Abstract
A partially conserved region spanning amino acids 142 to 191 of the Sindbis virus (SIN) nsP4 core polymerase is implicated in host restriction, elongation, and promoter recognition. We extended the analysis of this region by substituting Ser, Ala, or Lys for a highly conserved Arg183 residue immediately preceding its absolutely conserved Ser184-Ala-Val-Pro-Ser188 sequence. In chicken cells, the nsP4 Arg183 mutants had a nonconditionally lethal, temperature-sensitive (ts) growth phenotype caused by a ts defect in minus-strand synthesis whose extent varied with the particular amino acid substituted (Ser>Ala>Lys). Plus-strand synthesis by nsP4 Arg183 mutant polymerases was unaffected when corrected for minus-strand numbers, although 26S mRNA synthesis was enhanced at the elevated temperature compared to wild type. The ts defect was not due to a failure to form or accumulate nsP4 at 40°C. In contrast to their growth in chicken cells, the nsP4 Arg183 mutants replicated equally poorly, if at all, in mosquito cells. We conclude that Arg183 within the Pro180-Asn-Ile-Arg-Ser184 sequence of the SIN nsP4 polymerase contributes to the efficient initiation of minus strands or the formation of its replicase and that a host factor(s) participates in this event.
After the genome of an alphavirus is released into the cytoplasm of an infected cell, it is translated into the four nonstructural proteins (nsPs) that form the viral RNA-dependent RNA polymerase activities (47). The viral polymerase, probably also with host factors, forms a replication complex with the genome RNA and copies it into a minus-strand template. The minus strand, in turn, is used as a template to produce both the genome and the subgenomic 26S mRNA, the latter by recognition of an internal promoter in the minus strand. The 26S subgenomic mRNA is translated to produce the capsid and envelope proteins. Therefore, three polymerase activities are formed during the replication cycle as a result of the stepwise and regulated cleavage of the nsP polyprotein precursors (21, 23, 24, 36, 42, 51). The four nsPs are numbered according to their gene order and are translated initially as polyproteins P123 and P1234, both of which are subsequently cleaved by the viral papain-like protease activity present in nsP2. Briefly, cleavage of nsP4 from nascent P1234 occurs first and likely cotranslationally, activating the polymerase and forming an initial P123+nsP4 complex that is active in minus-strand synthesis. Second, cleavage at the 1/2 site produces nsP1+P23+nsP4 that is active in both minus-strand and genome synthesis. Finally, intermediate polyprotein P23 is cleaved, giving stable (fully processed) nsP1+nsP2+nsP3+nsP4 complexes that are active in the synthesis of genome and 26S subgenomic mRNA but not minus-strand RNA.
Each of the alphavirus nsPs has been assigned specific roles in viral replication based on the mapping of temperature-sensitive (ts) mutants, biochemical analyses, and protein homology comparisons (36, 47). How the nsP4 core polymerase functions in the formation of a replication complex, in polymerization, and in promoter recognition has been probed by determining sites of causal ts lesions affecting these aspects of viral RNA synthesis. Residues of nsP4 that affect viral RNA synthesis when modified include the N-terminal Tyr1 (41, 43), Gln93 when Val425 of nsP2 is also modified (11), Gly153 (3, 11), Gln191 (34, 39), and residues 142 and 187 (22). From such studies, it also became clear that nsP4 has intricate and complex interactions with other nsPs in addition to viral RNA and host factors.
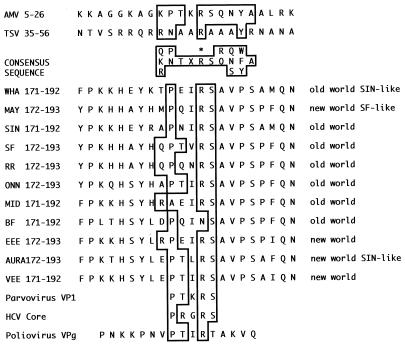
In the N-terminal third of nsP4 of most alphaviruses is a 5-amino-acid sequence, Pro180-Asn-Ile-Arg-Ser184 in Sindbis virus (SIN) and Pro-Thr-Val-Arg-Ser in Semliki Forest virus (SFV), which is in the middle of a region with highly to absolutely conserved individual residues or stretches of amino acids (Fig. 1). The Arg183 residue is near residues 187 and 191, which affect host range (22) and promoter recognition (34), respectively. Only Pro187 is part of the absolutely conserved Ser184-Ala-Val-Pro-Ser188 sequence present in all alphavirus nsP4 proteins (Fig. 1). In the sequence from residues 171 to 192 in SIN (its equivalent in other alphaviruses may differ by 1 nucleotide [nt]), absolutely conserved individual residues, in addition to Ser-Ala-Val-Pro-Ser, are Pro171, His175, Tyr177, Gln191, and Asn192, with nonpolar amino acids at positions 189 and 190. In addition, the SIN Pro180-Asn-Ile-Arg-Ser184 sequence was predicted to share similarities with a sequence motif (Pro-Thr-X-Arg-Ser) in ilarvirus and alfalfamovirus coat proteins, whose Arg residue is essential for their role as transcription activators (1, 48). Therefore, we focused on the Arg within this 5-amino-acid sequence in SIN nsP4 and created mutants that substituted Ser, Ala, or Lys for the Arg183 residue that is conserved in all known alphaviruses but Barmah Forest virus (20).
FIG. 1.
Amino acid alignment of the Pro-X-X-Arg-Ser sequence. Numbers denote the amino acid positions in the nsP4 proteins of the alphaviruses or for avian myeloblastosis virus (AMV) and tobacco streak virus (TSV) coat protein consensus sequence Pro-Thr-X-Arg-Ser (48). The substitution in SIN nsP4 conferring ts reactivation of minus-strand synthesis (34) is Gln191→Lys (34), and that conferring a host-restricted, ts RNA-negative phenotype in 35.1C2a virus (22) is Pro187→Arg (+). The consensus sequence determined for AMV and ilarvirus (1) coat protein is boxed, and the essential Arg (R∗) is indicated. References for the sequences are as follows: Whatora virus (WHA), Ellen Strauss, personal communication; Mayora virus (MAY), Ellen Strauss, personal communication; SIN, 46; SFV, 49; Ross River virus (RR), 8; O'nyong-nyong virus (ONN), 26; Middleburg virus (MID), 46; eastern equine encephalitis virus (EEE), 53; Aura virus, 33; Venezuelan equine encephalitis virus (VEE), 16; Barmah Forest virus (BF), 20; parvovirus VP1 and hepatitis C virus (HCV) core, 48; and poliovirus VPg, 30.
MATERIALS AND METHODS
Cells, virus, and plasmids.
