Abstract
Proteins encoded by the UL46 and UL47 genes of herpes simplex virus type 1 (HSV-1) constitute major components of the viral tegument. However, their functions have so far not been elucidated in detail. By use of monospecific antisera directed against bacterially expressed glutathione-S-transferase fusion proteins, the homologous UL46 and UL47 proteins of the alphaherpesvirus pseudorabies virus (PrV) were identified in virus-infected cells and in virions. The PrV UL46 gene product of 693 amino acids (aa) exhibits an apparent molecular mass of 95 kDa, whereas the UL47 product of 750 aa was identified as a 97-kDa protein. Both are present in purified virions, correlating with their role as tegument proteins. Immunofluorescence analysis by confocal laser scan microscopy showed that late in infection the UL46 product is detectable in the cytoplasm, whereas the UL47 product was observed to be diffuse in the cytoplasm and speckled in the nucleus. Virus mutants lacking either the UL46 or the UL47 gene or both were isolated on noncomplementing cells, demonstrating that these genes either singly or in combination are not required for productive viral replication. However, plaque sizes were decreased. Interestingly, in one-step growth analysis, UL47 deletion mutants exhibited an approximately 10-fold decrease in final titers, whereas the UL46 deletion mutant was not affected. This finding correlated with ultrastructural observations which showed unimpaired virion morphogenesis in the absence of the UL46 protein, whereas in the absence of the UL47 protein intracytoplasmic aggregates of partially tegumented capsids were observed. In summary, we identified the PrV UL46 and UL47 proteins and show that the UL47 protein plays an important role in virion assembly in the cytoplasm.
Herpesviruses assemble more than 30 proteins into mature virions which make up the nucleoprotein core, the capsid shell, the tegument, and constituents of the envelope (44). In particular, the tegument, which is situated in mature virions between the viral envelope and the capsid, is a complex structure containing numerous viral gene products (43). In the alphaherpesviruses, the tegument has been suggested to consist of more than 15 different proteins (37, 46). It has now been widely accepted that herpesviruses acquire their final tegument and envelope in the cytoplasm after having traversed the nuclear membrane in a process which includes acquisition of a primary envelope by budding of intranuclear genome-containing nucleocapsids at the inner leaflet of the nuclear membrane and subsequent fusion at the outer leaflet (reviewed in reference 37). However, the molecular interactions required for the formation of this complex structure are only incompletely understood.
Although the tegument has long been considered amorphous and unstructured, this may not be entirely true. In fact, the innermost layer of the tegument, which contacts the capsid, has been shown to exhibit icosahedral symmetry since its constituents specifically interact with the pentons of the capsid (11, 51). Experimental data suggest that this innermost layer of the tegument consists of the largest protein found in herpesviruses, homologs of the UL36 gene product of herpes simplex virus type 1 (HSV-1). This gene is conserved throughout the herpesviruses (1, 10, 32). The UL36 homologs of pseudorabies virus (PrV) (28) and human cytomegalovirus (4; M. E. Harmon and W. Gibson, Proc. Am. Soc. Virol., abstr. W35-4, 1996) have been demonstrated to physically interact with the product of the UL37 homologous genes, another conserved tegument protein. Thus, the second layer of tegument may consist of the UL37 protein. In fact, in the absence of the HSV-1 UL36 protein, apparently naked capsids accumulate dispersed in the cytoplasm (13), whereas in the absence of the PrV UL37 protein these intracytoplasmic capsids form ordered aggregates (27) which contain the UL36 protein. Moreover, the HSV-1 UL37 protein is also required for formation of fully tegumented and enveloped virions (14).
Unfortunately, the subsequent steps in virion formation are still largely unclear. As concerns the interaction between tegument proteins and viral envelope glycoproteins, thought to drive budding into trans-Golgi vesicles during secondary envelopment, it has recently been shown that the nonessential virion glycoproteins gE and gI, which form a complex, as well as gM are required for the formation of mature PrV virions (7). In the absence of these glycoproteins, capsids accumulate in the cytoplasm surrounded by electron-dense material, which, by immunolabeling, has been shown to consist of tegument protein (6). Thus, it appears that lack of gE/I and gM blocks secondary envelopment. The same phenotype was reproduced when only gM and the carboxy-terminal tail of gE were deleted, indicating a role for the cytoplasmic domain of gE in secondary envelopment (6). One tegument protein, the product of the UL49 gene of PrV, has been demonstrated to specifically interact with the carboxy termini of glycoproteins E and M, which may indicate a role for this interaction in virion morphogenesis (19). However, not only is UL49 nonessential for replication in cell culture of HSV-1 (41; G. Elliott and A. Whiteley, 26th Int. Herpesvirus Workshop, abstr. 8.09, 2001), BHV-1 (31), and PrV (12, 19), but also intensive studies so far have failed to assign any phenotype to the lack of PrV UL49 either in vitro or in vivo (12, 19). Therefore, the observed deletion of gE/I and gM has to affect more than the interaction with UL49.
It has recently been shown that the UL48 protein of HSV-1, which has primarily been characterized as a factor capable of trans-inducing the viral immediate-early (α-) genes (3) (hence its name α-trans-inducing factor or α-TIF; synonyms are VP16 and vmw65), is important for virion morphogenesis. An HSV-1 UL48 deletion mutant showed defects in nuclear egress as well as in secondary envelopment in the cytoplasm (38). In contrast, a UL48 deletion mutant of PrV (20) was not impaired in nuclear egress but accumulated nucleocapsids dispersed in the cytoplasm, indicating that the UL48 protein is required for efficient PrV virion formation in the cytoplasm (20). Interestingly, in cells infected with UL48-deleted PrV the intracytoplasmic capsids did contain UL36 and UL37 proteins but lacked the UL49 protein, whereas the capsidless particles produced in great numbers contained UL49. Therefore, we speculated that the UL48 protein may form an important bridge between the part of the tegument assembled on capsids, i.e., UL36 and UL37, and the part of the tegument, e.g., UL49, which may assemble at the future budding site by interaction with vesicle-bound viral glycoproteins (20, 37). A physical interaction between UL48 and UL49 proteins has been shown for the respective HSV-1 homologs (18).
However, there are clearly more molecular interactions required for formation of the herpesviral tegument. HSV-1 VP16 interacts with the viral host cell shut-off factor vhs, the product of the UL41 gene (45), which may secure its incorporation into nascent virus particles. This is supported by the fact that a mutated form of UL41 that does not bind UL48 is not present in virions (42). Moreover, the UL13 protein kinase of HSV-1 physically interacts with and phosphorylates the carboxy terminus of gE (39) which, like for UL49, may be important for its inclusion in the virion. However, the molecular details of how all the other tegument proteins (37, 46) find their way into the virus particle are unknown.
The products of the UL46 and UL47 genes (32) have initially been described as modulators of the UL48 transactivating function (33). The HSV-1 UL47 gene specifies two tegument proteins, VP13 and VP14 (34, 48), which represent differently processed forms of the primary translation product (48) with different phosphorylation and glycosylation (35). Similarly, two proteins, VP11 and VP12, arise from the UL46 gene (49). Surprisingly, neither of the UL46 and UL47 genes either singly or in combination is required for HSV-1 replication in cell culture (2, 49). However, analysis of specific deletion mutants confirmed their effect on the activity of α-TIF. In the absence of the UL47 gene, α-TIF activity is considerably reduced (50), and absence of UL46 or UL47 resulted in a delay in the appearance of thymidine kinase activity (49). Thymidine kinase is encoded by an early gene which requires immediate-early gene products for transactivation (49). Surprisingly, the absence of either UL46, or UL47, or both only slightly impaired viral replication, resulting in ca. 10-fold-reduced titers compared to an isogenic parental strain (49). In Marek's disease virus (MDV), an avian alphaherpesvirus, the UL46 and UL47 genes have also recently been shown to be dispensable for viral replication in cell culture, although their absence impairs viral growth (17).
