Abstract
The efficacy of adenovirus (Ad)-based gene therapy might be significantly improved if viral vectors capable of tissue-specific gene delivery could be developed. Previous attempts to genetically modify the tropism of Ad vectors have been only partially successful, largely due to the limited repertoire of ligands that can be incorporated into the Ad capsid. Early studies identified stringent size limitations imposed by the structure of the Ad fiber protein on ligands incorporated into its carboxy terminus and thus limited the range of potential ligand candidates to short peptides. We have previously identified the HI loop of the fiber knob domain as a preferred site for the incorporation of targeting ligands and hypothesized that the structural properties of this loop would allow for the insertion of a wide variety of ligands, including large polypeptide molecules. In the present study we have tested this hypothesis by deriving a family of Ad vectors whose fibers contain polypeptide inserts of incrementally increasing lengths. By assessing the levels of productivity and infectivity and the receptor specificities of the resultant viruses, we show that polypeptide sequences exceeding by 50% the size of the knob domain can be incorporated into the fiber with only marginal negative consequences on these key properties of the vectors. Our study has also revealed a negative correlation between the size of the ligand used for vector modification and the infectivity and yield of the resultant virus, thereby predicting the limits beyond which further enlargement of the fiber knob would not be compatible with the virion's integrity.
Adenoviruses (Ad) are among the best-characterized and most extensively used viruses. Elucidation of the morphologies of Ad virions and the mechanisms of viral DNA replication, gene expression, and interaction with the infected host, coupled with advances in biotechnology, have provided the rationale for the practical use of Ad as research tools and, most importantly, for considering these viruses as potential genetic therapeutic agents.
A number of biological properties of Ad made them a rational choice as gene delivery vehicles for human gene therapy. The extensive use of these viruses at the onset of gene therapy has been supported by their efficient transduction of a wide range of cell types (both dividing and quiescent), the capacity of an Ad genome to incorporate a substantial amount of heterologous DNA, the high stability of Ad virions in vivo, and the lack of oncogenicity in humans. However, some of the biological features of Ad vectors constitute real hurdles to their successful employment as gene therapeutic agents. The most serious limitations of Ad as gene therapy vectors proved to be the immunogenicity of Ad virions and transgene products, the suboptimal biodistribution of Ad particles in vivo, the lack of specificity of a vector for diseased tissues, and poor infectivity with a number of cell targets. These limitations define the goals for subsequent work aimed at improving this vector system.
In this regard, attempts to increase the selectivity of Ad vectors for target tissues have resulted in novel approaches in Ad vector design which exploit the concept of tissue-specific expression of therapeutic transgene or virus replication (for a review, see reference 11). By capitalizing on unique patterns of gene regulation that are characteristic of certain target tissues and tumors, it has been possible to design Ad vectors capable of selective cell killing or transgene expression. However, this previous work has not addressed the problem of efficient and selective transduction of target cells by Ad vectors, which is due to the inability of the virus to specifically recognize diseased cells as targets and infect them while avoiding normal cells. This lack of target specificity of Ad vectors is a direct consequence of the molecular recognition events taking place early in Ad infection. Specifically, human Ad of serotypes 2 and 5 (Ad2 and Ad5, respectively), which are most frequently used as delivery vectors for gene therapeutics, bind to cells via a cellular receptor known as the coxsackievirus and Ad receptor, or CAR (1, 41). The availability and accessibility of CAR molecules on the surface of a given cell are key factors that determine the efficiency of Ad infection. Furthermore, it appears that Ad binding to CAR is characterized by a threshold effect, in that expression of this receptor below a certain level renders the cells refractory to Ad infection (7). It has been shown that a number of normal cell types which might constitute targets for therapeutic Ad vectors express CAR rather inefficiently (19, 27, 33, 37, 43, 44, 46). Along the same lines, the vast majority of cancerous cells produce CAR at low levels, which makes their transduction by CAR-dependent Ad vectors inadequate (12, 20, 24). Most importantly, recent studies established an adverse correlation between the levels of CAR expression by certain tumor cell lines and their ability to actively metastasize (31, 32), thereby further highlighting the limited utility of Ad2- and Ad5-derived vectors for cancer gene therapy. On the other hand, molecular characterization of various diseased tissues and tumor types has revealed a number of disease-specific cell surface markers which can potentially be exploited as alternative receptors for Ad vectors should the strategies to alter Ad vector tropism be developed. In the aggregate, these findings have supported the concept of using tropism-modified or targeted Ad as a way to improve the efficiency and selectivity of Ad vectors by circumventing their dependency on CAR expression by a target cell.