Chicken embryo fibroblast (CEF) cells were prepared from 10-day-old embryos from the eggs of leukosis-free (specific-pathogen-free-COFAL/Marek-negative) flocks (Spafas, Roanoke, Ill.). CEF cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 6% (vol/vol) fetal bovine serum (FBS) and 5% (vol/vol) tryptose phosphate broth at 37°C. The Aedes albopictus C7-10 cell line was kindly provided by V. Stollar (45) and adapted to grow in DMEM supplemented with 6% FBS and 5% tryptose phosphate broth at 30°C.
The heat-resistant strain of SIN, SIN HR, and the ts RNA-negative mutants ts6 and ts11 were isolated by Burge and Pfefferkorn (4) and obtained from E. Pfefferkorn.
The infectious clone of SIN HR, pToto1101, was constructed by Rice and coworkers (31) and was a generous gift from C. Rice. The π34C shuttle vector contains the SIN sequence from PvuII (nt 5160) to NcoI (nt 8038) cloned into the plasmid πAN7 and was kindly provided by J. Lemm and C. Rice (Washington University, St. Louis, Mo.).
Construction of R183 mutants.
Plus-sense primers prepared by Operon Technologies Inc. (Alameda, Calif.) were complementary to nt 6294 to 6329 of the Toto1101 sequence. The primers differed at nt 6316 to 6318 and coded for Ala (GCA) or Lys (AAA) rather than the wild-type residue Arg (CGC). These primers and a downstream, minus-sense primer (nt 8081 to 8100) were used to amplify this region of the nsP4 gene of pToto1101 DNA by the PCR. The PCR products were separated on a 0.8% agarose gel in buffer containing 0.04 M Tris-acetate and 0.001 M EDTA. The desired fragment of approximately 1,800 bp in length was cut out of the gel and extracted (QIAquick PCR purification kit; Qiagen, Santa Clarita, Calif.). The PCR product was digested with BanII (sites at nt 5419 and 6305) and AatII (nt 7999). The fragments were separated on a 0.8% agarose-Tris-acetate-EDTA gel, and the desired fragment of about 1,700 nt was excised from the gel and purified. The mutated BanII (nt 6305)-AatII (nt 7999) fragment was swapped into pπ34C in place of the parental fragment by using T4 DNA ligase (Gibco BRL Products). The ligation products were used to transform competent Escherichia coli MC1061p3 cells, and the resulting colonies were screened for the presence of pπ34C-size DNA. π34C:Ala183 or Lys183 cDNA was extracted with the QIAprep Miniprep kit according to the instructions of the manufacturer (Qiagen) and sequenced to confirm that the mutagenesis was site specific and that there were no other changes from the parental sequence. One clone was found to have unexpectedly acquired the codon for Ser at the 183 position. Since Ser is an uncharged, polar amino acid, unlike Ala and Lys, the clone was processed along with the Ala183 and Lys183 clones. The pπ34C:Ala183, Ser183, or Lys183 DNA was digested with HpaI (nt 6919) and SpeI (nt 5262). The specific restriction fragments were purified and individually swapped into pToto1101. The ligated pToto1101:Ala183, Ser183, or Lys183 DNA was used to transform competent E. coli MC1061 cells, and the colonies were screened for Toto-size plasmids. Toto:Ala183, Ser183, or Lys183 cDNA was sequenced from nt 6214 to 6461 by using ThermoSequence or Sequenase 2.0 (Amersham Life Science) to confirm that the changes at nt 6316 to 6318 were present and were the only changes from the Toto1101 sequence in this region.
The mutant cDNAs were linearized at a unique site downstream of the viral sequence by using XhoI and transcribed by using MEGAscript SP6 (Ambion), according to the manufacturer's instructions except for the addition of 1.6 mM m7G(5′)ppp(5′)G cap. The transcription products were diluted in phosphate-buffered saline and immediately used to transfect CEF cells at 30°C. Cytopathic effect was seen usually within 2 to 3 days, at which time the virus was harvested. Site-specific mutagenesis was again confirmed by isolating virion RNA and determining the sequence of the genome in this region of nsP4 by amplifying nt 6245 to 6450 by reverse transcription-PCR and sequencing.
Infection and RNA labeling.
In all experiments CEF or C7-10 monolayers were infected with SIN HR, Toto1101, or the recombinant viruses as previously described (37). CEF cultures were shifted to 40°C by rinsing them with 40°C DMEM and incubating them at 40°C. A. albopictus C7-10 cells were shifted to 34.5°C in a similar manner. Cells were pulse-labeled with [3H]uridine (50 μCi/ml unless otherwise indicated) for 1 h in DMEM containing 20 μg of actinomycin D (AMD)/ml, 6% FBS, and 20 mM HEPES, pH 7.4. At the end of the labeling period, the cells were washed with ice cold phosphate-buffered saline and lysed with 5% lithium dodecyl sulfate in LET buffer (0.1 M LiCl, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4) containing 200 μg of proteinase K/ml. The DNA in the cell lysates was sheared by passage through a 27-gauge needle before acid-insoluble incorporation was determined.
Isolation of RF RNA and quantitation of minus strands.
The rate of minus-strand synthesis was determined as described by Dé et al. (5). Infected cultures were pulse-labeled for periods of 1 h at 30°C or 30 min at 40°C before harvest to determine the rate of viral RNA synthesis. For accumulation experiments, infected cultures were incubated in the presence of [3H]uridine beginning immediately after adsorption until the cultures were harvested. Deproteinized cell lysates were digested with RNase A and chromatographed on CF-11 cellulose (Whatman, Clifton, N.J.) as described previously (5), according to the procedures of Franklin (10), for the isolation of the replicative-form RNA (RF RNA), the double-stranded core of the viral replicative intermediates (RIs). Minus-strand RNA synthesized during the labeling period was measured by nuclease protection assays that determine the amount of [3H]uridine-labeled RF RNA that after denaturation will hybridize to an excess of unlabeled, virion 49S plus-strand RNA (5).
Protein labeling and immunoprecipitation.
CEF cells were infected at 30°C with a multiplicity of infection (MOI) of 100, as described above. After adsorption, cell monolayers were refed with complete 30°C DMEM and were subsequently shifted to 40°C when the rate of viral protein synthesis was at its peak. Cultures were refed with DMEM containing 1% of the normal methionine concentration, 335 mM NaCl, 5% dialyzed fetal calf serum, 2 μg of AMD/ml, and 20 mM HEPES (pH 7.4) for 90 min at 30°C prior to labeling at 30°C or for 40 min at 40°C prior to labeling at 40°C. The cultures were labeled for pulses of 30 min at 30°C or 15 min at 40°C with [35S]methionine in DMEM lacking methionine. One set of cultures was harvested immediately after the pulse by addition of 1% sodium dodecyl sulfate in 1/2× Laemmli buffer (19). The second set of dishes was chased in the presence of 20-fold excess cold methionine for 2 h at 30°C or 1 h at 40°C, after which the dishes were harvested.