The UL47 protein represents a major tegument protein in HSV-1 (34), bovine herpesvirus type 1 (BHV-1) (9, 29, 30, 47), and equine herpesvirus type 1 (48). The intracellular distribution of the HSV-1 UL47 gene products has been analyzed by fluorescent tagging. Data showed that they were found predominantly in the nucleus, which parallels their effect on UL48 activity (15). However, the UL47 proteins were also detected in the cytoplasm, in particular at late times after infection (16). Similarly, the BHV-1 UL47 protein was found in the cytoplasm as well as in the nucleus (47). The cytoplasmic localization late after infection by HSV-1 or BHV-1 may be indicative of a role for the UL47 protein in virion formation. However, so far, this assumption has not been analyzed.
During our studies to unravel the molecular basis for virion morphogenesis of the alphaherpesvirus PrV, the causative agent of Aujeszky's disease in pigs (36), we set out to identify the UL46 and UL47 proteins of PrV and functionally characterize their putative role in virion morphogenesis.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants were derived from the wild-type laboratory strain Kaplan (PrV-Ka) (24). Viruses were grown on rabbit kidney (RK13) or porcine kidney (PSEK) cells in Eagle's minimum essential medium supplemented with 10 or 5% fetal calf serum. A triple mutant lacking glycoproteins gE, gI, and gM (PrV-gE/I/M−) was propagated on gM-expressing cells (7) prior to infection of noncomplementing RK13 cells for analysis. RK13-UL46/47 cells constitutively express UL46 and UL47 genes under control of the human cytomegalovirus immediate-early promoter enhancer from vector pcDNA-3 (Invitrogen, Karlsruhe, Germany). They were established after cotransfection of pcDNA-UL46 and pcDNA-UL47 plasmids into RK13 cells followed by Geneticin (Life Technologies) selection and immunofluorescence screening with the monospecific antisera.
Preparation of monospecific anti-UL46 and anti-UL47 serum.
For generation of a UL46-specific antiserum, the complete UL46 open reading frame (8) was amplified by PCR using Platinum Pfx DNA polymerase (Invitrogen), primers UL46FOR and UL46REV (Table 1), and cloned genomic BamHI fragment 1 of PrV-Ka as a template. The 2.1-kbp amplification product contains EcoRI and NotI sites allowing insertion into the EcoRI/NotI-digested prokaryotic expression vector pGEX-4T-1 (Amersham-Biosciences, Freiburg, Germany). The obtained plasmid pGEX-UL46 was used for protein expression in Escherichia coli. A ca. 110- to 120-kDa glutathione-S-transferase (GST)-UL46 fusion protein was used for four immunizations of a rabbit after separation in a sodium dodecyl sulfate (SDS)-polyacrylamide gel and electroelution. Serum obtained after the fourth immunization was used in this study.
TABLE 1.
Primers used for PCR amplification
| Primer | Sequence | Restriction enzyme | Nucleotides | GenBank accession no. |
|---|---|---|---|---|
| UL48FOR | 5′-CACAGAATTCTCACCGTGTGCGAGGGGCTG-3′ | EcoRI | 755-774 | AJ437285 |
| UL48REV | 5′-CACAGGTACCCTCATGGTGGTCGCTGCGGC-3′ | KpnI | 185-166 | AJ010303 |
| UL47FOR | 5′-CACAGAATTCACCATGAGCGACGCGGGC-3′ | EcoRI | 178-195 | AJ010303 |
| UL47REV | 5′-CACAGCGGCCGCCGCGGCGCGAGGCATC-3′ | NotI | 2510-2494 | AJ010303 |
| UL46FOR | 5′-CACAGAATTCGCCTCCGCCATGCTCCGC-3′ | EcoRI | 2443-2460 | AJ010303 |
| UL46REV | 5′-CACAGCGGCCGCACGGGGGGTCCGCGG-3′ | NotI | 4553-4539 | AJ010303 |
| UL46REV2 | 5′-CACAAAGCTTCCGCCGTCCACCTCCAGCTC-3′ | HindIII | 3887-3871 | AJ010303 |
| UL27FOR | 5′-CACATCTAGACGAGACGTGTTGCCAAACAAGCG-3′ | XbaI | 3049-3032 | M17321 |
| UL27REV | 5′-CACAAAGCTTGGCGGCGCTACAACAGCACG-3′ | HindIII | 1571-1590 | M17321 |
To generate an antiserum specific for the UL47 protein, a 95-bp SalI/NotI fragment encoding codons 719 to 750 of the UL47 gene (8) was isolated from pcDNA-UL47 (see below), cloned into expression vector pGEX-4T-3 (Amersham-Biosciences), and expressed in E. coli. A ca. 30-kDa GST-UL47 fusion protein was electroeluted after SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and used for immunization of a rabbit as described above.
Isolation of UL46-, UL47-, and UL46/47-negative mutants.
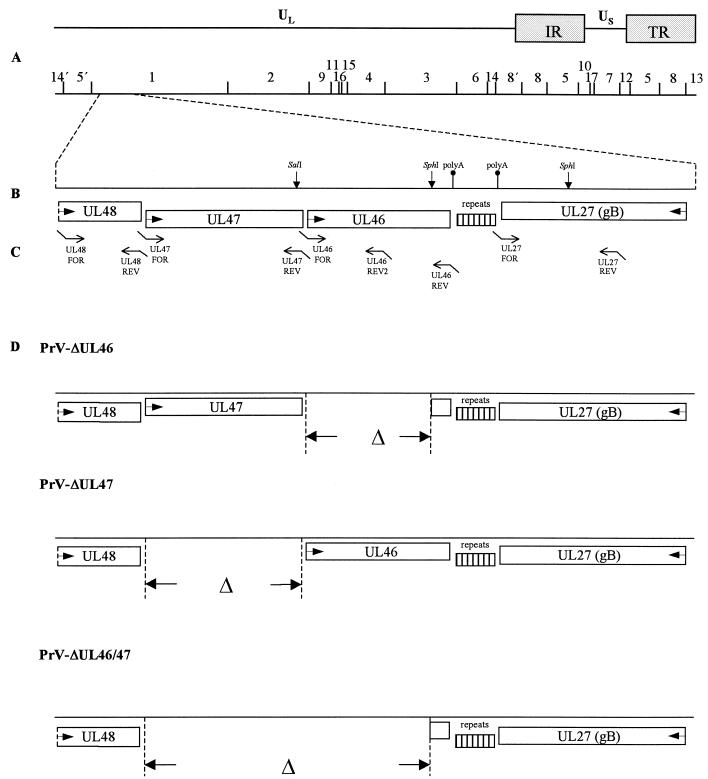
To generate a UL46-negative, a UL47-negative, and a UL46 and UL47 double-negative mutant, recombinant plasmids comprising 5′ and 3′ flanking sequences of the gene(s) to be deleted and a green fluorescence protein (GFP) marker gene cassette were assembled (23). To this end, parts of the genes UL27, UL46, UL47, and UL48 were amplified by PCR from cloned BamHI fragment 1 of viral DNA (Fig. 1 and Table 1). EcoRI, KpnI, NotI, HindIII, and XbaI restriction sites were introduced for convenient cloning. Fragments were amplified by using Platinum Pfx Polymerase (Invitrogen), and PCR products UL27, UL46, and UL48 were directly cloned into appropriately cleaved pUC19, whereas UL47 was first cloned into EcoRI/NotI-cleaved vector pcDNA3, giving rise to pcDNA-UL47. Then, the UL48 fragment was inserted into the EcoRI/KpnI site of pUC19-UL27, and UL46 was cloned into the XbaI/HindIII-cleaved pUC19-UL48. After cleavage of UL47 from pcDNA-UL47, it was inserted into the EcoRI site of pUC19-UL27. The resulting plasmids pUC19-UL46/48 and pUC19-UL27/48 were digested with BamHI and KpnI, plasmid pUC19-UL27/47 was cleaved with BamHI, and a GFP marker gene cassette was inserted into each plasmid (23). The resulting plasmids, designated pUC19-UL27/47GFP, pUC19-UL46/48GFP, and pUC19-UL27/48GFP, respectively, were sequenced to verify correct deletion and cotransfected with PrV-Ka DNA into RK13 cells. Marker gene-expressing mutants PrV-ΔUL46GFP, PrV-ΔUL47GFP, and PrV-ΔUL46/47GFP were isolated on RK13 cells. Correct deletion of the UL46 and/or UL47 genes and insertion of marker genes was verified by Southern blot analysis of mutant virus DNA.
