Abstract
Our recent study (C. L. Fata, S. G. Sawicki, and D. L. Sawicki, J. Virol. 76:8632-8640, 2002) found minus-strand synthesis to be temperature sensitive in vertebrate and invertebrate cells when the Arg183 residue of the Sindbis virus nsP4 polymerase was changed to Ser, Ala, or Lys. Here we report the results of studies identifying an interacting partner of the region of the viral polymerase containing Arg183 that suppresses the Ser183 codon mutation. Large-plaque revertants were observed readily following growth of the nsP4 Ser183 mutant at 40°C. Fifteen revertants were characterized, and all had a mutation in the Asn374 codon of nsP1 that changed it to either a His or an Ile codon. When combined with nsP4 Ser183, substitution of either His374 or Ile374 for Asn374 restored wild-type growth in chicken embryo fibroblast (CEF) cells at 40°C. In Aedes albopictus cells at 34.5°C, neither nsP1 substitution suppressed the nsP4 Ser183 defect in minus-strand synthesis. This argued that the nsP4 Arg183 residue itself is needed for minus-strand replicase assembly or function in the mosquito environment. The nsP1 His374 suppressor when combined with the wild-type nsP4 gave greater than wild-type levels of viral RNA synthesis in CEF cells at 40°C (∼140%) and in Aedes cells at 34.5°C (200%). Virus producing nsP1 His374 and wild-type nsP4 Arg183 made more minus strands during the early period of infection and before minus-strand synthesis ceased at about 4 h postinfection. Shirako et al. (Y. Shirako, E. G. Strauss, and J. H. Strauss, Virology 276:148-160, 2000) identified amino acid substitutions in nsP1 and nsP4 that suppressed mutations that changed the N-terminal Tyr of nsP4. The nsP4 N-terminal mutants were defective also in minus-strand synthesis. Our study implicates an interaction between another conserved nsP1 region and an internal region, predicted to be in the finger domain, of nsP4 for the formation or activity of the minus-strand polymerase. Finally, the observation that a single point mutation in nsP1 results in minus-strand synthesis at greater than wild-type levels supports the concept that the wild-type nsP sequences are evolutionary compromises.
Sindbis virus (SIN) is the type member of the Alphavirus genus of the Togaviridae family of viruses (reviewed in references 33 and 45). The 11,703-nucleotide (nt)-long SIN genome is of plus-sense polarity and possesses a 5′ methyl cap and a 3′ poly(A) tail. The four alphavirus nonstructural proteins (nsPs) are numbered according to their gene order and are translated from the 5′ two-thirds of the genome as polyprotein P123 or P1234. The polyproteins undergo a stepwise series of cleavages that in turn regulate their pattern of RNA synthetic activities (23, 25-27, 29, 37, 42, 47). As initially shown for SIN, intermediate polyproteins and then the cleaved, mature nsPs, together with host factors, are used to assemble viral replicases synthesizing genome-length minus and plus strands and a viral transcriptase synthesizing the 26S subgenomic mRNA that codes for the structural proteins. Only the minus-strand replicase activity is short lived and dependent on the continued translation of P1234 polyproteins to enable minus strands to be produced over the 4- to 5-h early part of the infectious cycle. After this time, minus-strand synthesis usually is not observed, at least in infected vertebrate cells.
In the N-terminal third of the nsP4 sequence is a highly conserved stretch of amino acids. Substitution of amino acids in this region produces mutants that show defects in critical virus-host interactions (24) or in polymerase elongation (3, 13) and, as described in the accompanying paper (9), in minus-strand synthesis. In the latter study (9), substitution of Ala, Ser, or Lys for the highly conserved Arg183 within the 5-amino-acid sequence Pro180-Asn-Ile-Arg-Ser in SIN nsP4 produced viruses that had temperature-sensitive (ts) minus-strand synthesis. Although the mutants were viable, their capacity to produce minus strands, and in turn plus strands and virus, at 40°C varied in chicken embryo fibroblast (CEF) cells depending on which amino acid replaced Arg183. Substitution of Arg183 with Lys caused only 40 to 50% of the wild-type level of minus-strand synthesis, indicating that a positive charge was important at this position. In Aedes albopictus cells, however, these same mutants were equally defective at 34.5°C, which indicated that minus-strand synthesis depended on the environment of the host cell. Similar results for nsP4 Pro187 mutant viruses were found by Lemm et al. (24), who concluded that it was the interaction with host cell factors and not the catalytic activity of the polymerase that was altered. Recently, regions of the SIN polymerase have been implicated in minus-strand synthesis: the absolutely conserved N-terminal Tyr residue (41) and residues Gln191 and Glu315, whose substitution formed suppressors able to rescue N-terminal nsP4 mutant polymerases (41). The substitution of Leu for Gln349 in nsP1 also formed a suppressor of the N-terminal nsP4 mutants. This amino acid in nsP1 is adjacent to the causal mutation in SIN ts11, a ts RNA-negative mutant specifically ts for minus-strand synthesis (14, 48).
We used the Ser (UCA)-183 virus to select second-site suppressors of the nsP4 Arg183 mutants, as it had the most severe ts defects in RNA synthesis. It also would require changes in two (CGA) or all three codon positions to restore an Arg codon (CGC, CGU, or CGG), favoring single compensatory changes if any could occur elsewhere in the genome. Large-plaque revertants were readily observed following growth at 40°C, and a total of 15 were characterized from six independent sets of 40°C passages.
MATERIALS AND METHODS
Cells, virus, and plasmids.
CEF cells, prepared from 10-day-old embryos of leukosis-free (specific-pathogen-free-COFAL/Marek-negative) flocks (Spafas, Roanoke, Ill.), were grown as described in the accompanying paper (9). The A. albopictus C7-10 cell line was kindly provided by V. Stollar and adapted to grow in Dulbecco's modified Eagle's medium supplemented with fetal calf serum and tryptose phosphate broth at 30°C.
The infectious clone of SIN HR, pToto1101, was constructed by Rice and coworkers (34) and was a generous gift from C. Rice (Washington University, St. Louis, Mo.). The construction of nsP4 Ser183 virus and its phenotype are described in the accompanying paper (9).
Isolation of Ser183 second-site revertants.
