonstrate that HIV-1 has the capacity to complete the reverse transcription reaction in resting cells to generate the full-length linear viral cDNA. Furthermore, a fraction of the viral DNA in resting T cells appears to have been processed by integrase. Taken together, these two observations lend support to a model in which one block to HIV-1 replication in resting T cells occurs after the reverse transcription event, perhaps due to an inability to enter the nucleus of a quiescent cell. We also demonstrate that rapid degradation of virus in the preintegration state occurs during the prolonged time that it takes for reverse transcription to be completed. Thus a more complete view of the fate of a resting CD4+ T cell encountering HIV-1 in the circulation might be as follows. Entry of R5 viruses is dependent on expression of CCR5. Only a subset of resting CD4+ T cells express CCR5 (30). In these cells, entry occurs and reverse transcription proceeds at a slow pace and requires several days for completion. During this period, competing degradative processes may lead to the irreversible loss of virus in many if not most infected resting CD4+ T cells. In some infected cells, reverse transcription eventually generates full-length viral cDNA, which is then processed by integrase. Despite the production of a suitable substrate for HIV-1 integrase, integration of the viral genome into the host cell chromosome and virus gene expression generally do not occur unless the cell receives an activating stimulus, and the linear viral genome may be subject to functional inactivation by nucleases. The result is that, in the absence of some form of stimulation provided by antigen, cytokines, or other microenvironmental factors, the decay of HIV-1 in the preintegration state is rapid. Therefore, the infection of resting CD4+ T cells is generally nonproductive.
Acknowledgments
T.C.P. and Y.Z. contributed equally to this work.
This work was supported by NIH grant AI43222 to R.F.S.
REFERENCES
- 1.Blankson, J. N., D. Finzi, T. C. Pierson, B. P. Sabundayo, K. Chadwick, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2000. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J. Infect. Dis. 182:1636-1642. [DOI] [PubMed] [Google Scholar]
- 2.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 4.Chun, T. W., K. Chadwick, J. Margolick, and R. F. Siliciano. 1997. Differential susceptibility of naive and memory CD4+ T cells to the cytopathic effects of infection with human immunodeficiency virus type 1 strain LAI. J. Virol. 71:4436-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 6.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crabtree, G. R. 1989. Contingent genetic regulatory events in T lymphocyte activation. Science 243:355-361. [DOI] [PubMed] [Google Scholar]
- 8.De Boer, R. J., M. Oprea, R. Antia, K. Murali-Krishna, R. Ahmed, and A. S. Perelson. 2001. Recruitment times, proliferation, and apoptosis rates during the CD8+ T-cell response to lymphocytic choriomeningitis virus. J. Virol. 75:10663-10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 10.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4+ T cells residing within human lymphoid tissues. Immunity 15:671-682. [DOI] [PubMed] [Google Scholar]
- 11.Esposito, D., and R. Craigie. 1998. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17:5832-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 13.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, M. S., S. Carteau, C. Hoffmann, L. Li, and F. Bushman. 1998. Retroviral cDNA integration: mechanism, applications and inhibition. Genet. Eng. 20:41-61. [DOI] [PubMed] [Google Scholar]
- 15.Hong, T., K. Drlica, A. Pinter, and E. Murphy. 1991. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J. Virol. 65:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins, M. K., A. Khoruts, E. Ingulli, D. L. Mueller, S. J. McSorley, R. L. Reinhardt, A. Itano, and K. A. Pape. 2001. In vivo activation of antigen-specific CD4 T cells. Annu. Rev. Immunol. 19:23-45. [DOI] [PubMed] [Google Scholar]
- 17.Ju, G., and A. M. Skalka. 1980. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell 22:379-386. [DOI] [PubMed] [Google Scholar]
- 18.Katz, R. A., C. A. Omer, J. H. Weis, S. A. Mitsialis, A. J. Faras, and R. V. Guntaka. 1982. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J. Virol. 42:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korin, Y. D., and J. A. Zack. 1998. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J. Virol. 72:3161-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korin, Y. D., and J. A. Zack. 1999. Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J. Virol. 73:6526-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkosky, J., R. A. Katz, and A. M. Skalka. 1990. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J. Acquir. Immune Defic. Syndr. 3:852-858. [PubMed] [Google Scholar]
- 22.Lori, F., A. Malykh, A. Cara, D. Sun, J. N. Weinstein, J. Lisziewicz, and R. C. Gallo. 1994. Hydroxyurea as an inhibitor of human immunodeficiency virus-type 1 replication. Science 266:801-805. [DOI] [PubMed] [Google Scholar]
- 23.McDougal, J. S., A. Mawle, S. P. Cort, J. K. Nicholson, G. D. Cross, J. A. Scheppler-Campbell, D. Hicks, and J. Sligh. 1985. Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen. J. Immunol. 135:3151-3162. [PubMed] [Google Scholar]
- 24.Meyerhans, A., J. P. Vartanian, C. Hultgren, U. Plikat, A. Karlsson, L. Wang, S. Eriksson, and S. Wain-Hobson. 1994. Restriction and enhancement of human immunodeficiency virus type 1 replication by modulation of intracellular deoxynucleoside triphosphate pools. J. Virol. 68:535-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder, L. C., and M. A. Muesing. 2000. Degradation of HIV-1 integrase by the N-end rule pathway. J. Biol. Chem. 275:29749-29753. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien, W. A., A. Namazi, H. Kalhor, S. H. Mao, J. A. Zack, and I. S. Chen. 1994. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J. Virol. 68:1258-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4+ T lymphocytes in infected children. J. Clin. Investig. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierson, T., T. L. Hoffman, J. Blankson, D. Finzi, K. Chadwick, J. B. Margolick, C. Buck, J. D. Siliciano, R. W. Doms, and R. F. Siliciano. 2000. Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J. Virol. 74:7824-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18:665-708. [DOI] [PubMed] [Google Scholar]
- 32.Polacino, P. S., H. A. Liang, and E. A. Clark. 1995. Formation of simian immunodeficiency virus long terminal repeat circles in resting T cells requires both T cell receptor- and IL-2-dependcent activation. J. Exp. Med. 182:617-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiser, J. 2000. Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 7:910-913. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J. M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoemaker, C., S. Goff, E. Gilboa, M. Paskind, S. W. Mitra, and D. Baltimore. 1980. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc. Natl. Acad. Sci. USA 77:3932-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoemaker, C., J. Hoffman, S. P. Goff, and D. Baltimore. 1981. Intramolecular integration within Moloney murine leukemia virus DNA. J. Virol. 40:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, J. S., S. Y. Kim, and M. J. Roth. 1990. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J. Virol. 64:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spina, C. A., J. C. Guatelli, and D. D. Richman. 1995. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J. Virol. 69:2977-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 42.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 43.Zack, J. A., A. M. Haislip, P. Krogstad, and I. S. Chen. 1992. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J. Virol. 66:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, H., O. Bagasra, M. Niikura, B. J. Poiesz, and R. J. Pomerantz. 1994. Intravirion reverse transcripts in the peripheral blood plasma of human immunodeficiency virus type 1-infected individuals. J. Virol. 68:7591-7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, H., Y. Zhang, T. Spicer, D. Henrard, and B. J. Poiesz. 1995. Nascent human immunodeficiency virus type 1 reverse transcription occurs within an enveloped particle. J. Virol. 69:3675-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, H., Y. Zhang, T. P. Spicer, L. Z. Abbott, M. Abbott, and B. J. Poiesz. 1993. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res. Hum. Retroviruses 9:1287-1296. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. [DOI] [PubMed] [Google Scholar]