Early attempts to target Ad to alternative cellular receptors which employed bifunctional protein complexes or fusion proteins (the so-called protein bridge approach) demonstrated the feasibility of Ad targeting and its potential utility for gene therapy. However, as the large-scale production of these multicomponent vectors has always been considered a major technological hurdle, an alternative strategy of genetic targeting of Ad has emerged (for a review, see reference 17). This strategy is based on the modification of Ad vector tropism by genetic incorporation of a targeting ligand into one of the proteins of the Ad capsid. The major advantage offered by the genetic approach is that the targeted vector is a one-component, self-sufficient agent which, once made and approved for clinical use, can then be propagated at any desired scale. However, the successes shown in the protein bridge-based studies have not been replicated in those attempts to use genetic targeting.
This lack of success is due primarily to the technical difficulties involved in the genetic modification of Ad capsid proteins. As binding of an Ad virion to CAR is mediated by the Ad fiber protein, this protein was the most logical choice as a carrier for targeting ligands. It is a homotrimeric protein which has a characteristic domain organization with an amino-terminal tail domain anchoring the fiber in Ad capsid; a carboxy-terminal globular domain, termed the knob, mediating binding to CAR; and a central shaft domain extending the knob away from the virion (reviewed in reference 4). The fibers are located at each of the 12 vertices of the icosahedral Ad capsid, where they associate with another capsid protein, called the penton base. While early efforts to alter Ad tropism via fiber modification involved incorporation of ligands into the carboxy terminus of the protein (14, 23, 43, 44), in subsequent studies, which took full advantage of the resolved three-dimensional structure of the fiber knob, we identified the so-called HI loop as an alternative locale for incorporation of targeting moieties (5, 16). Subsequent modifications of the HI loop of the fiber with peptide ligands were largely successful (5, 6, 26, 28, 45). However, in some instances, binding of the modified fibers to CAR was partially impaired (28, 45). To date, all Ad targeting endeavors have been limited to attempts to engraft short peptide ligands into the rather complex framework of the fiber knob, with no efforts being made to expand the repertoire of ligand candidates beyond peptides. As a result of the limited availability of high-affinity peptide ligands suitable for incorporation into the Ad fiber, only a few genetically targeted Ad prototypes have been derived to date. Thus, we reasoned that the practical utility of Ad targeting could be improved should other types of ligands prove to be compatible with the fiber structure.
This work represents an attempt to evaluate the possibility of genetic modification of Ad tropism by employing heterologous polypeptide sequences as ligand mimics. By incrementally increasing the size of the HI loop, we have shown that heterologous protein sequences of over 100 amino acid (aa) residues can be configured into Ad fiber and, for the first time, have demonstrated the possibility of incorporation of a polypeptide ligand into the Ad fiber protein.
MATERIALS AND METHODS
Cell lines.
293 human embryonal kidney cells (10) and U118MG human glioblastoma cells were purchased from Microbix (Toronto, Ontario, Canada) and the American Type Culture Collection (Manassas, Va.), respectively. Chinese hamster ovary (CHO) cells as well as their derivatives which express the VPAC1 and VPAC2 receptors, CHO/VPAC1 and CHO/VPAC2 (29), were provided by Mark Laburthe (Institut National de la Santé et de la Recherche Médicale, Paris, France). CHO cells were cultured in Ham's F-12 medium, while their VPAC-expressing derivatives were maintained in the same medium containing 100 μg of Geneticin (G418) per ml. All other cell lines were propagated in Dulbecco's modified Eagle's medium (DMEM)-F-12 medium. All media were supplemented with fetal calf serum (FCS) (10%), glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml). FCS was purchased from Gibco-BRL (Gaithersburg, Md.), and media and supplements were from Mediatech (Herndon, Va.). All cells were propagated at 37°C in a 5% CO2 atmosphere.
Antibodies.
Rabbit anti-Ad2 polyclonal antibodies were purchased from the American Type Culture Collection, and anti-mouse and anti-rabbit immunoglobulin polyclonal antibodies conjugated with horseradish peroxidase were from Amersham Pharmacia Biotech Inc. (Piscataway, N.J.) and DAKO (Carpinteria, Calif.), respectively. DAV-1 anti-Ad penton base (40) and 4D2 antifiber (13) monoclonal antibodies were provided by Glen Nemerow (The Scripps Research Institute, La Jolla, Calif.) and Jeffrey Engler (University of Alabama at Birmingham, Ala.), respectively.
Genetic engineering.
All recombinant DNA molecules generated in this study were designed by standard methods of genetic engineering (described in reference 35). Restriction endonucleases and T4 DNA ligase were from New England Biolabs (Beverly, Mass.). PCR was performed with Pfu DNA polymerase purchased from Stratagene (La Jolla, Calif.).