The stock of nsP4 Ser mutant virus used for the experiments described in this paper had a titer of 1010 PFU/ml at 30°C and an efficiency of plaquing (EOP; the titer at 40°C divided by the titer at 30°C) of 0.03 and produced small plaques (<1 mm) at 40°C. Three methods were used to isolate large-plaque revertants from a single stock of the Ser183 mutant. CEF cells were infected at 30°C with a multiplicity of infection (MOI) of 0.1 PFU/cell, and 1 h later the virus inoculum was removed, growth medium prewarmed to 40°C was added, and the infected cultures were incubated at 40°C for 2 days (40°C P-1). The second method was performed similarly to the first except that the nsP4 Ser183 virus stock was diluted 10−5 and four dishes were infected. The nsP4 Ser183 virus gave only small plaques at 40°C when dilutions of 10−5 or greater were plaque assayed directly (without growth at 40°C). Each dish was harvested separately, and virus from each was treated as a separate clonal pool (CP). The 40°C P-1 stock was used to infect another culture of cells in the same manner (40°C P-2). The 40°C P-1 and P-2 stocks were screened for large-plaque viruses at 40°C by plaque assay on CEF cells. Large plaques were picked, and the virus was twice plaque purified on CEF cells at 40°C. RNA was obtained following phenol-chloroform extraction and ethanol precipitation either from the virus in one plaque or from the virus in a stock made from the plaque-purified virus. For the third method, virus from five well-separated Ser183 virus plaques formed at 30°C (∼106 PFU/plaque) was used to infect CEF cells and the virus was grown at 40°C for 2 days. These stocks of virus were assayed at 40°C for the formation of large plaques. Large-plaque revertant viruses were isolated from well-separated plaques ∼5 mm in diameter. They were twice plaque purified before the viral RNA was sequenced. Statistical analysis using the Kruskal-Wallis test (44), a nonparametric method for single classification analysis of variance, gave a maximum value for H of 2.789, indicating that the reversion rate among the three groups (inocula prepared from cells infected with Ser183 mutant stock at an MOI of 0.1, the 10−5 dilution, or five individual 30°C plaques) was not significantly different. This argues against a contaminant being present in the mutant stock.
For selection of revertant viruses arising in Aedes cells, cultures of C7-10 cells were infected with nsP4 Ser183 virus (MOI of 50) at 34.5°C and maintained for 2 days at 34.5°C. Plaque assays were performed at 40°C on CEF monolayers, and virus from large plaques was harvested and plaque purified a second time on CEF cells at 40°C. SIN did not produce discrete plaque on these C7-10 cell monolayers. Small stocks of some of the twice-plaque-purified viruses were made in CEF cells at 40°C, and these gave 1- to 2-mm plaques at 40°C. Ten of 15 isolates had EOPs greater than 0.2 (ranging from 0.21 to 0.59); the other 5 isolates had EOP values of 0.04 to 0.12. Four isolates from the first plaque purification were sequenced.
The RNA encoding the nsP4 Ser183 region was copied into cDNA by reverse transcription and amplified by PCR with the Access reverse transcription-PCR (RT-PCR) system according to the instructions of the manufacturer (Promega Corporation). The cDNA was sequenced with Thermo Sequenase radiolabeled terminator cycle sequencing reagents (United States Biochemical Corporation).
Identification of compensatory second-site mutations.
The method that we used to map causal mutations with the infectious clone Toto1101 has been described previously (6). Briefly, revertant genome RNA was purified, copied into cDNA, amplified by RT-PCR with specific primers, and digested with SstI (nt 1) and Eco47III (nt 1401), which correspond to ∼90% of the nsP1 gene. This region was swapped into a cDNA copy of the Ser183 genome that had been cut with the same restriction enzymes and purified of the corresponding wild-type SstI-Eco47III fragment, by incubating both fragments with T4 DNA ligase (Gibco Technologies). Ligation products were transformed into competent MC1061 cells, colonies were screened for plasmid size, and the Toto1101-sized plasmid DNA was isolated with a QIAprep Miniprep kit (Qiagen). The mutant cDNAs were linearized at a unique XhoI restriction site immediately downstream of the viral sequence and transcribed with MEGAscript SP6 polymerase (Ambion), with the addition of 1.6 mM m7G(5′)ppp(5′)G cap. The transcription products were diluted in phosphate-buffered saline containing 50 μg of DEAE-dextran/ml and used to transfect CEF cells. Following transfection, the cultures were incubated at 30°C. Cytopathic effect was seen usually within 2 to 3 days, at which time the medium was harvested. Virus was screened for plaque size at 40°C, and the entire SstI-to-Eco47III region of all large-plaque isolates was sequenced.
Recombinants that contained the nsP1 Asn374-to-His or -Ile substitution (abbreviated as Asn374→His or -Ile) together with the wild-type nsP4 Arg183 gene were made by swapping the SstI-to-Eco47III region of the revertant genome into the wild-type Toto1101 background, as described above.
Infection and RNA labeling.
In all experiments CEF or C7-10 A. albopictus monolayers were infected with virus at an MOI of 100 and 300, respectively, as previously described (38). CEF cultures were shifted to 40°C by first rinsing them with 40°C medium, refeeding them with 40°C medium, and incubating the cultures at 40°C. A. albopictus C7-10 cells were shifted to 34.5°C in a similar manner. Cells were pulse-labeled with [3H]uridine (50 μCi/ml unless otherwise indicated) for 1 h in Dulbecco's modified Eagle's medium containing 20 μg of actinomycin D/ml, 6% fetal calf serum, and 20 mM HEPES, pH 7.4. At the end of the labeling period, the cells were washed with ice-cold phosphate-buffered saline and lysed with 5% lithium dodecyl sulfate in LET buffer (0.1 M LiCl, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4) containing 200 μg of proteinase K/ml. DNA in the cell lysates was sheared by passage through a 27-gauge needle before the acid-insoluble incorporation was determined.
Isolation of replicative-form (RF) RNA and quantitation of minus strands.
Minus-strand synthesis was determined as described in references 10 and 39 and in the accompanying paper (9). For accumulation experiments, all infected cultures were incubated in the presence of 200 μCi of [3H]uridine/ml plus 2 μg of actinomycin D/ml beginning immediately after adsorption.
RESULTS
Isolation of Ser183 second-site revertants.