FIG. 1.
Construction of PrV UL46 and UL47 deletion mutants. (A) A schematic map of the PrV genome shows the long (UL) and short (US) unique regions, the inverted repeat sequences (IR, TR), and the positions of BamHI restriction sites. Numbers indicate BamHI restriction fragments. (B) Coding regions of proteins in the genomic region relevant for this study. Arrows indicate transcriptional orientations. Also indicated is a region of repeated sequences between the UL27 and UL46 genes, as well as location of consensus sequences for polyadenylation (polyA) and relevant restriction sites. (C) Location of primers used for PCR amplification (see Table 1). (D) Genotypes of the constructed viral mutants.
To eliminate possible unwanted effects caused by insertion of the GFP cassette, mutants without marker genes were subsequently constructed. To obtain a UL46 deletion mutant, a 2-kbp SphI-subfragment of BamHI-fragment 1, comprising the polyadenylation consensus sequence of the UL46 gene and part of the following UL27 gene, and the 2.3-kbp UL47 fragment were cloned into pUC19, resulting in plasmid pUC19-SphI/UL47. After cotransfection of PrV-ΔUL46GFP DNA and pUC19-SphI/UL47 into RK13 cells by calcium phosphate precipitation (21), a nonfluorescing mutant virus (PrV-ΔUL46) (Fig. 1) was isolated. A UL47 deletion mutant without GFP insertion (PrV-ΔUL47) (Fig. 1) was isolated after cotransfection of PrV-ΔUL47GFP DNA and pUC19-UL46/48 plasmid DNA (see above) into RK13 cells. The UL46/47 double mutant without GFP (PrV-ΔUL46/47) (Fig. 1) was obtained after cotransfection of PrV-ΔUL46/47GFP DNA with plasmid pUC19-UL48/SphI. This plasmid contains part of the UL48 gene and the 2-kbp SphI-subfragment of BamHI-fragment 1 (see above). Correct recombination was verified by Southern blot analysis of mutant virus DNA (data not shown).
Virus purification.
For virus purification, cells were infected with PrV-Ka, PrV-ΔUL46, PrV-ΔUL47, or PrV-ΔUL46/47 and incubated until complete cytopathic effect developed. Remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), and cellular debris was removed by low-speed centrifugation. For reduction of volume, the virus-containing supernatant was centrifuged for 1 h at 22,000 rpm in a TST-28 rotor (Beckman). The pellet was resuspended in TBSal (200 mM NaCl, 2.6 mM KCl, 10 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 1.8 mM CaCl2), layered onto a discontinuous sucrose gradient (30, 40, and 50% sucrose), and centrifuged for 2 h at 20,000 rpm in a TST-28 rotor. Virions accumulated at the boundary between 40 and 50% sucrose and were harvested by aspiration, pelleted, and resuspended in TBSal.
Western blotting.
Virions were purified as described above. Infected cell lysates were harvested at different times after infection by scraping into the medium. Cells were collected by centrifugation at 13,000 rpm for 2 min in an Eppendorf centrifuge, washed twice with phosphate-buffered saline (PBS), and resuspended in 100 μl of PBS and the same volume of sample buffer. Three micrograms of purified virion preparation or 10 μl of cell lysate was separated by SDS-10% PAGE, electrotransferred onto nitrocellulose membranes, and reacted for 1 h at room temperature with monospecific antisera against the UL46 (dilution, 1:100,000), UL47 (dilution, 1:200,000), UL48 (dilution, 1:100,000 [20]), and UL49 (dilution, 1:100,000 [6]) tegument proteins, the UL19 major capsid protein (dilution, 1:200,000 [26]), or anti-glycoprotein B (gB) monoclonal antibody (MAb) b43-b5 (dilution, 1:500 [40]). After incubation with peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany), bound antibody was detected by enhanced chemiluminescence (SuperSignal; Pierce, Bonn, Germany) and recorded on X-ray film.
Indirect immunofluorescence and confocal microscopy.
RK13 cells grown on coverslips were infected for 20 h at a multiplicity of infection (MOI) of ca. 0.001 with PrV-Ka. Thereafter, they were fixed for 30 min with ethanol, washed twice with PBS, and subsequently incubated for 1 h each with anti-UL46 serum (dilution 1:300), anti-UL47 serum (dilution 1:500), or a monoclonal antibody against gE (40), and Alexa 488-conjugated secondary antibodies (Molecular Probes, Leiden, The Netherlands). Slides were washed repeatedly with PBS after each step. Fluorescence was preserved with a 9:1 mixture of glycerol and PBS, containing 25 mg of 1,4-diazabicyclooctane per ml and 1 μg of propidium iodide per ml for chromatin counterstaining. The slides were analyzed in a confocal laser scan microscope (LSM 510; Zeiss).
Determination of plaque size.
Plaque diameters were measured on RK13 cells 2 days after infection at 37°C under a methylcellulose medium. Thereafter, cells were fixed with 5% formaldehyde and stained with crystal violet. For each virus mutant 50 plaques were measured microscopically, and the average plaque diameter was determined. Values were calculated in comparison to PrV-Ka, which was set at 100%. Average percentages as well as standard deviations were determined from three independent experiments.
One-step growth analysis.
To monitor one-step growth, RK13 or RK13-UL46/47 cells were infected with PrV-Ka, PrV-ΔUL46, PrV-ΔUL47, and PrV-ΔUL46/47 at a MOI of 5 for 1 h at 4°C. Thereafter, the inoculum was replaced by prewarmed medium, and virus was allowed to penetrate for 90 min at 37°C. Then, remaining extracellular virus was inactivated by low-pH treatment. Cells and supernatants were harvested separately immediately thereafter (0 h), and after 4, 8, 12, 24, and 36 h of incubation at 37°C. Virus progeny was titrated on RK13 cells. Since no significant differences were observed in extra- and intracellular virus titers, the two were added and average values and standard deviations of two independent experiments were calculated.
Electron microscopy.
For ultrathin sectioning, RK13 or RK13-UL46/47 cells were infected at an MOI of 1 and fixed 16 h postinfection. Fixation and embedding for routine microscopy and for intracellular immunolabeling of viral proteins were performed as described previously (22, 26). Reactivities of monospecific antisera against the UL36 (28), UL37 (27), UL46 (this paper), UL47 (this paper), UL48 (20), and UL49 (6) tegument proteins were visualized with 10-nm gold-tagged secondary antirabbit antibodies (GAR10; British BioCell International). The counterstained ultrathin sections were analyzed with an electron microscope (EM400T, Tecnai 12; Philips, Eindhoven, The Netherlands).
RESULTS
Identification of PrV UL46 and UL47 proteins.