In order to generate recombinant Ad5 fiber genes encoding fibers with insertions of arginine-glycine-aspartic acid (RGD)-containing sequences, fragments of the Ad5 penton base gene corresponding to the RGD motif together with adjacent sequences were either assembled with oligonucleotides or generated by PCR and cloned into the EcoRV site of the fiber shuttle vector pXKΔHI. Vector pXKΔHI was derived from previously described pNEB.PK3.6 (18) to incorporate heterologous DNA within the region of the fiber gene corresponding to the HI loop of the knob domain. To generate this plasmid, an XbaI fragment corresponding to a segment of Ad5Luc3 (25) DNA of unknown sequence was deleted from pNEB.PK3.6, resulting in pXK3.1. To make pXKΔHI, a BstXI-MunI fragment in pXK3.1 was replaced with a BstXI-MunI fragment from pQE.KNOBΔHI (16). Because pXKΔHI contains a deletion of 8 nucleotides which affects the codons for Thr545, Thr546, and Pro547, all oligonucleotides subsequently used either as structural blocks or PCR primers were designed to contain at their 5′ termini additional sequences (below, italicized) to restore this deletion. The sequences corresponding to RGD-containing peptides of 13 aa (13RGD) and 23 aa (23RGD) were assembled with oligonucleotides RGD-loop.13T (ACA ACT AAC GAT CAT GCC ATT CGC GGC GAC ACC TTT GCC ACA CGG CC) and RGD-loop.13B (GGC CGT GTG GCA AAG GTG TCG CCG CGA ATG GCA TGA TCG TTA GTT GT) and with oligonucleotides RGD-loop.23T (ACA ACT CCG GTG GAG GAC ATG AAC GAT CAT GCC ATT CGC GGC GAC ACC TTT GCC ACA CGG GCT GAG GAG AAG CGC CC) and RGD-loop.23B (GGG CGC TTC TCC TCA GCC CGT GTG GCA AAG GTG TCG CCG CGA ATG GCA TGA TCG TTC ATG TCC TCC ACC GGA GTT GT), respectively, by annealing them in pairs as listed. The sequences corresponding to longer penton base-derived peptides, 33RGD, 43RGD, 53RGD, 63RGD, 73RGD, and 83RGD, were PCR amplified from Ad5 genomic DNA with the following pairs of primers: RGD-loop.33F (ACA ACT GCC GCG GCA ATG CAG CC) and RGD-loop.33R (GGT GCT TCG GCC TCA GCG CGC), RGD-loop.43F (ACA ACT AAC TCC AAC GCG GCA GCC) and RGD-loop.43R (GGG GCA GCT TCG GCC GCT G), RGD-loop.53F (ACA ACT AGC GGC GCG GAA GAG AAC TC) and RGD-loop.53R (GGT TGC GCA GCG GGG GC), RGD-loop.63F (ACA ACT AGC AAC AGC AGT GGC AGC G) and RGD-loop.63R (GGC TTC TCG ACC TCG GGT TGC G), RGD-loop.73F (ACA ACT GGT GGC GCA GGC GG) and RGD-loop.73R (GGC GGT TTC TTC TGA GGC TTC TCG), and RGD-loop.83F (ACA ACT ACC GAA CAG GGC GGG G) and RGD-loop.83R (GGC AGG GGT TTG ATC ACC GGT TT), respectively. The resultant plasmids with the correct structures have been designated pXK.FHI13RGD through pXK.FHI83RGD (in pXK.FHINNRGD, the NN indicates the number of amino acid residues of the penton base-derived polypeptide).
Recombinant Ad genomes incorporating modified fiber genes were derived by homologous DNA recombination in Escherichia coli BJ5183 with SwaI-linearized plasmid pVK700 essentially as described previously (16). pVK700 is a derivative of pTG3602 (3), which was modified to incorporate a firefly luciferase gene driven by the cytomegalovirus immediate early promoter in the place of the deleted E1 region. Also, pVK700 contains a nearly complete deletion of the fiber gene, which was replaced with a unique SwaI site.