The nsP4 Ser183 virus gave an EOP of 0.03 and formed only small plaques (<1 mm) at 40°C compared to the wild-type Toto1101 virus, which gave an EOP of 0.9 and formed plaques of 5 mm in diameter at 40°C (9). This original stock of mutant virus was used for all studies. No revertants producing large, wild-type-sized plaques (5 mm) were detected at dilutions of 10−5 to 10−10. Because the Ser183 virus is viable at 40°C, dilutions of 10−4 or lower produced extensive cell killing that prevented visualization of individual plaques. To isolate revertants of the Ser183 mutant virus, and particularly pseudorevertants, i.e., viruses that possessed compensatory changes elsewhere in the genome other than in codon 183 of nsP4, we used two 40°C selection strategies (Table 1). The first involved growing the Ser183 mutant at 40°C at an MOI of 0.1, and the second used a 10−5 dilution of the Ser183 stock. The infected CEF cells were maintained at 40°C for 2 days (P1-40°C), and then 1 μl (MOI of ∼0.1) of the resultant P1-40°C stock was used to infect a new CEF culture that was maintained at 40°C for 2 days (P2-40°C). Large-plaque variants arose readily at 40°C, as can be seen from the frequency of large plaques in Table 1. Progeny from the first 40°C passage were ∼50% or greater large-plaque viruses; progeny from the second 40°C passage were essentially all large-plaque viruses. Large-plaque revertants were picked, and each revertant was plaque purified twice before its genome RNA was sequenced through the region encoding residue 183 of nsP4. All revertants maintained the Ser183 UCA codon. Therefore, no true revertants or viruses possessing single-nucleotide changes in codon 183 were observed.
TABLE 1.
Isolation and characterization of nsP4 Ser183 large-plaque mutantsa
| Selection method | Frequency of lp (%) | Plaque size (30°C/40°C) | Location of second-site mutation
|
|
|---|---|---|---|---|
| Protein (no. analyzed) | Amino acid | |||
| Passage at 40°C | ||||
| Expt 1 | ||||
| CP1 40°C-P1 | 64 | lp/lp | nsP1 (1) | Asn374→His |
| CP2 40°C-P2 | 88 | lp/lp | nsP1 (1) | Asn374→Ile |
| Expt 2 | ||||
| CP3 40°C-P1 | 70 | lp/lp | nsP1 (1) | Asn374→His |
| CP4 40°C-P1 | 66 | lp/lp | nsP1 (1) | Asn374→His |
| CP5 40°C-P1 | 54 | lp/lp | nsP1 (1) | Asn374→His |
| CP6 40°C-P1 | 54 | lp/lp | nsP1 (1) | Asn374→His |
| Passage of 10−5 dilution at 40°C | ||||
| 10−5 dilution P0 | 0 (10/10, sp) | lp/sp | ||
| CP7 40°C-P1 | 68 | lp/lp | nsP1 (3) | Asn374→Ile |
| CP8 40°C-P1 | 83 | lp/lp | nsP1 (3) | Asn374→Ile |
| CP9 40°C-P1 | 92 | lp/lp | nsP1 (3) | Asn374→Ile |
| CP10 40°C-P1 | 0 | lp/sp | ||
| 30°C plaques passaged at 40°C | ||||
| CP11 40°C-P1 | 19 | lp/lp | NDb | ND |
| CP12 40°C-P1 | 44 | lp/lp | ND | ND |
| CP13 40°C-P1 | 28 | lp/lp | ND | ND |
| CP14 40°C-P1 | 15 | lp/lp | ND | ND |
| CP15 40°C-P1 | 74 | lp/lp | ND | ND |
lp is a large plaque of 5-mm diameter; sp is a small plaque of 1-mm diameter.
ND, not done.
Infection of CEF cells with a 105-fold dilution of the Ser183 mutant was done to determine if growth at high temperature provided an essential selective pressure. One milliliter each of the 10−5 dilution was used to infect four CEF cultures (CP7 to CP10) that were grown at 40°C for 2 days. Progeny from CP7, CP8, and CP9 but not CP10 produced large plaques after one passage at 40°C (Table 1). When the 10−5 dilution of the Ser183 mutant virus stock was assayed directly at 40°C for the presence of large plaques, 10 of 10 replicate petri plates of CEF cells revealed only small plaques. Because the 10 dishes with each dish being infected with 1.2 × 104 PFU had no large plaques, the reversion frequency was lower than 10−5.
We also analyzed virus derived from five well-separated Ser183 virus plaques isolated at 30°C and used to infect CEF cells which were grown for 2 days at 40°C (CP11, CP12, CP13, CP14, and CP15). Virus from each 30°C plaque retained the nsP4 Ser183 small-plaque, low-EOP phenotype (data not shown). After growth at 40°C, large-plaque variants arose, and 15 to 74% of the 40°C plaques were large, which indicated that growth at 40°C selected relatively quickly for the emergence of revertants. The results support the idea that growth at 40°C was needed for the selection of the pseudorevertants and that the pseudorevertants arose independently.
All of the large-plaque revertants retained the sequence coding for Ser at codon 183 of nsP4. The 3′ nontranslated regions (NTRs) of two revertant genomes were analyzed, and both had sequences identical to Toto1101 (data not shown), leading to the conclusion that the suppressor was located within the coding region of the genome or possibly within the 5′ NTR. It proved more difficult to isolate revertants of the nsP4 Ser183 virus from Aedes cells infected at 34.5°C because of the need to assay plaque size on CEF cells. It was apparent that large-plaque variants arose during growth at 34.5°C in C7-10 cells but that these accumulated additional mutations and exhibited a small-plaque phenotype when plaque purified in CEF cells more than once (data not shown).
Identification of nsP4 Ser183 compensatory changes.
The strategy used to locate the suppressor in the first two CEF cell-selected revertants is outlined in Fig. 1. Briefly, the causal mutations were mapped by amplifying specific regions of the viral genome by RT-PCR and digesting the resulting cDNAs with appropriate restriction enzymes. Then, the mutant-derived fragments were substituted for the corresponding region of an infectious Ser183 mutant cDNA. After linearization of the infectious cDNA, RNA transcripts were made and transfected into CEF cells, and the resulting progeny was screened for plaque size at 40°C in CEF cells. Recombinant cDNAs containing a revertant SstI-to-Eco47III fragment, which corresponds to nt 1 to nt 1401 of the nsP1 gene, produced large plaques at 40°C. When sequenced, this region contained single, unique changes. One revertant possessed an A-to-C change at nt 1179, predicting a change of Asn to His at amino acid 374 of nsP1; the second revertant had an A-to-T change at nt 1180, predicting a change of nsP1 Asn374 to Ile (Table 1). Given these results and as an initial screening approach, the entire SstI-to-Eco47III regions of the genome RNA of 13 other CEF cell-selected, large-plaque isolates were sequenced directly, without cloning. Four 40°C P-1 isolates had the nt 1179 A-to-C substitution that would change nsP1 Asn374 to His (Asn374→His), while nine 40°C P-1 isolates shared the nt 1180 A-to-T change, altering nsP1 Asn374 to Ile (Asn374→Ile). No additional mutations were identified in this region, and all revertants possessed a change within the SstI-to-Eco47III region.