In the second experiment, the cell monolayers were infected and then refed with DMEM containing 1% of the normal methionine concentration, 5% dialyzed fetal calf serum, 2 μg of AMD/ml, and 20 mM HEPES, pH 7.4, after adsorption. Cultures were shifted to 40°C when polyprotein synthesis was at its peak; labeled for 5 min; and chased for 5, 15, 30, or 60 min before harvest. Immunoprecipitation of the nsP4 proteins in infected cell extracts and analysis of the resultant immunoprecipitates on 5 to 10% polyacrylamide-Laemmli gels were performed as described previously (5, 19), with polyclonal, monospecific antibodies to nsP4 that were a generous gift from J. H. and E. G. Strauss (California Institute of Technology).
RESULTS
Construction of R183 recombinant viruses.
We used site-directed mutagenesis and the SIN infectious clone pToto1101 (31) to change Arg183 to Ala and Lys. In addition, a Ser183 mutant was found fortuitously from among the clones obtained from the Ala mutagenesis. Substitution of these amino acids for Arg183 was not lethal, with transcripts of all three nsP4 mutant infectious cDNAs yielding virus when transfected into CEF cells maintained at 30°C. The region of the genome RNA of each recombinant virus from nt 6245 to 6450 was sequenced and found to contain only the expected changes at nt 6316 to 6318 of codon 183 of nsP4 (data not shown).
Plaque phenotype of R183 mutants.
The size of the plaques and the efficiency of plaquing (EOP), which is the virus titer (PFU per milliliter) at 40°C divided by the titer at 30°C, of the Arg183 mutants were compared to those for SIN HR and ts6 and ts11, two ts RNA-negative mutants of SIN HR. At 30°C, the titers and sizes of plaques of virus produced in cells infected with the Arg183 mutants were essentially the same as those of virus produced by SIN HR (SIN HR and virus produced from pToto1101 are indistinguishable) (Table 1). The EOP was reduced in the Arg183 mutants and varied from 0.5 to 0.03, with Lys183 virus always having the highest EOP and Ser183 virus having the lowest. The <1-mm-diameter plaques formed at 40°C by Ala183 and Ser183 viruses were only 20% of the size of the 5-mm plaques formed by SIN HR at 40°C, and those formed by the Lys183 virus were 40% of the size of wild type. Thus, Arg183 mutants had a ts but nonlethal phenotype that differed from traditional single-base, conditionally lethal, ts mutants that give EOPs of 10−4 to 10−5 and whose 40°C plaques are large and are the products of revertants.
TABLE 1.
Plaque phenotype of Arg183 mutant viruses
| Virus | 30°C titer (109 PFU/ml) | EOP (40°C PFU/ ml/30°C PFU/ml) | 40°C plaque sizea (mm) |
|---|---|---|---|
| SIN HR | 4.4 | 0.91 | 5 |
| ts6 | 1.7 | 0.0016 | 5 |
| ts11 | 8.3 | 0.00013 | 5 |
| Ser183 | 3.9 | 0.03 | <1 |
| Ala183 | 6.1 | 0.11 | <1 |
| Lys183 | 2.3 | 0.48 | 2 |
When infected at time zero or shifted to 40°C at 1 h p.i., ts6 and ts11 do not form plaques even after 2 days of incubation. However, revertants arise at low dilutions and are wild type in plaque size (5 mm).
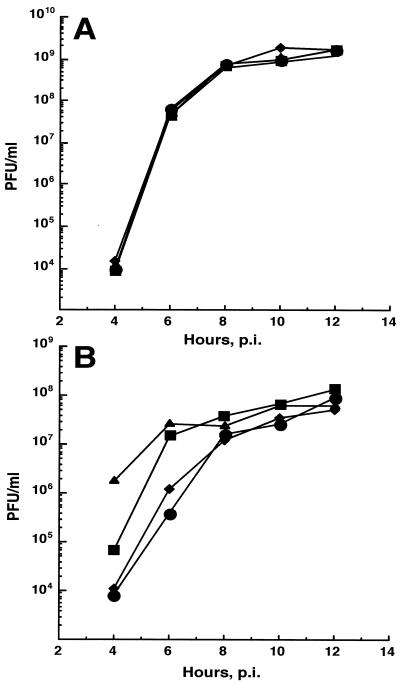
The 40°C small plaque phenotype of the Arg183 mutants was associated with delayed virion release (Fig. 2). Cells infected with SIN Toto1101 virus (Arg183) at 40°C attained a maximum rate of virion release by 6 h postinfection (p.i.), compared to 10 to 12 h for the Arg183 mutant-infected cells. The extent of the delay in release of PFU at 40°C was proportional to the plaque sizes generated by the individual mutants, which provided an explanation for their smaller plaque size at 40°C. At 30°C there was no difference in virion release among the Arg183 mutants and SIN Toto1101 (Fig. 2).
FIG. 2.
Release of virions by Arg183 mutant viruses and by parental virus. CEF cultures were infected at an MOI of 100 at 30°C and maintained at 30°C (A) or shifted to 40°C at 1 h p.i. (B). At 2-h intervals, the medium was removed and the cells were refed with the same volume of fresh medium. The viral titer was determined by plaque assays performed at 30°C on CEF cells. ▴, parental Toto virus; ▪, Lys183 virus; ♦, Ala183 virus; •, Ser183 virus.
RNA phenotype of R183 mutants.
We determined if the Arg183 mutants had ts defects in viral RNA synthesis. CEF cells were infected at 30°C with either mutant or parental virus, maintained at 30°C or shifted to 40°C at the indicated times, and labeled with [3H]uridine for 1-h periods at 30 or 40°C in the presence of AMD (Fig. 3). At 30°C (Fig. 3A), the time that it took for all three Arg183 mutant-infected cells to reach maximal rates of RNA synthesis was delayed about 1 h compared to SIN Toto1101-infected cultures. Nevertheless, RNA synthesis by the three Arg183 mutants at 30°C attained close to (50 to 70%) the same maximal rate as SIN Toto1101. When the infection was initiated and maintained at 40°C, cells infected with either Ala183 or Ser183 virus failed to incorporate levels of [3H]uridine during 30-min pulse-labeling above mock levels (data not shown). When shifted to 40°C between 2 and 4 h p.i., each of the mutants failed to increase viral RNA synthesis to the level that was attained at 30°C by 6 h p.i. (Fig. 3 and data not shown). Rather, viral RNA synthesis increased during the first hour after temperature shift and then leveled off at a constant rate. Once again, Ser183 was slightly more ts than Ala183, and Lys183 was the least ts, attaining the highest, final rate of RNA synthesis after temperature shift.