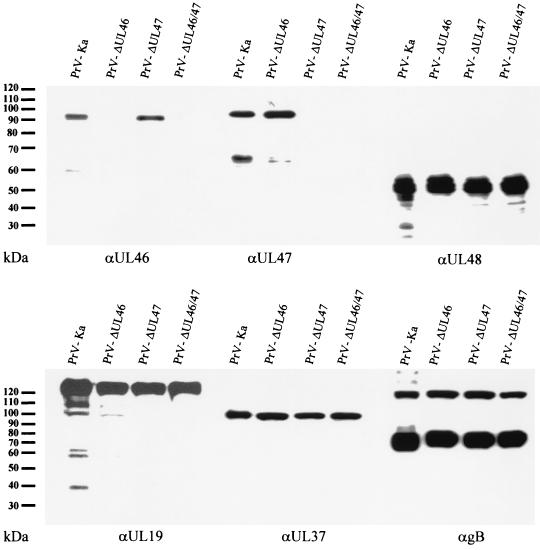
For identification of the UL46 gene product the complete UL46 open reading frame was expressed as a GST fusion protein and used for immunization of a rabbit. The obtained antiserum specifically recognized in preparations of purified PrV-Ka virions a 95-kDa protein which was absent in PrV-ΔUL46 virions (Fig. 2). It was also detected in similar amounts in PrV-ΔUL47 virions but not in PrV-ΔUL46/47 virus preparations. The UL47-specific antiserum recognized a prominent 97-kDa protein in wild-type PrV-Ka virions as well as in PrV-ΔUL46 virion preparations (Fig. 2), a protein which was not detected in PrV-ΔUL47 and PrV-ΔUL46/47 virions. A second protein of 65 kDa also reacted with the antiserum, although its intensity varied between different virion preparations (Fig. 2 and data not shown). In addition to the prominent 95- and 97-kDa proteins, both antisera also detected lower-molecular-weight proteins which may represent breakdown products, processed forms of the proteins, or internal translation products. However, the specificity of the antisera could clearly be demonstrated by their reactivity in the viral deletion mutants.
FIG. 2.
Identification and virion localization of PrV UL46 and UL47 proteins. Purified virions of PrV-Ka, PrV-ΔUL46, PrV-ΔUL47, and PrV-ΔUL46/47 were analyzed by immunoblotting using monospecific antisera against the UL46, UL47, UL48, UL19, and UL37 proteins or a monoclonal antibody against gB.
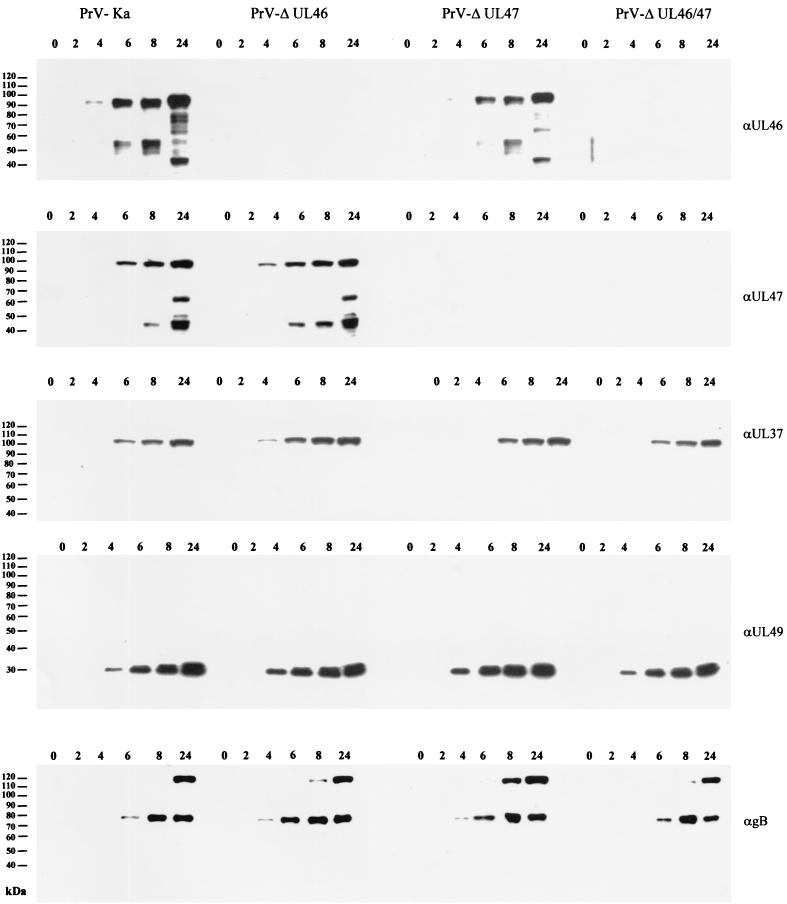
Expression kinetics of PrV UL46 and UL47 proteins.
In lysates of PrV-Ka-infected cells (Fig. 3) the UL46-specific antiserum also recognized a 95-kDa major protein as well as several lower-molecular-weight protein bands. The UL46 protein was first detectable at 4 h postinfection and increased in amount until 24 h p.i. The lower-molecular-weight proteins also increased in amount and number until 24 h p.i,. which supports our assumption that they constitute breakdown products. A similar pattern of UL46 expression was detected in PrV-ΔUL47-infected cells, whereas no UL46-related proteins were found in cells infected by PrV-ΔUL46 or PrV-ΔUL46/47.
FIG. 3.
Expression kinetics of PrV UL46 and UL47 proteins. RK13 cells were infected at an MOI of 5 with PrV-Ka, PrV-ΔUL46, PrV-ΔUL47, or PrV-ΔUL46/47 and harvested and analyzed at the indicated hours after infection. Blots were probed with monospecific antisera against the UL46, UL47, UL37, and UL49 proteins or a monoclonal antibody against gB.
Expression kinetics for the UL47 protein were similar. It became first detectable at 4 to 6 h p.i. and increased in amount until 24 h p.i. A lower-molecular-weight protein of 45 kDa was first observed at 8 h p.i., and a 65-kDa protein appeared only at 24 h p.i. The latter protein could also be detected in purified virions (Fig. 2). No UL47-related proteins were detectable in cells infected with PrV-ΔUL47 or PrV-ΔUL46/47, proving the specificity of the reactions.
Expression of the tegument proteins UL37 and UL49 occurred with kinetics similar to those of the UL46 and UL47 proteins, whereas gB appeared to be expressed later in the replication cycle, being first detectable at 6 h p.i.
Intracellular localization of the UL46 and UL47 proteins of PrV.