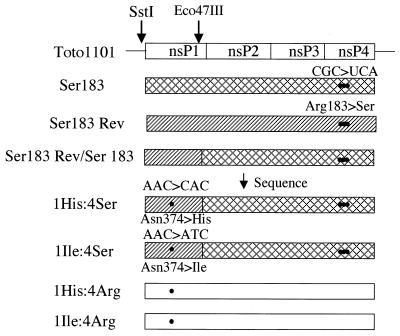
FIG. 1.
Schematic representation of the strategy used to construct hybrid genomes. cDNA copies of the SstI-to-Eco47III region of revertant genomes (hatched boxes) were exchanged for the corresponding fragment in the nsP4 Ser183 (crosshatched boxes) or the Toto1101 (open boxes) genomic backgrounds.
Following selection in Aedes C7-10 cells at 34.5°C and plaque purification on CEF cells at 40°C, four of four large-plaque isolates were found that possessed the Asn374→His change (data not shown), which was identical to one of the suppressors selected at 40°C in CEF cells. It is possible that novel suppressors may have arisen in C7-10 cells but that growth in the vertebrate host led to their being selected against in favor of suppressors that were more efficient in CEF cells.
Recombinants whose genomes encoded the nsP1 Asn374→His or →Ile change together with wild-type nsP2, nsP3, and nsP4 (Arg183) proteins also were constructed (Fig. 1). To simply the nomenclature, recombinant viruses will be identified by the nature of the nsP1 and nsP4 proteins (e.g., the nsP1 Asn374→His plus nsP4 Arg183→Ser double mutant [pseudorevertant] virus is referred to as 1His:4Ser).
Plaque phenotype on CEF cells.
Mutant, pseudorevertant, and nsP1 recombinant plaque sizes and titers were compared with Toto1101, which has an EOP closer to 1 and produces 5-mm plaques at 40°C. Viruses 1His:4Ser, 1His:4Arg, 1Ile:4Ser, and 1Ile:4Arg produced wild-type-sized plaques and wild-type titers at 30°C (Table 2). At 40°C, they gave 5-mm plaques after 2 days and EOP values ranging from 0.54 to 1.2. Interestingly, the 1His:4Arg virus consistently gave an EOP greater than 1.0. Thus, a change of Asn374 of nsP1 to either Ile or His compensated for the defects of the nsP4 Ser183 mutant that cause overall growth and plaque formation in CEF cells at 40°C.
TABLE 2.
Plaque phenotype of recombinant viruses on CEF cells
| Virus | 30°C titer (PFU/ml) | EOPa (PFU/ml at 40°C/PFU/ml at 30°C) | Diam of 40°C plaques (mm) |
|---|---|---|---|
| Toto1101 | 6.6 × 109 | 0.90 | 5 |
| Ser183 | 1.0 × 1010 | 0.03 | 1 |
| 1His:4Ser | 3.4 × 109 | 0.66 | 5 |
| 1His:4Arg | 4.2 × 109 | 1.3 | 5 |
| 1Ile:4Ser | 1.5 × 109 | 0.54 | 5 |
| 1Ile:4Arg | 1.5 × 109 | 0.69 | 5 |
EOP value represents the averages of two independent plaque assays.
RNA phenotype in CEF cells.
Viral RNA synthesis by the pseudorevertants and recombinants was analyzed in vertebrate cells. CEF cultures were labeled with [3H]uridine in the presence of actinomycin D for 1-h periods at 30 or 40°C in cultures shifted up to 40°C at 1 h postinfection (p.i.). The nsP4 Ser183 mutant and its pseudorevertants had the same maximum viral transcription rates at 30°C, which were about half the rates observed for Toto1101 (Table 3). At 40°C, both pseudorevertants synthesized RNA at 10- to14-fold-greater levels than the nsP4 Ser183 mutant, confirming that the suppressive activity resided within nsP1 and functioned in vertebrate cells. But their higher transcription levels at 40°C were only half that observed for Toto1101 virus with the nsP4 Arg183 polymerase. The pseudorevertants also retained the enhanced synthesis of 26S mRNA relative to genome RNA at 40°C of the nsP4 Ser183 mutant (Table 4). The nsP1 recombinant viruses, 1His:4Arg and 1Ile:4Arg, achieved the same levels of viral transcription as Toto1101 at 30°C. It is particular noteworthy that the 1His:4Arg virus synthesized RNA at about 25 to 40% greater rates than wild-type virus at 40°C, and it was only this virus that consistently gave an EOP value greater than 1.0 (Table 3).
TABLE 3.
Relative efficiency of viral RNA synthesis in CEF cells at 30 and 40°C
| Virus | 30°Ca | 40°Ca | 40°C/30°Cb |
|---|---|---|---|
| Toto1101 | 1.00 | 1.00 | 1.00 |
| Ser183 | 0.65 | 0.04 | 0.06 |
| 1His:4Ser | 0.52 | 0.57 | 1.10 |
| 1His:4Arg | 0.93 | 1.39 | 1.49 |
| 1Ile:4Ser | 0.53 | 0.40 | 0.75 |
| 1Ile:4Arg | 0.89 | 1.16 | 1.30 |
CEF cells were infected at 30°C at an MOI of 100 and maintained at 30°C or shifted to 40°C 1 h p.i. Cultures were labeled for 1-h pulses with 50 μCi of [3H]uridine/ml in the presence of 20 μg of actinomycin D/ml between 6 and 9 h p.i., a period in the replication cycle when the rate of viral RNA synthesis was maximal. The average, acid-precipitable incorporation for the three time periods is expressed relative to that for parental Toto1101 virus.
The relative level of RNA synthesis at 40°C was divided by the relative level of RNA synthesis at 30°C.
TABLE 4.
Relative 49S and 26S mRNA synthesis by nsP Ser183 pseudorevertants and the nsP1 recombinantsa
| Virus | Molar ratio of 49S/26S RNA
|
40°C/30°C ratio | |
|---|---|---|---|
| 30°C | 40°C | ||
| Toto1101 | 0.11 | 0.43 | 3.9 |
| 1His:4Ser | 0.09 | 0.23 | 2.6 |
| 1Ile:4Ser | 0.07 | 0.23 | 3.3 |
| 1His:4Arg | 0.09 | 0.40 | 4.4 |
| 1Ile:4Arg | 0.08 | 0.39 | 4.9 |
CEF monolayers were infected at an MOI of 100 and maintained at 30°C or shifted to 40°C at 1 h p.i. Cultures were labeled with [3H]uridine (50 μCi/ml) for 1-h pulses between 6 and 9 h p.i. The RNA was obtained and analyzed by electrophoresis on 0.8% agarose gels. The incorporation in each species was expressed as moles by dividing by the difference in molecular weights of the 49S and 26S RNAs. The molar ratios shown are the average values of the three 1-h samples for each virus.