In order to replace the RGD-encoding sequence in the plasmids of the pXK.FHINNRGD series with a recognition sequence for the restriction exonuclease BaeI, BstXI-MfeI fragments in these plasmids were replaced with homologous sequences assembled as follows. First, DNA sequences adjacent to the RGD-encoding region in the Ad5 penton base gene were amplified by sticky-end PCR (sePCR) (47) using genomes of the Ad vectors Ad5Luc.13RGD through Ad5Luc.83RGD (Ad5Luc.NNRGD). The sequence located upstream from the RGD-encoding region of Ad5Luc.13RGD DNA was amplified with the primers BstXI.F.short (TTG GAA CTT TAG AAA TGG AGA TC) and RGD.R.long (AAT GGC ATG ATC GTT AGT TGT ATC) and the primer pair BstXI.F.long (AAT ATT GGA ACT TTA GAA ATG GAG) and RGD.R.short (CAT GAT CGT TAG TTG TAT CTC CTG). The sequence located downstream of this site was amplified with the primers RGD.F.short (TGC CAC ACG GCC ATC TG) and MfeI.R.short (GAA AAA TAA ACA CGT TGA AAC ATA AC) and with primers RGD.F.long (ACC TTT GCC ACA CGG CCA) and MfeI.R.long (AAT TGA AAA ATA AAC ACG TTG AAA C). Similarly, the sequences which flank the codons of the RGD tripeptide in all other Ad5Luc.NNRGD genomes were amplified by employing the primer pairs BstXI.F.short and RGD20.R.long (AAT GGC ATG ATC GTT CAT GTC), BstXI.F.long and RGD20.R.short (CAT GAT CGT TCA TGT CCT CCA), RGD20.F.short (TGC CAC ACG GGC TGA GG) and MfeI.R.short, and RGD20.F.long (ACC TTT GCC ACA CGG GCT) and MfeI.R.long. PCR products representing the same RGD-flanking sequence were mixed in equimolar amounts, denatured, and annealed. The products of sePCR which contained BstXI- or MfeI-compatible cohesive ends were then mixed with a duplex comprising the BaeI recognition site, BaeI-stuffer.T (TCA ACT ACT GAC GCG AGT ACC TCC TAA AAC CTT) and BaeI-stuffer.B (TTT AGG AGG TAC TCG CGT CAG TAG TTG AAA TGG), and ligated with pXK.3.1 cleaved with BstXI and MfeI. The resultant plasmids were designated pHI.PBNN (pHI.PB10 through pHI.PB80).
In order to generate recombinant Ad5 fiber genes encoding the proteins with insertions of vasoactive intestinal polypeptide (VIP) within the extended HI loops, the duplexes made with oligonucleotides VIP.T (CAC TCA GAT GCA GTC TTC ACT GAC AAC TAT ACC CGC CTT AGA AAA CAA ATG GCT GTA AAG AAA TAT TTG AAC TCA ATT CTG AAT GGA AAG AGG ACC TT) and VIP.B (CCT CTT TCC ATT CAG AAT TGA GTT CAA ATA TTT CTT TAC AGC CAT TTG TTT TCT AAG GCG GGT ATA GTT GTC AGT GAA GAC TGC ATC TGA GTG AAT GG) were cloned into the BaeI-digested plasmids of the pHI.PBNN series, thereby resulting in plasmids designated pHI.PBNNVIP. Additional details on this plasmid construction scheme are available upon request.
To design the fiber gene coding for the protein with the insertion of mutated VIP sequence VIP18-23 within the extended HI loops, the duplex made of oligonucleotides VIP18-23.T (CAC TCA GAT GCA GTC TTC ACT GAC AAC TAT ACC CGC CTT AGA AAA CAA ATG TTG GTA AAG AAA TAT GCT AAC TCA ATT CTG AAT GGA AAG AGG ACC TT) and VIP18-23.B (CCT CTT TCC ATT CAG AAT TGA GTT AGC ATA TTT CTT TAC CAA CAT TTG TTT TCT AAG GCG GGT ATA GTT GTC AGT GAA GAC TGC ATC TGA GTG AAT GG) was cloned into the BaeI site of pHI.PB10VIP. The newly designed plasmid was named pHI.PB10VIP18-23.
Sequencing of recombinant DNA derived in this study was done on a model CEQ2000XL DNA analysis system from Beckman-Coulter (Fullerton, Calif.).
Viruses.
Ad vectors containing recombinant fiber proteins and expressing the firefly luciferase as a reporter were generated by transfection of 293 cells with PacI-digested derivatives of plasmid pVK700 as described previously (16). The viruses were propagated on 293 cells and purified by equilibrium centrifugation in CsCl gradients by a standard protocol (9). Determination of virus particle titer was accomplished spectrophotometrically by the method of Maizel et al. (22) by using a conversion factor of 1.1 × 1012 viral particles per absorbance unit at a wavelength of 260 nm. The infectious titers of the vectors were determined on 293 cells in a spot assay developed by Bewig and Schmidt (2). The modifications of the fiber gene were confirmed by PCR analysis with the primers F5.F1596 (AAC GAT TAC ACT AAA CGG TAC ACA GG) and F5.R1658 (CCA GAC CAG TCC CAT GAA AAT G), which anneal up- and downstream of the site of the insertion within the fiber open reading frame.