FIG. 3.
Viral RNA synthesis and effect of inhibiting protein synthesis with cycloheximide. CEF cultures were infected with Ala183 (•), Ser183 (♦), Lys183 (▪), Toto1101 Arg183 (▴), or SIN nsP4 elongation mutant ts6 (▾) at an MOI of 100 at 30°C and maintained at 30°C (A) or shifted to 40°C at 3 h p.i., at which time one set of dishes was incubated in the absence of cycloheximide (B) and a duplicate set was incubated with 100 μg of cycloheximide/ml (C). At hourly intervals, cultures were labeled for 1-h periods with 50 μCi of [3H]uridine/ml in the presence of 20 μg of AMD/ml and in the continued presence or absence of cycloheximide. The acid-insoluble incorporation in 50,000 cells was determined.
It was clear that the ts defect in the Arg183 mutants did not affect viral RNA synthesis that had begun at 30°C and thus was not an elongation defect (3). The slightly increased transcription rates observed for the Arg183 mutants after the shift to 40°C were due to formation of new replication complexes. After the shift to 40°C, cells infected with the Arg183 mutants had rates of RNA synthesis in the absence of cycloheximide (Fig. 3B) that were two- to fivefold greater than in its presence (Fig. 3C), which is consistent with the formation of only a limited number of new complexes after the temperature shift. The extent of this difference varied in the order Ser<Ala<Lys.
The Arg183 mutants were specifically defective in minus-strand synthesis. To enhance our ability to detect the very small amount of viral RNA synthesized at 40°C by the Arg183 mutants, labeled minus strands were allowed to accumulate over time by labeling infected cells continuously with [3H]uridine beginning at 1 h p.i. at 30 or 40°C and in the presence or absence of cycloheximide. As shown in Table 2, the Ser183, Ala183, and Lys183 mutants yielded only 7, 13, and 27% as many labeled minus strands, respectively, after shift to 40°C at 1 h p.i. as at 30°C but accumulated as many as did Toto1101-infected cells when kept at 30°C. Less than 1% of the 30°C amount of viral minus strands accumulated if cycloheximide was included in the medium at the time of shift to 40°C. Thus, the 7 to 27% new minus strands made at 40°C were products of new complexes formed at 40°C.
TABLE 2.
Accumulation of minus-strand RNA at 30 and 40°C in the absence or presence of cycloheximidea
| Virus | RNA at 30°C (cpm)
|
% of RF cpm in minus strands | Minus strand RNA (cpm)
|
40°C/30°C (−CH) (%) | ||
|---|---|---|---|---|---|---|
| RFs | Minus strands | 40°C + CH | 40°C | |||
| Arg183 (Toto1101) | 318,797 | 109,028 | 34.2 | 862 | 127,725 | 117.2 |
| Ser183 | 314,746 | 194,829 | 45.0 | 1,635 | 14,453 | 7.4 |
| Ala183 | 373,236 | 141,635 | 52.2 | 889 | 18,735 | 13.2 |
| Lys183 | 311,907 | 152,522 | 48.9 | 1,050 | 41,437 | 27.2 |
CEF cells were infected at an MOI of 100 and maintained at 30°C or shifted to 40°C 1 h p.i. One set of 40°C cultures (40°C + CH) was incubated with 100 μg of cycloheximide (CH)/ml beginning at the time of shift to 40°C; a duplicate set of cultures was incubated at 40°C in the absence of cycloheximide (“40°C”). All cultures were labeled with 250 μCi of [3H]uridine/ml in the presence of 20 μg of AMD/ml, from 1 to 7 h p.i., when they were harvested. The RF RNA cores of the viral RIs were isolated, and the amount of labeled minus-strand RNA was determined.
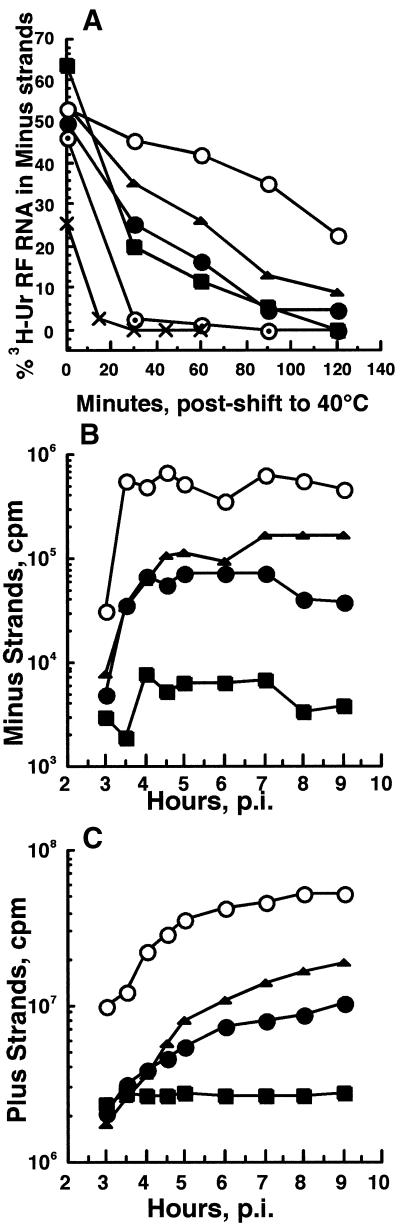
Although fewer minus strands were made at 40°C in the Arg mutant-infected cells, their synthesis followed a normal pattern in that minus-strand synthesis stopped at the same time at 40°C as in the Toto1101-infected cells (Fig. 4A). The analysis was done by shifting infected cells to 40°C when minus-strand synthesis was very active and the rates of viral RNA synthesis were increasing exponentially, i.e., at 2 h p.i. for Toto1101-infected cultures and 3 h p.i. for mutant-infected cultures. This is when only ∼10% of the 30°C maximal rate was observed. Minus-strand synthesis in Toto1101-infected cells continued at high rates after shift to 40°C, and only after peaking did it decline. Because 40 to 45% of the total radiolabeled uridine incorporated into Toto1101 RI RNA was in minus strands, 80 to 90% of all minus strands were newly made within the pulse period. Minus-strand synthesis with Ala183, Ser183, and Lys183 viruses also continued for up to 2 h after shift to 40°C, but it was less and decreased faster with time at 40°C than for Toto1101 virus. The Arg183 mutant defect differed from that of nsP3 mutant SIN ts4, which abruptly ceased minus-strand synthesis after temperature shift. At 30°C, the kinetics of minus-strand synthesis and cessation by the Arg183 mutants were similar to those for parental virus (data not shown).