Enhanced RNA synthesis by nsP1-374 variants at 40°C.
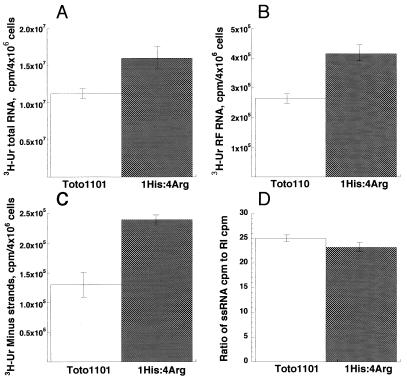
To probe whether greater-than-wild-type rates of transcription at 40°C by the 1His:4Arg virus were due to the synthesis of more plus strands from each template or to the production of more minus-strand templates, CEF cultures were infected in triplicate with each virus and labeled continuously with [3H]uridine from 1 to 8 h p.i. The accumulated incorporation in viral plus strands (as total RNA), in purified cores of the replicative intermediates (RIs), and in viral minus strands was determined (Fig. 2). Compared to the average values obtained for similar triplicate CEF cultures infected with Toto1101, the 1His:4Arg-infected cultures accumulated ∼40% more total RNA (Fig. 3A) and ∼40% more RF RNA (Fig. 2B) and minus-strand RNA (Fig. 2C). This is precisely the pattern expected if greater numbers of minus strands were being produced and used as templates.
FIG. 2.
RNA accumulation at 40°C by Toto1101 and 1His:4Arg at 40°C. Triplicate CEF cultures were infected with Toto1101 (open boxes) or 1His:4Arg (shaded boxes) at an MOI of 100 at 30°C and were shifted to 40°C at 1 h p.i. Cultures were labeled continuously with 100 μCi of [3H]uridine/ml in the presence of 20 μg of actinomycin D/ml from 1 to 8 h p.i., when the cells were harvested. (A) Total RNA. Aliquots of each total cell lysate were analyzed for acid-insoluble incorporation. (B) RF RNA. RF RNA cores of the viral RIs were isolated as described in Materials and Methods, and their incorporation of radiolabel was determined. (C) Minus-strand RNA. The amount of radiolabeled RF RNA that was in minus strands was determined in nuclease protection assays. (D) Synthesis of plus-strand 49S and 26S RNA relative to RI RNA. The migration positions on agarose gels of RI RNA and 49S and 26S plus-strand RNA were visualized by electrophoresis and autoradiography of total cell lysates as described in Materials and Methods. After the gel areas containing each species were cut and counted, the ratio of the incorporation in 49S plus 26S plus-strand RNA to the incorporation in RI RNA was calculated and is shown.
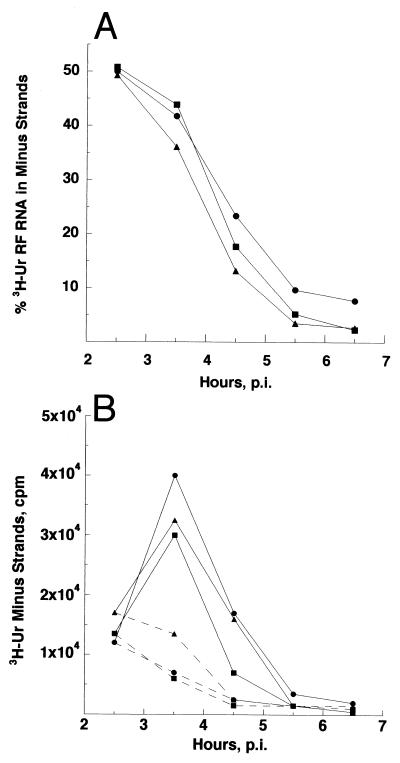
FIG. 3.
Kinetics of Toto1101 and 1His:4Arg minus-strand syntheses at 40°C. (A) CEF cultures were infected with Toto1101 (•), 1Ile:4Arg (▴), or 1His:4Arg (▪) at an MOI of 100 at 30°C and were shifted to 40°C at 1 h p.i. Cultures were labeled for 1-h pulses from 1 to 7 h p.i. with 200 μCi of [3H]uridine/ml in medium containing 20 μg of actinomycin D/ml. Cells were harvested at the end of the pulse period. Viral RF RNA was isolated, and the percentage of radiolabeled RF RNA in minus strands was determined as described in Materials and Methods. (B) Cycloheximide treatment prevents minus-strand synthesis by nsP1 recombinant viruses. Cultures infected as described for panel A were incubated in the absence of cycloheximide (solid lines) or with 100 μg of cycloheximide/ml (dashed lines) beginning at 2.5 h p.i. and during the labeling period. Incorporation in the minus-strand RNA component of the viral RI and native RF was determined as described in Materials and Methods.
To measure the relative amounts of plus strands made from each minus-strand template, the single-stranded 49S and 26S species and the viral RIs were electrophoretically separated. The individual bands were cut out of the gel, and the radioactivity in each was determined. As seen in Fig. 2D, the accumulated incorporation in 49S plus 26S RNA relative to that in the RIs was similar for both 1His:4Arg virus and Toto1101. The same relative amounts of 49S and 26S plus-strand RNA were produced by the two viruses (Table 4). Thus, while more minus strands were made, the activities of the 1His:4Arg plus-strand replicase and transcriptase were not altered detectably.
Increased numbers of minus strands could have arisen because minus strands were synthesized continuously, i.e., cessation did not occur at the end of the early phase. To determine whether the normal kinetics of minus-strand synthesis and cessation occurred, 1His:4Arg- or 1Ile:4Arg-infected CEF cells were shifted to 40°C at 1 h p.i. and radiolabeled at 40°C for 1-h periods until 7 h p.i. As shown in Fig. 3, transcription rates during the first 3 h p.i. increased rapidly and ∼50% of the total labeled RI and RF core RNA was in minus strands, as expected when 90% or more of the intermediates are newly formed each hour. Beginning at about 4 h p.i., minus-strand synthesis in both 1His:4Arg- and Toto1101-infected cells began to decline, essentially ceasing after 7 h p.i. Addition of cycloheximide to the culture medium at 2 h p.i. stopped minus-strand synthesis (Fig. 3B), indicating that the 1His:4Arg and 1Ile:4Arg minus-strand replicase activities turned over with time as did that of the wild-type virus (40). Thus, the increased numbers of minus strands arose from enhanced activity or efficiency of the 1His:4Arg minus-strand replicase and not from a change in its longevity (37).