Gene transfer experiments.
Cells grown in the wells of 24-well plates to 90 to 100% confluence were washed with growth medium containing 2% FCS and then were infected for 30 min at room temperature with an Ad vector diluted in 0.4 ml of the same medium at a multiplicity of infection (MOI) of 40 virus particles per cell. The infected monolayers were then incubated at 37°C in an atmosphere of 5% CO2. Twenty hours postinfection, the virus-containing medium was aspirated and the cells were washed with phosphate-buffered saline and lysed in 0.25 ml of luciferase reporter lysis buffer (Promega, Madison, Wis.). The luciferase activity in the cell lysates was then measured according to the manufacturer's protocol. Each data point was set in triplicate and calculated as the mean of three determinations.
To show the receptor specificity of an Ad vector, some experiments included incubation of cells with a blocking agent, either the recombinant Ad5 fiber knob (18) or synthetic VIP, purchased from American Peptide Company (Sunnyvale, Calif.). The knob protein and VIP were diluted in the serum-free medium to concentrations ranging from 0.1 to 100 μg/ml and from 10 ng/ml to 10 μg/ml, respectively, and then added to cells in 0.2-ml aliquots for 10 min prior to addition of the virus. Incubation with the virus was continued at room temperature for another 30 min. The medium containing the virus and the inhibitor was removed, and the cell monolayers were washed with the medium containing 10% FCS, overlaid with fresh medium, and incubated further at 37°C. Determination of luciferase activity and data processing were done as described above.
Labeling of Ad vectors with [methyl-3H]thymidine.
Ad virions were labeled with [methyl-3H]thymidine as described in detail elsewhere (36). Briefly, 293 cells grown in 175-cm2 flasks were infected with Ad (MOI of 100 viral particles per cell) diluted in 15 ml of DMEM-F-12 medium-2% FCS. Sixteen hours postinfection, 1 mCi of [methyl-3H]thymidine (Amersham, Arlington Heights, Ill.) was added to each flask and the cells were further incubated at 37°C until complete cytopathic effect was observed. Cells were then harvested, and the virus was purified from the cell lysates by centrifugation in CsCl gradients according to a standard protocol. Specific radioactivities of labeled virus preparations measured in a liquid scintillation counter were in the range of 10−5 to 10−4 cpm per virion.
Ad binding assay.
Binding of 3H-labeled Ad to CHO and CHO/VPAC1 cells was assayed in a procedure described previously (36). Specifically, target cells (4 × 105 cells per aliquot) were incubated for 1 h on ice with 3H-labeled Ad at an MOI of 3,500 viral particles per cell in 200 μl of Ham's F-12 medium-2% FCS. The cells were then pelleted by centrifugation for 5 min at 2,000 × g and washed twice with 0.5 ml of ice-cold Ham's F-12 medium-2% FCS. After the last wash, the cell-associated radioactivity was determined in a scintillation counter. The number of viral particles bound per cell was calculated using the specific radioactivity of the viral preparation, the total radioactivity of the sample, and the number of cells.
Western blot analysis.
Aliquots of Ad vectors equal to 1010 viral particles were denatured by boiling for 5 min in Laemmli buffer and loaded onto a sodium dodecyl sulfate-7.5% polyacrylamide gel. Upon separation, viral proteins were electroblotted onto a polyvinylidene difluoride membrane and detected with either 4D2 antifiber or DAV-1 anti-Ad penton base monoclonal antibody. The blots were developed with an ECL Plus detection kit using horseradish peroxidase-conjugated secondary anti-mouse immunoglobulin antibody, both purchased from Amersham Pharmacia Biotech.
RESULTS
The HI loop of the Ad5 fiber knob domain can accommodate heterologous polypeptides.
In order to assess the capacity of the HI loop of the Ad5 fiber knob domain to accommodate polypeptide ligands, its length was incrementally increased by genetic incorporation of fragments of the RGD-containing loop of the Ad5 penton base. All these fragments were engineered to contain the RGD motif in the middle, flanked by penton base-derived sequences of equal lengths. The length of each flanking sequence in the shortest construct was 5 aa residues; this was increased by 5-aa increments in other constructs. DNA sequences corresponding to the fragments of the penton base protein were either assembled with oligonucleotides or amplified by PCR and then cloned between codons Thr546 and Pro547 of the Ad5 fiber gene contained in shuttle vector pXKΔHI. A total of eight fragments of the penton base protein ranging in size from 13 to 83 aa were thus incorporated into the Ad5 fiber knob. The largest of these fragments included the entire loop of the penton base protein. The newly designed fiber genes were transferred by homologous DNA recombination in E. coli into the genome of the E1-deleted, firefly luciferase-expressing Ad5 genome contained in rescue vector pVK700. Subsequent transfection of 293 cells with the resultant recombinant genomes led to the rescue of all eight viral vectors designated Ad5Luc.NNRGD, where NN indicates the length of an insert in amino acid residues.