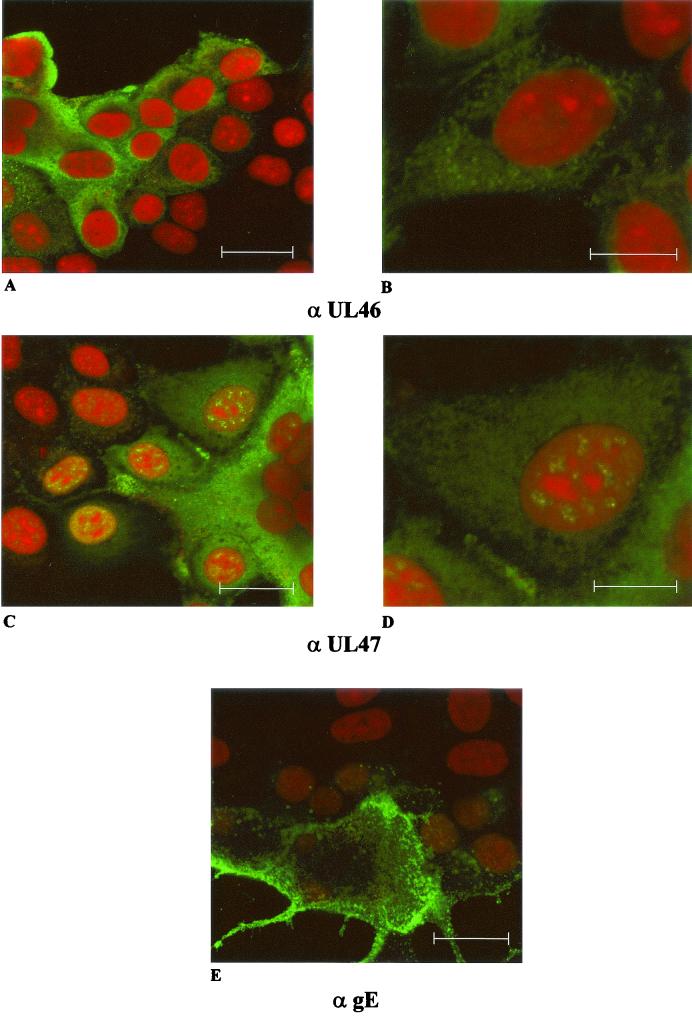
To analyze the intracellular localization of the UL46 and UL47 proteins, confocal laser scan immunofluorescence microscopy was performed on RK13 cells infected with PrV-Ka under plaque assay conditions. As shown in Fig. 4, the UL46-specific fluorescence was located in the cytoplasm. In contrast, UL47 fluorescence was detectable diffusely in the cytoplasm as well as in speckles in the nucleus. Thus, the UL47 protein might have a dual functional role in the nucleus and in the cytoplasm, whereas this appears not to be the case for the UL46 protein. For control, the envelope glycoprotein gE was detectable in the cytoplasm with preferential accumulation at the plasma membrane.
FIG. 4.
Intracellular localization of PrV UL46 and UL47 proteins. Immunofluorescence analysis by confocal laser scan microscopy of RK13 cells infected with PrV-Ka under plaque assay conditions was performed by using monospecific antisera against the UL46 (upper row) or UL47 proteins (middle row) or a monoclonal antibody against gE (bottom). For UL46 and UL47 detection, two different magnifications are shown (bars represent 25 μm in left panels and 50 μm in right panels). Red, propidium iodide stain; green, reactivity of specific antibodies.
In vitro replication of UL46 and UL47 deletion mutants of PrV.
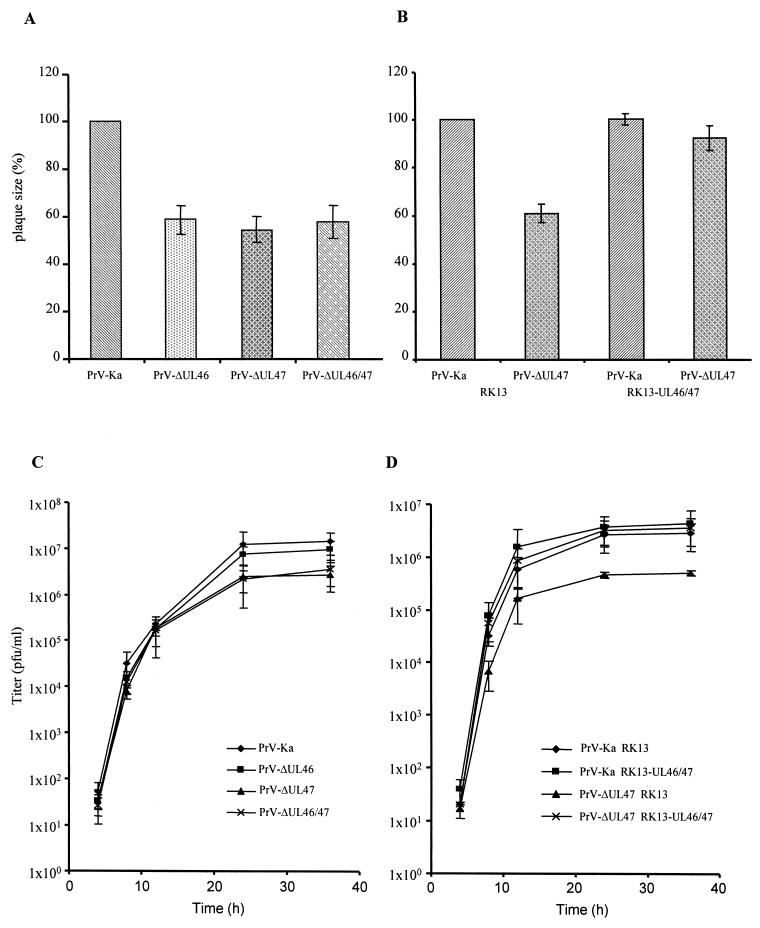
Deletion mutants lacking either the UL46 gene or the UL47 gene or both were isolated on noncomplementing RK13 cells, which already demonstrates that both proteins, either singly or in combination, were not required for productive viral replication in tissue culture. To analyze growth properties of the viral mutants in more detail, RK13 cells were infected under plaque assay conditions and plaque sizes were analyzed 48 h p.i. Plaques produced by PrV-ΔUL46, PrV-ΔUL47, and PrV-ΔUL46/47 were of similar size, which amounted to ca. 60 to 70% of the size of wild-type PrV-Ka-induced plaques (Fig. 5A). This indicates that deletion of either gene or of both had a significant effect on plaque formation but that no additive or synergistic effects occurred when both genes were simultaneously deleted. Moreover, the plaque sizes of PrV-ΔUL47 (Fig. 5B) and PrV-ΔUL46 (not shown) were restored to wild-type levels on RK13-UL46/47 cells.
FIG. 5.
Plaque size and one-step growth kinetics of PrV UL46 and UL47 deletion mutants. Growth properties of deletion mutants in UL46 (PrV-ΔUL46) or UL47 (PrV-ΔUL47) or both (PrV-ΔUL46/47) were assessed by analyzing relative plaque size on noncomplementing (A) or RK13-UL46/47 (B) cells compared to that of PrV-Ka, which was taken as 100%. (C and D) One-step replication kinetics on noncomplementing and RK13-UL46/47 cells.
In one-step growth kinetics, wild-type PrV-Ka and viral deletion mutants exhibited similar increases in the production of infectious progeny between 4 and 12 h p.i. (Fig. 5C). However, final titers of both mutant viruses lacking the UL47 protein, PrV-ΔUL47 and PrV-ΔUL46/47, were consistently ca. 10-fold lower than final titers of PrV-Ka or PrV-ΔUL46. However, titers of PrV-ΔUL47 reached wild-type levels on complementing RK13-UL46/47 cells (Fig. 5D). Thus, deletion of the UL47 protein had an effect on viral replication as analyzed in one-step growth kinetics whereas deletion of UL46 did not influence viral replication.
Absence of UL47 interferes with virion morphogenesis in the cytoplasm.