Host restriction in C7-10 cells is not rescued.
The nsP1 pseudorevertants selected in vertebrate cells did not compensate for the ts PFU and RNA synthesis phenotypes of the nsP4 Ser183 mutant in C7-10 cells (Table 5). At 30°C, all viruses caused the release of full yields of virus (108 PFU/ml). At 34.5°C, the nsP4 Ser183 mutant and pseudorevertants 1His:4Ser and 1Ile:4Ser released 15- to 50-fold fewer PFU than Toto1101 and 1His:4Arg and 1Ile:4Arg recombinant viruses. Thus, the presence of nsP1 His or Ile374 did not restore wild-type growth in C7-10 cells to nsP4 Ser183 viruses.
TABLE 5.
Virion release by C7-10-infected cells at 30 and 34.5°Ca
| Virus | 30°C titer (PFU/ml) | 34.5°C titer (PFU/ml) | 34.5°C titer/ 30°C titer |
|---|---|---|---|
| Toto1101 | 9.4 × 108 | 6.3 × 108 | 0.67 |
| Ser183 | 7.4 × 108 | 1.1 × 107 | 0.01 |
| 1His:4Ser | 6.0 × 108 | 1.2 × 107 | 0.02 |
| 1His:4Arg | 9.3 × 108 | 6.7 × 108 | 0.72 |
| 1Ile:4Ser | 1.4 × 108 | 3.5 × 107 | 0.25 |
| 1Ile:4Arg | 4.2 × 108 | 2.3 × 108 | 0.55 |
C7-10 cells were infected at 30°C at an MOI of 300 and maintained at 30°C or shifted to 34.5°C after a 1-h adsorption period. At 22 h p.i., the medium was harvested and assayed for virus titer on CEF cells at 30°C.
Nor did it restore full viral RNA synthesis in C7-10 cells (Table 6). At 30°C, the same approximately twofold-lower level of RNA synthesis compared to Toto1101 was observed for the nsP4 Ser183 mutant and its pseudorevertants. While lower, the level of viral RNA synthesis was sufficient to produce full yields of PFU. Of the two recombinant viruses, only in 1His:4Arg virus-infected cells was viral RNA synthesized as efficiently as in Toto1101-infected cells at 30°C. At 34.5°C, neither pseudorevertants nor the 1Ile:4Arg virus synthesized RNA as efficiently, indicating that the ability to rescue the nsP4 Ser183 defective function was host restricted. Only nsP4 proteins containing Arg at residue 183 functioned fully in mosquito cells when combined with nsP1 proteins containing Asn or His at position 374. In mosquito cells, the presence of Ile at position 374 of nsP1 was inhibitory at either temperature. Enhanced RNA synthesis by 1His:4Arg virus was also seen in C7-10 cells at 34.5°C, where levels were 200% that of Toto1101-infected cells. Thus, transcription enhancement occurred in both vertebrate and invertebrate cell environments with 1His:4Arg virus and was observed at elevated temperature for a particular host cell (i.e., 40°C for CEF cells and 34.5°C for C7-10 cells).
TABLE 6.
Viral RNA synthesis in C7-10 cells at 30 and 34.5°C
| Virus | 30°Ca | 34.5°Ca | 34.5°C/30°Cb |
|---|---|---|---|
| Toto1101 | 1.00 | 1.00 | 1.00 |
| Ser183 | 0.65 | MLc | ML |
| 1His:4Ser | 0.48 | 0.16 | 0.33 |
| 1His:4Arg | 0.88 | 2.0 | 2.3 |
| 1Ile:4Ser | 0.31 | ML | ML |
| 1Ile:4Arg | 0.49 | 0.3 | 0.61 |
C7-10 cells were infected at 30°C at an MOI of 300 and maintained at 30°C or shifted to 34.5°C at 1 h p.i. Cultures were labeled for 1-h pulses with 50 μCi of [3H]uridine/ml in the presence of 20 μg of actinomycin D/ml between 17 and 20 h p.i., a time after infection when RNA synthesis was at a constant, maximal rate. The average, acid-insoluble incorporation for the three time periods was obtained and expressed relative to the incorporation by cells infected with parental Toto1101.
The efficiency of RNA synthesis relative to Toto1101 at 34.5°C was divided by the efficiency of RNA synthesis relative to Toto1101 at 30°C.
ML, incorporation was at mock-infected cell levels.
DISCUSSION
Minus-strand synthesis is the first transcription event that initiates plus-strand virus replication. With alphaviruses, minus-strand synthesis requires a newly formed replicase whose activity apparently requires its association with membranes; polyprotein processing to free the nsP4 core polymerase; recognition of the genome as a template; and perhaps its recruitment from the translation machinery, assembly of the polymerase complex and host cell cofactors on the template, and completion of the first phosphodiester bond of the nascent 5′ poly(U) sequence. In this process, interactions likely form between different domains of the nsP4 protein as well as between nsP4 and other nsPs, essential host cofactors, and the viral template RNA. Our accompanying study (9) identified amino acid 183 of nsP4 as important in the efficient synthesis of minus strands. This amino acid is likely to be outside the polymerase palm domain and perhaps part of the finger domain region (31). We report here the first evidence for an interaction between the region containing amino acid 183 of nsP4 and a region of nsP1 including amino acid 374. Together, the results emphasize the importance of interactions of nsP4 with nsP1, especially in the minus-strand synthesis that involves nsP1 as part of the uncleaved P123 intermediate. These interactions affected only plus-strand synthesis because they caused an increase or decrease in the numbers of minus-strand templates made. Substituting His or Ile in place of the wild-type Asn374 in nsP1 suppressed the nsP4 Ser183 mutant in vertebrate (CEF) cells but not in invertebrate (Aedes) cells. In CEF cells, the pseudorevertants containing His- or Ile374 in nsP1 together with Ser183 in nsP4 restored minus-strand synthesis at 40°C to wild-type levels. In replication complexes made of wild-type nsP4 proteins, the presence of modified nsP1 His374 caused the production of 140% of wild-type numbers of minus strands at 40°C in CEF cells. Because plus-strand synthesis at 34.5°C in Aedes cells was 200% of that caused by wild-type virus, we think that a similar enhancement of minus-strand synthesis would occur in the mosquito environment. The low level of overall transcription in mosquito cells hindered the direct determination of minus strands. We favor the interpretation that such modified nsP1 proteins enhance the efficiency of initiation of minus-strand synthesis.