Rescued viruses were further amplified on 293 cells, and their identities were confirmed by PCR using DNA released from CsCl-purified virions as a template. As seen in Fig. 1A, this test confirmed the presence of inserts within the fiber genes of all rescued Ad vectors. These data were further corroborated by partial sequencing of the fiber genes contained in the Ad5Luc.NNRGD genomes.
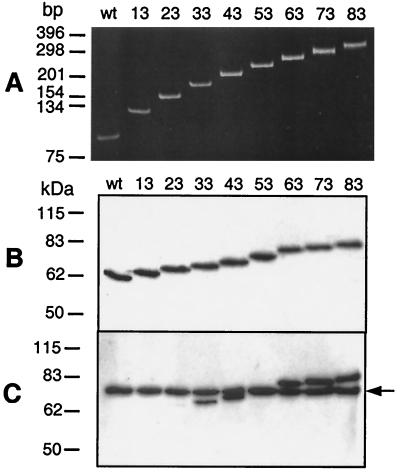
FIG. 1.
Confirmation of incorporation of the penton base-derived polypeptides into the fibers of Ad5Luc.NNRGD vectors. (A) Polyacrylamide gel electrophoresis of PCR products obtained with genomic DNAs of Ad5Luc.NNRGD vectors and the primers flanking the site of insertion. (B and C) Western blot analyses of Ad5Luc.NNRGD capsid proteins. Aliquots of CsCl-purified Ad vectors (1010 viral particles) were denatured by boiling them in Laemmli sample buffer and separated on a sodium dodecyl sulfate-polyacrylamide gel. Viral proteins transferred to a polyvinylidene difluoride membrane were probed with either 4D2 antifiber tail (B) or DAV-1 anti-penton base (C) monoclonal antibody. DAV-1 recognizes the epitope IRGDTFATR, which is present in the fibers of all Ad5Luc.NNRGD vectors. Numbers at the top show the sizes of the ligand-mimicking polypeptides in amino acid residues. The arrow on the right of panel C shows the position of the penton base protein. “wt” corresponds to the Ad5Luc1 control vector incorporating wild-type Ad5 fibers.
Subsequent analysis of Ad5Luc.NNRGD virions by Western blotting showed that the lengths of the fibers incorporated into the capsids of these vectors correspond to those expected for fibers with extended HI loops (Fig. 1B). By using the monoclonal antibody DAV-1, which recognizes an RGD-containing epitope within the Ad5 penton base, we also demonstrated the presence of this epitope in most of the vectors derived (Fig. 1C). Although all the fragments of the penton base protein incorporated into Ad5Luc.NNRGD fibers contained a minimal epitope for DAV-1, IRGDTFATR (40), this antibody produced a faint signal on Ad5Luc.23RGD fiber and no signal at all on Ad5Luc.13RGD fiber. This finding suggests that the presence of the minimal epitope for DAV-1, as determined by affinity-directed mass spectrometry, is insufficient to ensure high-affinity binding of this antibody to the epitope-bearing protein in a Western blot assay.
Incorporation of the penton base-derived polypeptides into the Ad5 fiber knob has marginal effects on the yields and infectivities of the resultant viruses.
Having established the identities of the newly rescued Ad, we next wished to test whether the incorporation of the penton base-derived polypeptides into the fibers of these vectors had any effect on the productivity of infection and the infectivity of these viruses.
To determine the yields of the vectors, 293 cells were infected with each Ad5Luc.NNRGD virus at an MOI of 3 infectious units per cell and the infection was allowed to proceed for 48 h. The progeny virions were then released from the infected cells and purified by double banding on CsCl gradients. As shown in Table 1, the yields of Ad5Luc.NNRGD vectors per 108 cells were very similar (6.3 × 1011 to 2.3 × 1012 viral particles) to that of the control Ad5Luc1 virus incorporating wild-type Ad5 fibers (2 × 1012 viral particles). The most significant changes in vector yields were noticed for the viruses incorporating the longest inserts. Specifically, Ad5Luc.63RGD, Ad5Luc.73RGD, and Ad5Luc.83RGD amplified two- to threefold less efficiently than Ad5Luc1.