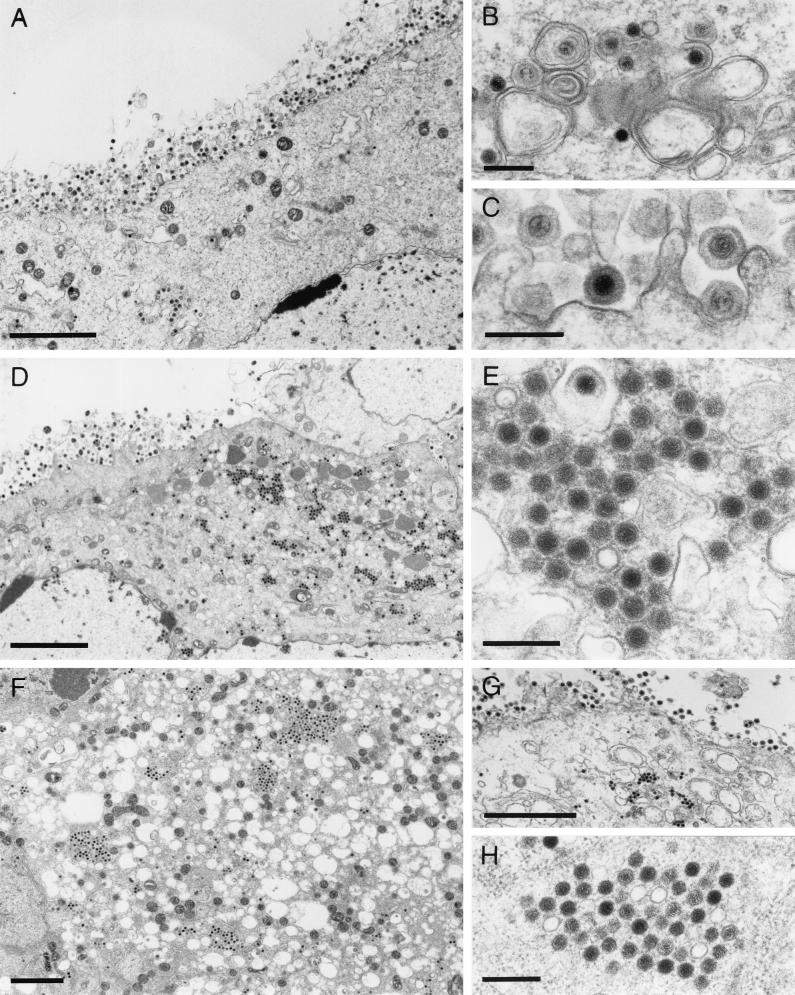
To analyze in more detail the replication defect observed in the absence of UL47, RK13 cells were infected with PrV-ΔUL46, PrV-ΔUL47, or PrV-ΔUL46/47 and examined by electron microscopy 16 h after infection. As shown in Fig. 6, cells infected with PrV-ΔUL46 showed typical herpesvirus morphogenesis (Fig. 6A) with no apparent deficiency. Capsid assembly in the nucleus as well as virion formation in the cytoplasm (Fig. 6B) appeared to occur at wild-type levels, and numerous extracellular virions lined the plasma membrane (Fig. 6A and C). This correlates well with the replicative ability measured by one-step growth kinetics. In contrast, cells infected with PrV-ΔUL47 or PrV-ΔUL46/47, besides also showing all stages of virion maturation (Fig. 6D and F) including extracellular mature virions (Fig. 6D and G), exhibited local clusters of capsids (Fig. 6E and H) in an ordered arrangement. Whereas in the PrV-ΔUL47-infected cells these capsids appeared to be surrounded by electron-dense material, this was less evident in PrV-ΔUL46/47-infected cells. Most intracytoplasmic capsids contained viral genome as indicated by their electron dense core. However, empty electron-lucent capsids could also be observed (Fig. 6E and H). An occasional translocation of empty capsids through the nuclear membranes has previously been demonstrated (22). Frequently, in the absence of the UL46 protein in either PrV-ΔUL46- (not shown) or PrV-ΔUL46/47-infected cells (Fig. 6F), intracytoplasmic vesiculation was observed. The importance of this phenomenon is unclear at present but did not seem to significantly interfere with viral replication since PrV-ΔUL46 was not impaired in one-step growth (Fig. 5C). Normal replication of PrV-ΔUL47 was restored in RK13-UL46/47 cells (Fig. 7A to C).
FIG. 6.
Electron microscopy of mutant virus-infected cells. RK13 cells were infected with PrV-ΔUL46 (A to C), PrV-ΔUL47 (D and E), or PrV-ΔUL46/47 (F to H) and analyzed by electron microscopy 16 h after infection. (A to C) Unimpaired virion morphogenesis in the absence of the UL46 protein including secondary envelopment in the cytoplasm (B) as well as numerous extracellular virions (A and C) are shown. (D) Overview of a PrV-ΔUL47-infected cell demonstrating that besides all stages of normal virion morphogenesis, intracytoplasmic aggregations of capsids can be observed (shown at higher magnification in panel E). Similar aggregations were also observed in PrV-ΔUL46/47-infected cells (F to H). Bars, 3 μm in panels A, D, F, and G and 300 nm in panels B, C, E, and H.
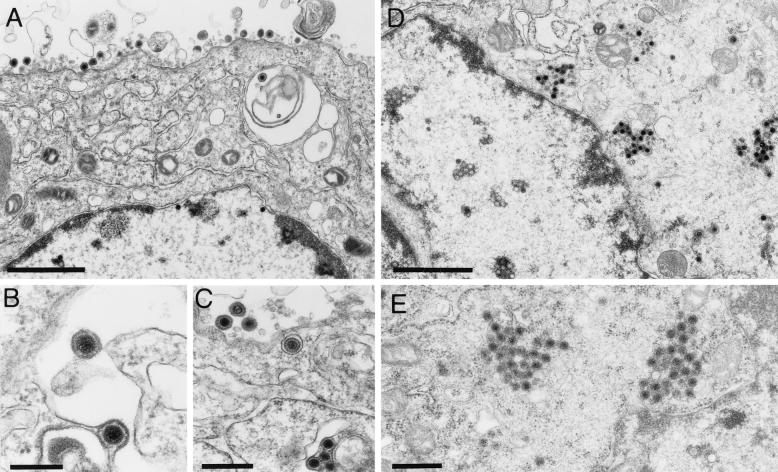
FIG. 7.
Electron microscopy of infected RK13-UL46/47 cells. Transcomplementing RK13-UL46/47 and normal RK13 cells were infected with PrV-ΔUL47 and analyzed by electron microscopy 16 h after infection. Panels A to C show unimpaired virion formation on complementing cells, whereas panels D and E demonstrate, in a parallel assay, the formation of capsid aggregates in the cytoplasm of noncomplementing cells. Bars, 1.5 μm in panels A and D, 100 nm in panel B, and 500 nm in panels C and E.