We do not know if these domains in nsP4 and nsP1 interact physically or through another partner(s). Both the nsP4 Ser183 ts defect and its suppression by the nsP1 His- or Ile374 protein were host-dependent processes. Therefore, a host factor(s) is likely to play some role in nsP1-nsP4 interactions that are required for minus-strand replicase activity. Only those viruses producing nsP4 containing Arg183 replicated efficiently in invertebrate cells at 34.5°C, and thus, the presence of an Arg residue at this position would appear to be a requirement for Aedes host factor interactions during the formation of the minus-strand replicase. Interactions of host factors that appear to be essential for the formation of the minus-strand replicase showed less dependence on the presence of an Arg at position 183 of nsP4 in CEF cells (9). When the virus made nsP4 with Arg183 and nsP1 with His374, it had enhanced overall transcriptional levels in Aedes cells at 34.5°C. Based on these results, we hypothesize that the formation of the minus-strand replicase requires the interaction of the host factor(s) with nsP4 before nsP1 associates with nsP4. The host factor may function to bring nsP4 to nsP1 or act as a chaperone to fold nsP4 into a conformation that will permit its interaction with nsP1.
Our present results extend the experimental data that support a role for a region of nsP1 in minus-strand synthesis. Asn374 of nsP1 is located in the carboxy-terminal half of the protein (Fig. 4). It is in a domain conserved among alphaviruses and associated with the methyltransferase domain of alphavirus-like viruses (19); it is predicted to be outside of the catalytic region (A. Gorbalenya, personal communication). The viral capping mechanism of action differs from that of cellular capping, as the alphavirus activity first binds and methylates the GTP to form a 7-methyl-GTP moiety and then transfers it to the 5′ terminus of nascent 49S and 26S plus strands (2, 21, 30). The nsP1 protein is the N-terminal polypeptide in the two viral nsPs (P1234 and P123) initially translated from the 49S genome. Its cleavage from nascent P1234 polyproteins is thought to occur in trans and only after cleavage in cis forms P123 and nsP4 (7, 16, 17). Combinations of P123+nsP4 and nsP1+P23+nsP4 are active polymerases, and both can function as minus-strand replicases (23, 27, 43, 47). Fully cleaved nsP1+nsP2+nsP3+nsP4 complexes normally express solely plus-strand replicase and transcriptase activities. Thus, theoretically, a suppressor in nsP1 could act to restore high levels of transcription by restoring minus-strand replicase activity of Ser183 nsP4 or by increasing the synthesis of capped plus strands from each template. Our results support the former possibility.
FIG. 4.
Alphavirus nsP1 sequence comparison in the region containing SIN amino acids 335 to 397. The relative positions of the nsP1 suppressors, the causal lesion for ts11 (14), and the suppressor for nsP4 Tyr1→Ala (41) are indicated. Residues identical to the SIN sequence are denoted by asterisks. Sources for sequence data are as follows: SIN and Semliki Forest virus (SFV), 46; Whatora virus (WHA), Ellen Strauss, personal communication; Mayora virus (MAY), Ellen Strauss, personal communication; Venezuelan equine encephalitis virus (VEE), 18; eastern equine encephalitis virus (EEE), 49; Aura virus, 36; O'nyong-nyong virus (ONN), 28; Ross River virus (RR), 8; and Barmah Forest virus (BF), 22.
As seen in Fig. 4, within this same domain of nsP1 are two residues that have been implicated elsewhere in nsP4 interactions for the initiation of SIN minus-strand synthesis, Ala348 (14, 48) and Thr349 (41); the present study extends this region to amino acid 374. Of these nsP1 changes, only the Ala348-to-Thr and Thr349-to-Lys substitutions conferred ts minus-strand synthesis (14, 48). Only the substitutions at amino acids 349 and 374 of nsP1 created suppressors for mutations in the gene for the nsP4 polymerase, e.g., amino acid substitutions for the N-terminal Tyr and for Arg183, respectively, that caused ts minus-strand synthesis. The nsP1 Thr349-to-Lys proteins, as well as nsP4 Asp315-to-Gly, -Val, or -Lys proteins, were efficient suppressors of lethal amino acid substitutions for the N-terminal Tyr in nsP4. Based on such results, Shirako et al. (41) suggested that the N-terminal nsP4 suppressors were eliminating the need for an interaction that normally occurred between the N-terminal ring structure and a component of the minus-strand replicase complex. Whether this involves a new, direct interaction between the nsP1-349 region and some other part of nsP4 is not known. It is known that the role of nsP1-348 in minus-strand initiation is distinct from any functional requirements for P1234 processing and overall polymerase complex assembly (47).
Suppression by nsP1 Lys349 or nsP4 Gly315 of N-terminal nsP4 mutants occurred essentially only at 30°C; at 40°C, minus-strand synthesis was delayed and reduced (41). This was in contrast to findings for the Ser183 nsP4 defect (9), whose suppression by nsP1 His374 or Ile374 occurred readily at 40°C in CEF cells. Both nsP1 His374 and Ile374 pseudorevertants accumulated with about equal frequency but only when the Ser183 mutant was grown under 40°C selective pressure. Their facile generation is expected for a single-base substitution, given the high level of replication occurring in these cells (producing 1010 PFU/ml or 104 PFU/cell). Either a positively charged or a nonpolar amino acid in nsP1 suppressed the substitution for Arg183 in nsP4. This suggests that a basic His residue could rescue minus-strand synthesis by restoring important charge properties to the nsP1-nsP4 microenvironment, while the Ile could do so by providing additional noncovalent, hydrophobic, and van der Waals contacts. While His183 of nsP1 functioned well with nsP4 Arg183 in mosquito cells, the presence of Ile at amino acid 183 of nsP1 inhibited overall transcription at either 30 or 34.5°C, arguing that it interfered with assembly or activity of the polymerase in the mosquito cell environment. Shirako et al. (41) showed that, in one case, mutants with diverse amino acids substituted for the N-terminal Tyr of nsP4 were rescued by the same amino acid substitution in nsP1 (41). In our case, two different amino substitutions in nsP1 suppressed the same amino acid substitution in nsP4. We think that this suggests that the same or a related step in minus-strand initiation is being affected in both cases. The nsP1-374 suppressor involved a change of AAC (Asn) to CAC (His) or AUC (Ile). These are tranversions and are less likely to occur than transitions, supporting the notion that they were specifically selected. Not found were other single-base changes that would replace Asp (GAC), Tyr (UAC), Ser (AGC), or Thr (ACC). Not finding these among the suppressor population argues that such amino acids could not restore functionality to the nsP4-nsP1 interaction.