FIG. 4.
Effect of shift from 30 to 40°C on minus-strand and plus-strand synthesis. (A) Ability to continue minus-strand synthesis. CEF cultures were infected with Ser183 (▪), Ala183 (•), Lys183 (▴), ts4 (×), or Toto1101 (○) at an MOI of 100 at 30°C and were shifted to 40°C at 2 h p.i. (Toto1101 and ts4) or 3 h p.i. (Arg183 mutants), when ∼10% of each virus's 30°C maximal transcription rate was observed. Cultures maintained at 30°C were labeled at 30°C for 1 h beginning the hour before the time of shift-up (zero time point); cultures shifted to 40°C were labeled for 30-min periods beginning at the time of shift. Labeling was performed with 250 μCi of [3H]uridine/ml in the presence of 20 μg of AMD/ml. A duplicate set of Ala183-infected cultures was treated with 100 mg of cycloheximide/ml beginning at the time of shift to 40°C and labeled in the presence of the drug (⊙). Cells were harvested at the end of the pulse period, and the viral RF RNA was isolated and analyzed for its content of radiolabeled minus strands. These results are expressed as percentages of the total incorporation in the cores of RI-native RFs (active also in plus-strand synthesis) that was in minus-strand RNA (see Materials and Methods). (B) Accumulation of minus-strand RNA. The viral RF RNA was purified from the lysates analyzed in panel C, and their total amount of radiolabeled minus-strand RNA was determined in RNase protection assays, as described in Materials and Methods. (C) Accumulation of plus-strand RNA. CEF cells, infected with each virus at an MOI of 100 at 30°C, were shifted to 40°C at 1 h p.i. and labeled continuously with 250 μCi of [3H]uridine/ml in the presence of 20 μg of AMD/ml. Cells were harvested at the times indicated, and acid-precipitable incorporation into viral RNA per 3.7 × 106 cells was determined.
When minus-strand templates were accumulating, the level of plus strands increased exponentially (Fig. 4B and C). When minus-strand synthesis ceased (4 to 5 h p.i. for both mutant and parental viruses) and the number of minus-strand templates stopped increasing, the accumulation of plus strands assumed a constant linear rate. At 40°C, the Arg183 mutants formed fully functional plus-strand polymerase complexes. The lower number of templates produced a lower overall level of plus strands, but the ratio of 49S and 26S plus strands relative to the number of templates equaled that produced by Toto1101 and differed little from the ratios found at 30°C (Table 3). They were also similar to the ratio of total plus strands to RI made at 7 to 8 h p.i. at 40°C by nsP complexes formed at 30°C (Table 3).
TABLE 3.
Synthesis of 49S RNA, 26S mRNA, and RI RNA by alphavirus nsP complexes made at 30 or 40°Ca
| Virus | Ratio of cpm in plus strands/cpm in RIs
|
Molar ratio of 49S to 26S RNAb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 30°C | Shift to 40°C
|
nsP complexes made at 40°C
|
nsP complexes made at 30°C
|
||||||
| 1 h p.i. | 6 h p.i. | 30°C | Shift to 40°C 1 h p.i. | 40/30°C | 30°C | Shift to 40°C 6 h p.i. | 40/30°C | ||
| Ser183 | 8.1 | 7.1 | 8.6 | 0.22-0.25 (0.21) | 0.24-0.30 (0.27) | 1.1 | 3.2 | ||
| Ala183 | 9.0 | 8.5 | 5.9 | 0.17-0.27 (0.21) | 0.29-0.33 (0.30) | 1.4 | 2.7 | ||
| Lys183 | 7.3 | 8.6 | 7.1 | 0.16-0.20 (0.18) | 0.26-0.40 (0.31) | 1.7 | 3.9 | ||
| Arg183 | 7.4 | 10.9 | 9.9 | 0.20-0.27 (0.24) | 0.55-0.63 (0.58) | 2.4 | 3.3 | ||
| SFV ts4 | 0.16 | 2.46 | 15.4 | ||||||
| SFV ts1 | 1.40 | 5.15 | 3.6 | ||||||
| SIN ts17b | 0.20 | 1.40 | 7.0 | ||||||
CEF cells were infected with each virus at an MOI of 100 and maintained at 30°C or shifted up to 40°C after adsorption for 1 h at 30°C or were shifted up at 6 h p.i. The latter conditions allowed examination of the activity at 40°C of replication complexes made at 30°C. The cultures kept at 30°C or shifted at 1 h p.i. to 40°C, in triplicate, were labeled from 6 to 7 h p.i. with 50 μCi of [3H]uridine/ml (at 30°C) or 300 μCi/ml (at 40°C), after which the RNA was analyzed by electrophoresis on 0.8% agarose gels and labeled RI, 49S, and 26S RNA bands were cut out and counted (see Materials and Methods). The low levels of RNA synthesis by Arg183 mutants at 40°C necessitated the use of this higher amount of [3H]uridine/ml. Cultures shifted at 6 h p.i. were labeled at 40°C from 7 to 8 h p.i. Averages of the values obtained for each of the triplicate cultures are shown in parentheses.
The counts per minute in each species were corrected for the differences in molecular weights of the 26S and 49S RNAs and are expressed as the molar ratio of 49S to 26S RNA. Molar ratios for SIN ts17 and the SFV mutants were from reference 36.
The Arg183 mutants showed enhanced 26S mRNA synthesis compared to wild-type virus (Table 3). Usually with alphaviruses the ratio of the synthesis of 49S genomes to 26S mRNA increased two- to threefold when cells were shifted from 30 to 40°C and decreased when shifted from 40 to 30°C, i.e., more genomes and less subgenomic mRNA were made at the higher temperature and vice versa at the lower temperature (35). The enhanced ability to recognize the subgenomic promoter for 26S mRNA synthesis was a property only of complexes formed at 40°C. Complexes of nsPs that were synthesized and assembled by the Arg183 mutants at 30°C and then shifted to 40°C behaved like parental ones (Table 3).
R183 mutants are host restricted.
The phenotype expressed by the Arg183 mutants also varied with the type of host cell (Table 4). While Toto1101 produced the same high numbers of PFU in C7-10 cells at 34.5°C as at 30°C, the Arg183 mutants produced less than 2% of the PFU at 34.5°C but gave full yields at 30°C. The defect in progeny production at 34.5°C was not due to the effects of temperature alone as the Arg183 mutants produced full yields when propagated on CEF cells at 34.5°C.