TABLE 1.
Productivity of Ad5Luc.NNRGD vectors in 293 cells
| Virus | Particles per 108 cellsa |
|---|---|
| Ad5Luc1 | 2.0 × 1012 ± 2.4 × 1011 |
| Ad5Luc.13RGD | 2.0 × 1012 ± 3.6 × 1011 |
| Ad5Luc.23RGD | 2.3 × 1012 ± 1.5 × 1011 |
| Ad5Luc.33RGD | 2.0 × 1012 ± 4.9 × 1010 |
| Ad5Luc.43RGD | 1.8 × 1012 ± 5.3 × 1010 |
| Ad5Luc.53RGD | 2.2 × 1012 ± 7.3 × 1010 |
| Ad5Luc.63RGD | 1.2 × 1012 ± 6.3 × 1010 |
| Ad5Luc.73RGD | 6.3 × 1011 ± 1.1 × 1011 |
| Ad5Luc.83RGD | 1.1 × 1012 ± 9.5 × 1010 |
The values shown are the means of values for two independent viral preparations ± standard deviations.
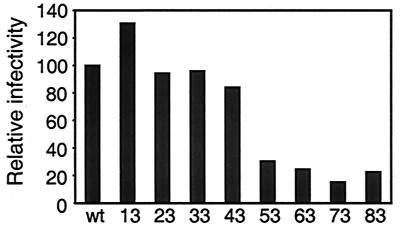
The infectivity of Ad5Luc.NNRGD on 293 cells was determined in a spot assay and compared with the infectivity of unmodified control virus. This comparison showed an inverse correlation between the size of the insert within the HI loop of the knob and the capacity of a given Ad5Luc.NNRGD vector to infect 293 cells (Fig. 2). Overall, the infectivities of Ad5Luc.NNRGD vectors ranged from 15% (Ad5Luc.73RGD) to 130% (Ad5Luc.13RGD) of that of Ad5Luc1.
FIG. 2.
Relative infectivities of Ad5Luc.NNRGD viruses. Infectious titers of the purified viruses were determined in a spot assay on 293 cells and were then compared to that of the Ad5Luc1 control vector incorporating unmodified Ad5 fibers. Bars represent infectious unit/viral particle ratios for Ad5Luc.NNRGD vectors divided by the ratio obtained for Ad5Luc1 (wt). Numbers at the bottom show the sizes of the penton base-derived polypeptides incorporated into the HI loop of a given Ad vector.
Therefore, these data suggest that the extensions of the HI loop in the knob domains of Ad5Luc.NNRGD fibers did not dramatically affect the functional structures of these fibers and did not compromise the overall architecture of modified virions either. These findings were further corroborated by the fact that during the rescue and subsequent amplification of Ad5Luc.NNRGD viruses, we did not observe any significant variations in the dynamics of the development of cytopathic effect among the vectors.
The RGD tripeptide contained in the modified Ad5Luc.NNRGD virions may serve as a targeting ligand as it retains its natural capacity to bind integrins.
The incorporation of the penton base-derived sequences into the CAR-binding domain of the fiber might lead to different outcomes with respect to the tropism of the newly derived Ad vectors. One possibility would be fibers possessing dual receptor specificities. First, these molecules could retain their CAR-binding properties; second, the RGD sequence, which is present in the fibers of all Ad5Luc.NNRGD vectors, could preserve its natural capacity to bind cellular integrins. Alternatively, the resultant fibers could possess only one of these two binding specificities with either the RGD tripeptide being incapable of binding to integrins or the site of CAR attachment being destroyed in the modified proteins. A third outcome would represent a combination of these two scenarios, whereby some of the modified viruses would have dual receptor tropism while others would bind only to one of the two potential targets. Whereas the studies of the infectivities of the vectors suggested that the infectivities of Ad5Luc.NNRGD vectors had not been significantly changed compared to that of the vector with wild-type fibers, the relative contributions of each of the two receptor-binding mechanisms to the overall infectivity were not clear.
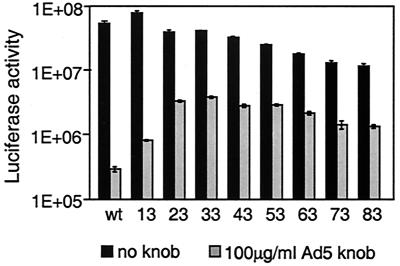
This lack of clarity was addressed by infecting 293 cells with Ad5Luc.NNRGD vectors in the presence of the recombinant Ad5 fiber knob protein, which was employed as a selective inhibitor of the interaction between cellular CAR and the fibers of the modified Ad vectors. Although the knob efficiently blocked the infection of both the control vector and the vectors bearing the fibers with extended HI loops, this inhibition differed qualitatively for the various Ad vectors (Fig. 3). While the knob blocked gene transfer by Ad5Luc1 and Ad5Luc.13RGD by 2 orders of magnitude, gene delivery by the majority of the fiber-modified vectors in the presence of this inhibitor was decreased only 10-fold.