These results and those of Shirako et al. (41) argue that interactions between nsP4 and nsP1 are essential for minus-strand synthesis. The nsP1-nsP4 interaction identified in this study may play one of several roles in alphavirus replication. Only nsP1 associates with membranes when expressed in the absence of the other nsPs, and thus, nsP1 likely directs nsP polyprotein precursors to sites where replication complexes are assembled and eventually function (20). The finger domain of RNA-dependent RNA polymerases is hypothesized to determine the preference for RNA as a template (15). As shown for other viral systems, an effect on template specificity (4, 12) or template recruitment (1, 5) and the switch from translation to transcription (11) are an intriguing possibility since the nsP4 Ser183 polymerase defect in minus-strand synthesis indicates that the interaction is not one required for plus-strand synthesis by either immature (nsP1-P23-nsP4) or mature (fully cleaved, nsP1-nsP2-nsP3-nsP4) replication complexes. Thus, suppression is a property of the P123 form of nsP1, and its action affects the earliest polymerase complex. Another possibility is that nsP1 stimulates initiation activity by nsP4 as seen previously for poliovirus 3Dpol (32, 35) and mutations disrupting 3AB-3Dpol interaction (50). Future experiments will attempt to distinguish between these possibilities and to determine the specific step blocked in the SIN Arg183 polymerase mutants.
Acknowledgments
We thank Farida Attar and Isabel Novella for helpful discussions.
We acknowledge support from Public Health Service grant AI-15123 from the National Institutes of Health.
REFERENCES
- 1.Ahola, T., J. A. den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., and L. Kaariainen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1988. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J. Virol. 62:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canter, D. M., and J. Perrault. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219:376-386. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dé, I., S. G. Sawicki, and D. L. Sawicki. 1996. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J. Virol. 70:2706-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot, R. J., W. R. Hardy, Y. Shirako, and J. H. Strauss. 1990. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 9:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faragher, S. G., A. D. Meek, C. M. Rice, and L. Dalgarno. 1988. Genome sequences of a mouse-avirulent and a mouse-virulent strain of Ross River virus. Virology 163:509-526. [DOI] [PubMed] [Google Scholar]
- 9.Fata, C. L., S. G. Sawicki, and D. L. Sawicki. 2002. Alphavirus minus-strand RNA synthesis: identification of a role for Arg183 of the nsP4 polymerase. J. Virol. 76:8632-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin, R. M. 1966. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc. Natl. Acad. Sci. USA 55:1504-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, Y., and J. Lenard. 1995. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: pathways of assembly. J. Virol. 69:7718-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, Y. S., A. Grakoui, C. M. Rice, E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 63:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 63:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 16.Hardy, W. R., Y. S. Hahn, R. J. de Groot, E. G. Strauss, and J. H. Strauss. 1990. Synthesis and processing of the nonstructural polyproteins of several temperature-sensitive mutants of Sindbis virus. Virology 177:199-208. [DOI] [PubMed] [Google Scholar]
- 17.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinney, R. M., K. R. Tsuchiya, J. M. Sneider, and D. W. Trent. 1992. Genetic evidence that epizootic Venezuelan equine encephalitis (VEE) viruses may have evolved from enzootic VEE subtype I-D virus. Virology 191:569-580. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 20.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laakkonen, P., M. Hyvonen, J. Peranen, and L. Kaariainen. 1994. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J. Virol. 68:7418-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, E., C. Stocks, P. Lobigs, A. Hislop, J. Straub, I. Marshall, R. Weir, and L. Dalgarno. 1997. Nucleotide sequence of the Barmah Forest virus genome. Virology 227:509-514. [DOI] [PubMed] [Google Scholar]
- 23.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levinson, R. S., J. H. Strauss, and E. G. Strauss. 1990. Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees. Virology 175:110-123. [DOI] [PubMed] [Google Scholar]
- 29.Merits, A., L. Vasiljeva, T. Ahola, L. Kaariainen, and P. Auvinen. 2001. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease. J. Gen. Virol. 82:765-773. [DOI] [PubMed] [Google Scholar]
- 30.Mi, S., R. Durbin, H. V. Huang, C. M. Rice, and V. Stollar. 1989. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 170:385-391. [DOI] [PubMed] [Google Scholar]
- 31.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 32.Plotch, S. J., and O. Palant. 1995. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 69:7169-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75:10118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards, O. C., and E. Ehrenfeld. 1998. Effects of poliovirus 3AB protein on 3D polymerase-catalyzed reaction. J. Biol. Chem. 273:12832-12840. [DOI] [PubMed] [Google Scholar]
- 36.Rumenapf, T., E. G. Strauss, and J. H. Strauss. 1995. Aura virus is a New World representative of Sindbis-like viruses. Virology 208:621-633. [DOI] [PubMed] [Google Scholar]
- 37.Sawicki, D., D. B. Barkhimer, S. G. Sawicki, C. M. Rice, and S. Schlesinger. 1990. Temperature sensitive shut-off of alphavirus minus strand RNA synthesis maps to a nonstructural protein, nsP4. Virology 174:43-52. [DOI] [PubMed] [Google Scholar]
- 38.Sawicki, D. L., and S. G. Sawicki. 1985. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology 144:20-34. [DOI] [PubMed] [Google Scholar]
- 39.Sawicki, D. L., and S. G. Sawicki. 1980. Short-lived minus-strand polymerase for Semliki Forest virus. J. Virol. 34:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawicki, D. L., S. G. Sawicki, S. Keranen, and L. Kaariainen. 1981. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J. Virol. 39:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2000. Suppressor mutations that allow sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology 276:148-160. [DOI] [PubMed] [Google Scholar]
- 42.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirako, Y., and J. H. Strauss. 1998. Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase. J. Virol. 72:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokal, R. R., and F. J. Rohlf. 1980. Biometry, 2nd ed. W. H. Freeman & Co., San Francisco, Calif.
- 45.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takkinen, K. 1986. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 14:5667-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 68:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1991. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 65:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver, S. C., A. Hagenbaugh, L. A. Bellew, S. V. Netesov, V. E. Volchkov, G. J. Chang, D. K. Clarke, L. Gousset, T. W. Scott, D. W. Trent, et al. 1993. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology 197:375-390. [DOI] [PubMed] [Google Scholar]
- 50.Xiang, W., A. Cuconati, A. V. Paul, X. Cao, and E. Wimmer. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892-904. [PMC free article] [PubMed] [Google Scholar]