TABLE 4.
Virion release by C7-10 cells and CEF cells at 30 and 34.5°C
| Virus | PFU/ml at temp (°C) in cell type:
|
||||
|---|---|---|---|---|---|
| C7-10a
|
CEFb
|
||||
| 30 | 28 | 34.5 | 30 | 34.5 | |
| Ser183 | 2.5 × 109 | 2.6 × 106 | 5.0 × 109 | 6.0 × 109 | |
| Ala183 | 1.2 × 109 | 1.2 × 107 | 5.8 × 109 | 9.5 × 109 | |
| Lys183 | 1.1 × 109 | 6.0 × 107 | 1.1 × 1010 | 2.9 × 109 | |
| Arg183 (Toto1101) | 3.2 × 108 | 3.6 × 108 | 1.4 × 1010 | 6.4 × 109 | |
| 35.1C2ac | 1.2 × 107 | 1.3 × 106 | 9.0 × 107 | ||
C7-10 cells were infected at 30°C with an MOI of 300 and maintained at 30°C or shifted to 34.5°C after a 1-h adsorption at 30°C. At 21 h p.i., the cells were refed with new medium, and the harvested medium was assayed for PFU production on CEF cells at 30°C.
CEF cells were infected at 30°C with an MOI of 100 and maintained at 30°C or shifted to 40°C after adsorption. At 21 h p.i., the medium was harvested and assayed for PFU production on CEF cells at 30°C.
The results for 35.1C2a are from the work of Lemm et al. (22). Infected C7-10 cells were maintained at 28°C or shifted to 34.5°C at 1 h p.i. At 28 h p.i., the C7-10 medium was harvested and assayed for PFU production on CEF cells at 28°C. CEF cells were infected and maintained at 34.5°C until 6 h p.i., when the medium was harvested and assayed for PFU production on CEF cells at 28°C.
In C7-10 cells, the three Arg183 mutants were less efficient at synthesizing RNA than was parental Toto1101, even at 30°C (Table 5). Only when the mutant-infected cells were shifted to 34.5°C late in infection, after the full complement of viral RIs had formed, did we observe a rate of RNA synthesis that was the same as their maximal 30°C rates. Thus, replication complexes assembled at 30°C in C7-10 cells remained fully active in plus-strand synthesis at 34.5°C, but few or no new ones formed at 34.5°C.
TABLE 5.
Viral RNA synthesis in C7-10 cells and CEF cells at 30 and 34.5°C
| Virus | C7-10 cellsa
|
CEF cellsb
|
|||
|---|---|---|---|---|---|
| 30°C | 34.5°C Early | 34.5°C Late | 30°C | 34.5°C Early | |
| Ser183 | 0.43 | NDc | 0.59 | 0.71 | 0.87 |
| Ala183 | 0.38 | ND | 0.62 | 0.64 | 1.0 |
| Lys183 | 0.40 | ND | 0.63 | 0.66 | 0.86 |
| Arg183 (Toto1101) | 1.0 | 1.5 | 1.7 | 1.0 | 1.5 |
C7-10 cells were infected at 30°C at an MOI of 300 and maintained at 30°C, shifted to 34.5°C after adsorption (Early), or shifted to 34.5°C at 16 h p.i. (Late). Cultures were labeled for 1-h pulse periods from 16 to 21 h p.i. with 50 μCi of [3H]uridine/ml in the presence of 20 μg of AMD/ml. The acid-precipitable incorporation was determined and is expressed relative to Toto1101 at 30°C.
CEF cells were infected at 30°C at an MOI of 100 and maintained at 30°C or shifted to 34.5°C after adsorption. Infected cultures were labeled for 1-h pulse periods with [3H]uridine in the presence of AMD as described above. The acid-precipitable incorporation is expressed relative to Toto1101 at 30°C.
ND, not detected above the level of acid-precipitable incorporation by mock-infected cells.
Polyprotein processing by R183 mutants.
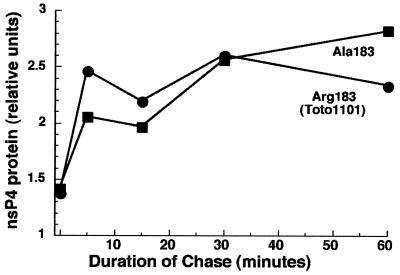
Active alphavirus polymerase requires the processing of nsP4 from polyprotein precursors. We determined whether replacement of Arg183 by other amino acids affected the cleavage of nsP4 from P1234 or its stability once cleaved. Infected CEF cultures were shifted to 40°C when protein synthesis was at its peak (4 h p.i. for Toto1101 and 5 h p.i. for the Arg183 mutants). Cultures were pulse-labeled with [35S]methionine for 15 min and then chased in the presence of a 20-fold excess of unlabeled methionine to allow nascent proteins to be processed. At the end of a 1-h chase period, similar amounts of labeled nsP4 were immunoprecipitated from extracts of cells infected with either the Arg183 mutants or the parental Arg183 virus (data not shown). Analysis of the kinetics of nsP4 processing during shorter chase intervals found that the rate of cleavage of nsP4 from P1234 also was the same for Toto1101 and Ala183 viruses (Fig. 5).
FIG. 5.
Processing of nsP4 by Ala183 and Arg183 (Toto1101) viruses. Cells were infected with Ala183 or Toto1101 at an MOI of 100 and were shifted to 40°C at 5 and 3.5 h p.i., respectively, when the maximum amount of nsP translation was occurring (data not shown). Cultures were pulse-labeled at 40°C immediately following shift with 100 mCi of [35S]methionine/ml for 5 min and chased in the presence of a 20-fold excess of unlabeled methionine for 5, 15, 30, or 60 min before the samples were harvested. Cell extracts were immunoprecipitated with anti-nsP4 antibodies, and the immunoprecipitates were analyzed by polyacrylamide gel electrophoresis (Materials and Methods). The amount of cleaved nsP4 in each sample, quantified by scanning radioautographs of fluorographed gels, is expressed in relative units.