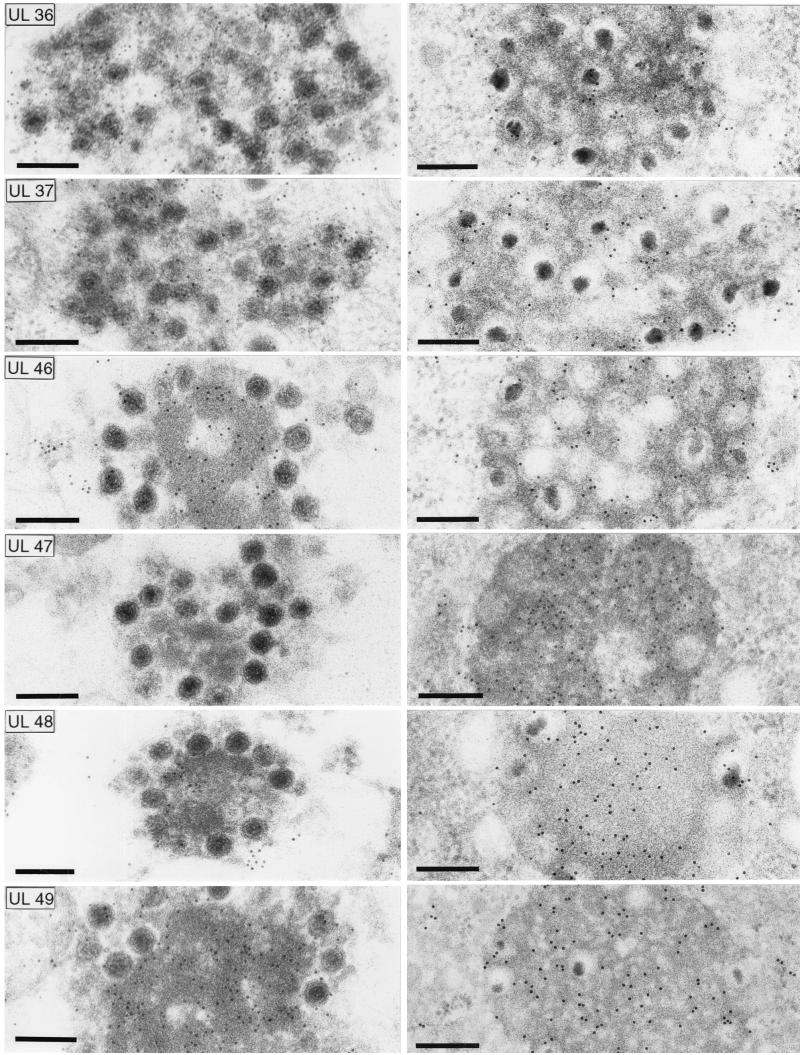
To analyze the composition of the capsid aggregates formed in the cytoplasm of noncomplementing cells infected by PrV-ΔUL47 as concerns their content of viral tegument proteins, immunoelectron microscopy was performed to compare them to the aggregations formed in the absence of glycoproteins gE, gI, and gM, which have previously been shown to be composed of capsids and tegument proteins (6, 7). As demonstrated in Fig. 8, the capsid aggregates of PrV-ΔUL47 (Fig. 8, left row) labeled with antisera against the UL36, UL37, UL46, UL48, and UL49 proteins indicating that they indeed contain bona fide tegument. In comparison, all tested antisera reacted with the aggregations formed in the absence of gE, gI, and gM (Fig. 8, right row). In most cases, intensity of labeling was similar in PrV-ΔUL47 and PrV-gE/I/M− clusters. There were two exceptions: the UL47-specific antiserum labeled the PrV-gE/I/M− clusters but, as expected, failed to react with the PrV-ΔUL47 aggregates. Moreover, the UL48 antiserum labeled the PrV-gE/I/M− clusters quite heavily, while label was only relatively sparse on the PrV-ΔUL47 aggregations, suggesting that the presence of UL47 protein enhances incorporation of the UL48 protein into the aggregates.
FIG. 8.
Immunoelectron microscopy. Intracytoplasmic aggregates formed in the absence of the UL47 protein (left panel) or in the absence of glycoproteins gE/I and gM (right panel) (7) were analyzed by immunoelectron microscopy using monospecific antisera against the tegument proteins UL36, UL37, UL46, UL47, UL48, and UL49. Bar, 250 nm.
DISCUSSION
In this paper we report the identification and characterization of the PrV homologs of HSV-1 UL46 gene products (VP11/12) and UL47 gene products (VP13/14) as well as construction and functional analysis of viral mutants defective in expression of either or both of the two genes.
Whereas absence of UL46 resulted in only moderately reduced plaque sizes in cell culture but did not affect one-step growth kinetics and virion morphogenesis, deletion of UL47 did impair virus replication. In the absence of the UL47 protein, titers obtained in one-step growth analyses were consistently ca. 10-fold lower than those of wild-type PrV-Ka. Moreover, electron microscopic analyses revealed that although all stages of normal virion morphogenesis were observed, lack of UL47 in addition resulted in the formation of cytoplasmic aggregates containing capsids surrounded by amorphous protein material. Since these inclusions labeled with antibodies against the tegument proteins UL36, UL37, UL46, UL48, and UL49, we assume that they constitute tegumented capsids. Although similar in appearance to the aggregates detected in the absence of glycoproteins gE/I and gM (6, 7), the tegumented capsids that accumulated in the absence of UL47 were smaller and contained less electron-dense material around the capsid shell. Moreover, the absence of UL47 did not impair secondary envelopment nearly as drastically as the absence of gE/I and gM. Whereas in the triple-mutant PrV-gE/I/M− viral titers were decreased ca. 1,000- to 10,000-fold (7), absence of UL47 resulted in an only 10-fold decrease in viral titers. However, in both mutants secondary envelopment seems to be impaired whereas tegumentation of capsids still occurred. This is in contrast to the situation in the absence of the UL37 protein. Here, the observed intracytoplasmic clusters of capsids contained only the UL36 protein but not the UL49 protein (27). Of note, neither a reduction in plaque size nor viral titer nor any aggregates of intracytoplasmic capsids were observed on RK13-UL46/47 cells infected with PrV-ΔUL47. Therefore, the data suggest that absence of the UL47 protein defines another intermediate in virion formation and that the PrV UL47 protein plays a role in secondary envelopment.
It has been shown by yeast-two hybrid analysis that gE and gM interact via their cytoplasmic tails with the tegument protein UL49. This interaction is required for incorporation of UL49 into nascent virions since in the absence of gE and gM no UL49 protein is found in the purified-virion fraction whereas it is present in normal amounts in infected cells (19). Surprisingly, deletion of UL49 did not detectably influence replication of PrV either in cell culture (12, 19) or in the rat model host (12). Therefore, other tegument-envelope protein interactions which drive secondary envelopment are presumably present. The UL47 protein may be one of these additional tegument proteins which make contact with the future envelopment site via direct or indirect interaction with viral envelope glycoproteins. However, so far no specific interaction between the UL47 protein and PrV glycoproteins has been observed.
Glycoproteins gE and gM have been designated so-called nonessential glycoproteins (36) due to the fact that they can be deleted singly from the virus without drastic impairment of viral replication in cell culture. In contrast, simultaneous deletion of both resulted in a severe growth defect (7). A similar functional redundancy may be present in the tegument proteins. However, simultaneous deletion of UL46 and UL47 did not inhibit viral replication or reduce plaque sizes beyond the influence of the UL47 deletion alone. It is conceivable that other tegument proteins act as functional analogs for UL46 and/or UL47, securing virion formation even in the absence of these two prominent tegument constituents. Since more than 15 proteins have been demonstrated or suggested to be part of the alphaherpesvirus tegument (37, 46), there are plenty of candidates.
The UL46 and UL47 genes of HSV-1 specify proteins which are posttranslationally modified by glycosylation and/or phosphorylation (35) or nucleotidylylation (5), resulting in the appearance of at least two products for each gene (34, 35, 49). These have been designated VP11/12 and VP13/14, respectively. With the monospecific antisera, we were able to identify the UL46 and UL47 homologous proteins of PrV both in infected cells and as constituents of purified virions. In PrV, the UL46 gene product of 693 amino acids (aa) has a calculated mass of 75 kDa, whereas it migrates in SDS-PAGE with an apparent molecular mass of 95 kDa. A similar situation applies to the UL47 gene product, whose primary translation product of 750 aa should exhibit a molecular mass of 80 kDa, but the UL47 protein identified in infected cells and purified virions runs as a 97-Da protein in SDS-PAGE. It remains to be determined whether the PrV gene products are also modified like their HSV-1 counterparts, which could at least partly explain their aberrant migration in SDS-PAGE.
The clustered genes UL46 to UL49 all encode prominent tegument proteins. However, only the absence of the UL48 protein appears to have a drastic effect on virion morphogenesis in PrV (20) and HSV-1 (38), whereas the absence of UL49 did not detectably impair PrV replication (12, 19). This is in contrast to the situation in MDV, in which deletion of UL48 did not significantly impair viral growth whereas absence of UL49 proved to be lethal for the virus (17). Thus, there appear to be functional differences between tegument protein homologs in alphaherpesviruses. However, in PrV, HSV-1, and MDV, the UL46 and UL47 proteins were not required for productive viral replication. Deletion of UL46 in PrV resulted in only a moderate reduction in plaque size (this paper) and resulted in a delay in the onset of HSV-1 replication (50). Lack of UL47 showed a somewhat intermediate phenotype in PrV with ca. 10-fold reduced titers and a clearly discernible ultrastructural phenotype in infected cells (see above), whereas in HSV-1 the early phenotype was similar to that of a UL46 mutant (50). Whether deletion of UL47 in HSV-1 also impaired late steps in virion morphogenesis has not yet been analyzed. However, it is evident that within this gene cluster the encoded proteins contribute to strikingly different degrees to the process of virus maturation.
The UL46 and UL47 gene products of HSV-1 have first been identified by their influence on the transactivating function of the UL48 protein on viral immediate-early promoters (33), a function which has been confirmed by using specific viral deletion mutants (50). However, it is unclear how this influence is being exerted, whether directly by physical interaction with the UL48 protein, or indirectly involving other viral and/or cellular proteins. We recently showed that the PrV UL48 protein also influences expression of immediate-early genes (20), although the early stages of infection proceed quite efficiently in the absence of virion-bound UL48 protein. It remains to be analyzed whether the PrV UL46 and UL47 proteins also influence a possible transactivating function of the PrV UL48 protein, which, however, does not in any case appear to be critical for PrV replication, at least in cell culture. A physical interaction between the UL46 and UL48 gene products has been proposed for the HSV-2 homologs (25), but the functional importance of this observation is unclear. We show here that absence of either UL46 or UL47 or both did not impair intracellular expression or incorporation of the UL48 or UL37 tegument proteins into extracellular virions. However, immunoelectron microscopic analyses indicate that less UL48 protein may be present in the aggregations of tegumented capsids found in the absence of the UL47 protein than in the aggregations formed in the absence of the gE/I and gM glycoproteins. This suggests that UL47 influences incorporation of UL48 into the viral tegument. In contrast, no obvious difference was detected in the other tested tegument proteins, i.e., UL36, UL37, UL46, and UL49.
Consistent with the current view that herpesviruses acquire their final tegument and envelope in the cytoplasm (37), both the UL46 and UL47 proteins of PrV were detected in the cytoplasm late after infection, as analyzed by immunofluorescence microscopy. Whereas UL46 could not be found in the nucleus, UL47 was present in the nucleus in punctate areas. Nuclear localization has also been observed for the HSV-1 UL47 protein fused to a GFP reporter (15, 16), which may correlate with its influence on UL48 transactivating function in the nucleus. A prolonged eclipse phase in cells infected with a UL48-deletion mutant of PrV (20) may be indicative of a delayed onset of viral replication attributable to the absence of the UL48 transactivating function. However, in contrast to HSV-1 (49), deletion of neither UL46 nor UL47 resulted in a delay in the production of infectious PrV progeny, which suggests the absence of a modulating effect on α-TIF function. In keeping with the prediction from the primary envelopment-deenvelopment-secondary envelopment pathway, UL46 and UL47 proteins were not detectable by immunolabeling in perinuclear primary enveloped virions, whereas intracytoplasmic and extracellular mature virus particles were heavily labeled (Fig. 7 and data not shown). We also show that the absence of the UL47 protein did not affect virion localization of the UL46 gene product and vice versa. Therefore, these two proteins appear to be incorporated into nascent virions independent of each other.
In summary, we identified the UL46 and UL47 gene products of PrV and demonstrate a role for the UL47 protein in virion morphogenesis at late stages of infection. In the absence of UL47 secondary envelopment is impaired, resulting in intracytoplasmic aggregation of tegumented capsids. Future studies will be directed at elucidating the putative viral and/or cellular proteins which may interact with UL47 to secure its incorporation into nascent virions and at uncovering the function of the UL46 protein.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/5-1).
We are grateful to Egbert Mundt for preparation of the antisera and to Klaus Osterrieder for help with the confocal microscope. We thank Uta Hartwig, Petra Meyer, and Nadine Müller for expert technical assistance and E. Zorn and H. Stephan for photographic help.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Barker, D., and B. Roizman. 1990. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology 177:684-692. [DOI] [PubMed] [Google Scholar]
- 3.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bechtel, J., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47 and UL49 gene products. J. Biol. Chem. 269:17401-17410. [PubMed] [Google Scholar]
- 6.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bras, F., S. Dezélée, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29-40. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, D., and V. Misra. 1991. The most abundant protein in bovine herpes 1 virions is a homologue of herpes simplex virus type 1 UL47. J. Gen. Virol. 72:3077-3084. [DOI] [PubMed] [Google Scholar]
- 10.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 12.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, P., G. Sexton, J. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the UL37 polypeptide of herpes simplex virus type 1 abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnelly, M., and G. Elliott. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 75:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly, M., and G. Elliott. 2001. Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 75:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorange, F., K. Tischer, J.-F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, F. L., and A. J. van der Eb. 1973. A new technique for assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 22.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jöns, A., and T. C. Mettenleiter. 1997. Green fluorescent protein expressed by recombinant pseudorabies virus as an in vivo marker for viral replication. J. Virol. Methods 66:283-292. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 25.Kato, K., T. Daikoku, F. Goshima, H. Kume, K. Yamaki, and Y. Nishiyama. 2000. Synthesis, subcellular localization and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch. Virol. 145:2149-2162. [DOI] [PubMed] [Google Scholar]
- 26.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. The pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaBoissiere, S., M. Trudel, and C. Simard. 1992. Characterization and transcript mapping of a bovine herpesvirus type 1 gene encoding a polypeptide homologous to the herpes simplex virus type 1 major tegument proteins VP13/14. J. Gen. Virol. 73:2941-2947. [DOI] [PubMed] [Google Scholar]
- 30.LaBoissiere, S., M. Trudel, and C. Simard. 1996. The bovine herpesvirus type 1 major tegument protein VP8 expressed in recombinant vaccinia virus does not induce significant immunity in mice. Virus Res. 40:191-198. [DOI] [PubMed] [Google Scholar]
- 31.Liang, X., B. Chow, Y. Li, C. Raggio, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the unique long region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 33.McKnight, J., P. Pellett, F. Jenkins, and B. Roizman. 1987. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate α-trans-inducing factor-dependent activation of α genes. J. Virol. 61:992-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLean, G., F. Rixon, N. Langeland, L. Haarr, and H. Marsden. 1990. Identification and characterization of the virion protein products of herpes simplex virus type 1 UL47 gene. J. Gen. Virol. 71:2953-2960. [DOI] [PubMed] [Google Scholar]
- 35.Meredith, D., J. Lindsay, I. Halliburton, and G. Whittaker. 1991. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J. Gen. Virol. 72:2771-2775. [DOI] [PubMed] [Google Scholar]
- 36.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 37.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mossman, K., R. Sherburne, C. Lavery, J. Duncan, and J. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, T., W. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 40.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomeranz, L., and J. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptides. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 44.Roizman, B., and P. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 45.Smibert, C., B. Popova, P. Xiao, J. Capone, and J. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 47.Van Drunen Littel-van den Hurk, S., S. Garzon, J. van den Hurk, L. A. Babiuk, and P. Tijssen. 1995. The role of the major tegument protein VP8 of bovine herpesvirus-1 in infection and immunity. Virology 206:413-425. [DOI] [PubMed] [Google Scholar]
- 48.Whittaker, G., M. Riggio, I. Halliburton, R. Killington, G. Allen, and D. Meredith. 1991. Antigenic and protein sequence homology between VP13/14, a herpes simplex virus type 1 tegument protein, and gp10, a glycoprotein of equine herpesvirus 1 and 4. J. Virol. 65:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y., and J. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, Y., D. Sirko, and J. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, Z., D. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]