DISCUSSION
Our long-term interest has focused on the mechanism controlling the number of minus strands that accumulate in alphavirus-infected cells because, ultimately, it is the number of minus strands that determines viral RNA synthesis. The present study was undertaken to probe further a region in the nsP4 polymerase that is between residues 142 and 191 and that might functionally resemble the alfalfamovirus coat protein Pro-Thr-X-Arg-Ser sequence motif (1, 48). Replacement of the Arg residue at position 183 within the nsP4 sequence of SIN conferred a nonlethal, ts defect in viral minus-strand synthesis that varied in extent depending upon the amino acid substituted and the cells in which the virus was replicating. Thus, a positive charge at this position appears to be important in the acquisition of efficient minus-strand replicase activity by the nsP4-containing complex. Arg is a preferred or optimal residue at this position, both from its conservation among alphavirus nsP4 proteins and from the twofold-increased level of minus-strand synthesis that it gave compared to Lys183 nsP4 proteins. The “leakiness” of the Arg183 mutants was responsible for their relatively high EOP compared to conditionally lethal, ts mutants that fail completely to replicate at nonpermissive temperatures. It also made analysis of their phenotype difficult. The Arg183 mutant ts defects in minus-strand synthesis were not due to any failure of nsP4 to form or stably accumulate at 40°C. In CEF cells, reduced rates of minus-strand synthesis during the crucial, early phase of virus infection reduced the final number of viral templates, which in turn reduced the rate of plus-strand synthesis and delayed virion release. The latter was associated with and likely caused the 40°C small plaque phenotype.
Substitutions for Arg183 in nsP4 affected only polymerase assembled at 40°C and lowered its transcription efficiency at 40°C. The observed phenotype indicated that the Arg183 region of nsP4 is particularly important for the activity of unstable P123+nsP4 (active only in minus-strand synthesis) or nsP1+P23+nsP4 (active in both minus-strand and genome synthesis) replicases. Of particular note, minus-strand synthesis ceased in all of the Arg183 mutant-infected cells at about the same time (4 to 5 h p.i. at 40°C) as with the parental virus and well before parental (maximal) amounts of minus strands accumulated. Had Arg183 mutant minus-strand synthesis been able to continue, eventually normal, parental levels of minus strands and replication-transcription complexes would have formed. Because minus-strand synthesis shut off at the same time after infection as in parental virus-infected cells, fewer Arg183 mutant minus strands and replication-transcription complexes were made at 40°C.
In addition to the major defect in minus-strand synthesis, transcription complexes formed at 40°C in cells infected with the Arg183 mutants, and especially the Ser183 virus, were different from those formed at 30°C or from those formed at 30 or 40°C by the parental virus. The Ser183 transcription complexes assembled and assayed at 40°C retained higher levels of 26S mRNA synthesis that were close to levels observed at 30°C. The two- to threefold loss of 26S mRNA synthesis and the corresponding increase in 49S plus-strand synthesis seen with parental virus upon shift to 40°C did not occur. We argue that this is not a reflection merely of the lower level of plus-strand synthesis because thermal instability of 26S mRNA synthesis is observed for SIN and SFV throughout the infectious cycle and at both early (low RNA levels) and late (high RNA levels) times. The ability to retain efficient subgenomic mRNA synthesis at high temperature was related inversely to the level of minus-strand synthesis at 40°C. This inverse relationship between 26S and minus-strand synthesis, seen before only for certain nsP2 mutants (21, 34, 36, 38, 42), supports the existence of a switch from minus-strand synthesis to 26S mRNA synthesis associated with cleavage of P23 polyproteins. One prediction is that this region of nsP4 acts in differential 26S and minus-strand promoter recognition. Finally, the nsP4 Arg183 mutant function was different than that affected by a change of nsP4 Gln191→Lys. The latter substitution kept minus-strand synthesis going or reactivated it at 40°C and did not alter subgenomic promoter recognition (39).
The Arg183 mutants possessed a host-dependent ts defect similar to, but more accentuated than, SIN 35.1C2a (22), a mutant in which Pro187 within the absolutely conserved Ser-Ala-Val-Pro-Ser sequence motif in nsP4 was changed to Arg. While the Arg183 and Pro187 mutants grew efficiently at 34.5°C in CEF cells, both were severely inhibited at this temperature, and partially defective at 30°C, in mosquito cells. Mosquito host factors may be more stringent in their interactions or more limited in number or kind compared to CEF factors, as also found by others (17, 18, 22). Lemm et al. (22) hypothesized that it was the interaction of nsP4 with host components necessary for replication that was rendered ts and not nsP4's enzymatic activity. Our studies agree with this conclusion and show that such host factors specifically affect SIN minus-strand synthetic events. The type of host factor might resemble the bacterial host factor hfq required by bacteriophage Qβ only for minus-strand synthesis (2, 28, 40). Alternatively, folding of the nsPs or the chaperones involved in their folding may be different in invertebrate cells than in vertebrate cells.
Crystal structures of RNA and DNA polymerases indicate that they share common structural features (13, 15, 25) and a common catalytic mechanism (44). The nucleotide binding sites formed by finger, palm, and template base interactions “open” in the absence of a template, enclose or clamp around the template upon its binding (15, 44), and flexibly open and close during base pair formation (6, 7). The nsP4 Arg183 defects are suggestive of polymerase initiation or template binding alterations. However, because little is known about promoter recognition, higher-order assembly of the replicase and transcriptase, and the means of modulation of template selection, a specific role(s) cannot be assigned yet to this region. As shown in Fig. 1, sequences with apparent similarity to the SIN Pro180-Thr-X-Arg-Ser184 nsP4 sequence also have upstream sequences rich in Lys and other basic amino acids. The SIN nsP4 Arg183 region did not resemble the ilarvirus and alfalfamovirus coat protein motif's role in plus-strand synthesis (50). The Arg residue in the poliovirus VPg sequence is proposed to stabilize a UTP moiety for transfer to the terminal Tyr residue of VPg (30) and to function during plus- and minus-strand initiation (14, 32). Within SIN nsP4, changes at amino acid 287 in a loop at the tip of the predicted finger domain (29) and at amino acids 592 and 609 at the base of the thumb domain increased affinity for CTP and UTP by altering possibly the geometry of the nucleoside triphosphate binding pocket (27).
To begin to identify protein regions contacting the SIN nsP4 Arg183 region, we selected pseudorevertants of the Arg183 mutants that suppress its defective interactions. Possibilities could include compensatory changes within other regions of nsP4 or within other nsPs, such as nsP1 (12, 41, 52) or nsP3 (51), that are known to specifically affect minus-strand synthesis. The companion paper (9) reports results of this screening and the identification of a new region of nsP1 as a potential interaction partner for the Arg183 region of the alphavirus polymerase.
Acknowledgments
We acknowledge Lee Gehrke, Harvard Medical School, for discussions that formed the impetus for this study. We also gladly acknowledge the generous gift of Toto1101 cDNA from C. M. Rice and of monospecific nsP4 antiserum from E. G. Strauss and J. H. Strauss.
Support for this study was derived from Public Health Service grant AI-15123 from the National Institutes of Health.
REFERENCES
- 1.Ansel-McKinney, P., S. W. Scott, M. Swanson, X. Ge, and L. Gehrke. 1996. A plant viral coat protein RNA binding consensus sequence contains a crucial arginine. EMBO J. 15:5077-5084. (Erratum, 15:7188-7189.) [PMC free article] [PubMed] [Google Scholar]
- 2.Barrera, I., D. Schuppli, J. M. Sogo, and H. Weber. 1993. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J. Mol. Biol. 232:512-521. [DOI] [PubMed] [Google Scholar]
- 3.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1988. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J. Virol. 62:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge, B. W., and E. R. Pfefferkorn. 1966. Isolation and characterization of conditional-lethal mutants of Sindbis virus. Virology 30:204-213. [DOI] [PubMed] [Google Scholar]
- 5.Dé, I., S. G. Sawicki, and D. L. Sawicki. 1996. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J. Virol. 70:2706-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doublie, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 391:251-258. [DOI] [PubMed] [Google Scholar]
- 7.Eom, S. H., J. Wang, and T. A. Steitz. 1996. Structure of Taq polymerase with DNA at the polymerase active site. Nature 382:278-281. [DOI] [PubMed] [Google Scholar]
- 8.Faragher, S. G., A. D. Meek, C. M. Rice, and L. Dalgarno. 1988. Genome sequences of a mouse-avirulent and a mouse-virulent strain of Ross River virus. Virology 163:509-526. [DOI] [PubMed] [Google Scholar]
- 9.Fata, C. L., S. G. Sawicki, and D. L. Sawicki. 2002. Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant. J. Virol. 76:8641-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin, R. M. 1966. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc. Natl. Acad. Sci. USA 55:1504-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn, Y. S., A. Grakoui, C. M. Rice, E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 63:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 63:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 14.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 16.Kinney, R. M., B. J. Johnson, J. B. Welch, K. R. Tsuchiya, and D. W. Trent. 1989. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology 170:19-30. [DOI] [PubMed] [Google Scholar]
- 17.Kowal, K. J., and V. Stollar. 1981. Temperature-sensitive host-dependent mutants of Sindbis virus. Virology 114:140-148. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn, R. J., Z. Hong, and J. H. Strauss. 1990. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J. Virol. 64:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Lee, E., C. Stocks, P. Lobigs, A. Hislop, J. Straub, I. Marshall, R. Weir, and L. Dalgarno. 1997. Nucleotide sequence of the Barmah Forest virus genome. Virology 227:509-514. [DOI] [PubMed] [Google Scholar]
- 21.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 26.Levinson, R. S., J. H. Strauss, and E. G. Strauss. 1990. Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees. Virology 175:110-123. [DOI] [PubMed] [Google Scholar]
- 27.Lin, Y. H., P. Yadav, R. Ravatn, and V. Stollar. 2000. A mutant of Sindbis virus that is resistant to pyrazofurin encodes an altered RNA polymerase. Virology 272:61-71. [DOI] [PubMed] [Google Scholar]
- 28.Miranda, G., D. Schuppli, I. Barrera, C. Hausherr, J. M. Sogo, and H. Weber. 1997. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 267:1089-1103. [DOI] [PubMed] [Google Scholar]
- 29.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 30.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 31.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rueckert, R. R. 1996. Picornaviridae and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 33.Rumenapf, T., E. G. Strauss, and J. H. Strauss. 1995. Aura virus is a New World representative of Sindbis-like viruses. Virology 208:621-633. [DOI] [PubMed] [Google Scholar]
- 34.Sawicki, D., D. B. Barkhimer, S. G. Sawicki, C. M. Rice, and S. Schlesinger. 1990. Temperature sensitive shut-off of alphavirus minus strand RNA synthesis maps to a nonstructural protein, nsP4. Virology 174:43-52. [DOI] [PubMed] [Google Scholar]
- 35.Sawicki, D. L., L. Kaariainen, C. Lambek, and P. J. Gomatos. 1978. Mechanism for control of synthesis of Semliki Forest virus 26S and 42s RNA. J. Virol. 25:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawicki, D. L., and S. G. Sawicki. 1994. Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes. Arch. Virol. Suppl. 9:393-405. [DOI] [PubMed] [Google Scholar]
- 37.Sawicki, D. L., and S. G. Sawicki. 1985. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology 144:20-34. [DOI] [PubMed] [Google Scholar]
- 38.Sawicki, D. L., and S. G. Sawicki. 1993. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J. Virol. 67:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawicki, S. G., and D. L. Sawicki. 1986. The effect of loss of regulation of minus-strand RNA synthesis on Sindbis virus replication. Virology 151:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuppli, D., G. Miranda, H. C. Tsui, M. E. Winkler, J. M. Sogo, and H. Weber. 1997. Altered 3′-terminal RNA structure in phage Qbeta adapted to host factor-less Escherichia coli. Proc. Natl. Acad. Sci. USA 94:10239-10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2000. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology 276:148-160. [DOI] [PubMed] [Google Scholar]
- 42.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirako, Y., and J. H. Strauss. 1998. Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase. J. Virol. 72:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steitz, T. A. 1998. A mechanism for all polymerases. Nature 391:231-232. [DOI] [PubMed] [Google Scholar]
- 45.Stollar, V., B. D. Stollar, R. Koo, K. A. Harrap, and R. W. Schlesinger. 1976. Sialic acid contents of sindbis virus from vertebrate and mosquito cells. Equivalence of biological and immunological viral properties. Virology 69:104-115. [DOI] [PubMed] [Google Scholar]
- 46.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1983. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl. Acad. Sci. USA 80:5271-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. (Erratum, 58:806.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson, M. M., P. Ansel-McKinney, F. Houser-Scott, V. Yusibov, L. S. Loesch-Fries, and L. Gehrke. 1998. Viral coat protein peptides with limited sequence homology bind similar domains of alfalfa mosaic virus and tobacco streak virus RNAs. J. Virol. 72:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takkinen, K. 1986. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 14:5667-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thole, V., M. L. Garcia, C. M. van Rossum, L. Neeleman, F. T. Brederode, H. J. Linthorst, and J. F. Bol. 2001. RNAs 1 and 2 of alfalfa mosaic virus, expressed in transgenic plants, start to replicate only after infection of the plants with RNA 3. J. Gen. Virol. 82:25-28. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 68:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1991. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 65:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weaver, S. C., A. Hagenbaugh, L. A. Bellew, S. V. Netesov, V. E. Volchkov, G. J. Chang, D. K. Clarke, L. Gousset, T. W. Scott, D. W. Trent, et al. 1993. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology 197:375-390. (Erratum, 202:1083, 1994.) [DOI] [PubMed] [Google Scholar]