FIG. 3.
Infectivity and receptor specificity of Ad5Luc.NNRGD viruses. 293 cells grown in monolayer culture were preincubated for 10 min with either DMEM-F-12 medium or the same medium containing Ad5 knob at a concentration of 100 μg/ml. Ad5Luc.NNRGD viruses were then added to the cells at an MOI of 40 viral particles per cell and incubated for 30 min. The virus-containing medium was aspirated and replaced with fresh medium, and the cells were incubated for an additional 20 h. The graph shows the luciferase activities in relative light units detected in the lysates of infected cells. Each data point corresponds to an average of three measurements. Numbers at the bottom indicate the lengths of the inserts within the fibers. wt, the Ad5Luc1 control vector.
This variability in the effect caused by the knob indicates that while binding to CAR remained the preferred and more efficient of the two cell entry pathways for the Ad5Luc.NNRGD virions, the contribution of CAR-independent, RGD-mediated attachment to cellular integrins to the overall cell binding of the vectors becomes significant when the size of the RGD-containing insert within the HI loop exceeds 23 aa.
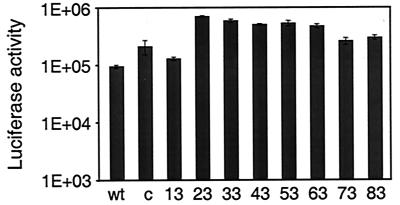
The integrin-binding capacity of the RGD peptide contained in the fibers of the Ad5Luc.NNRGD virions was further demonstrated in a gene transfer experiment using U118MG human glioblastoma cells, which are characterized by moderate expression of αv integrins while being low in CAR (24). Additionally, the previously reported Ad5LucFHIRGD vector (5), which contains within its fiber knob the RGD peptide in an optimized configuration, CDCRGDCFC (RGD-4C), was also used in these studies for the purpose of comparison. As expected based on the results obtained with 293 cells, the presence of the RGD peptide in the fibers of Ad5Luc.NNRGD viruses resulted in augmentation of the transgene expression in U118MG cells by these vectors (Fig. 4). Compared to the control vector Ad5Luc1, Ad5Luc.NNRGD viruses transduced CAR-deficient U118MG cells two- to eightfold more efficiently. Notably, these levels of transgene expression were similar to or even higher than those detected in Ad5LucFHIRGD-infected cells.
FIG. 4.
Ad-mediated gene transfer to U118MG cells. U118MG cells were infected for 30 min with Ad5Luc.NNRGD viruses at an MOI of 40 viral particles per cell. Luciferase activity was determined in cell lysates 20 h postinfection. Each data point corresponds to an average of three determinations. Numbers below the graph show the length of the HI loop extensions in amino acids. wt, negative control vector Ad5LucI incorporating wild-type Ad5 fibers; c, the control vector Ad5LucFHIRGD bearing fibers modified with the RGD-4C peptide.
In the aggregate, completion of this phase of our work resulted in the demonstration of the feasibility of very significant enlargements of the HI loop of the fiber knob by incorporation of heterologous polypeptides. Furthermore, it has been shown that the receptor-binding moieties contained within such inserts may function as targeting ligands, thereby altering Ad vector tropism.
Extended HI loops in the fibers of Ad5Luc.NNRGD vectors may serve as frameworks for incorporation of targeting ligands.
The successful generation of Ad5Luc.NNRGD vectors and the demonstration of their functional integrity and expanded tropism supported the subsequent use of the Ad5Luc.NNRGD fibers as a framework for the incorporation of other targeting polypeptides. Indeed, as the RGD tripeptide was shown to be functional in the context of the extended HI loops, it was reasonable to expect that other peptides or polypeptides possessing receptor-binding capabilities could also be displayed in a functional configuration by the modified fibers.
With these considerations in mind, we modified the fiber shuttle vectors of the pXK.FHINNRGD series, which contain the genes encoding the fiber-penton base fusion proteins. This was done to generate a series of the fiber shuttle vectors allowing easy incorporation of the ligand-encoding sequences in place of the deleted RGD codons. The rationale behind this strategy was that in the resultant fibers the fragments of the flexible loop derived from the penton base protein, which normally flank the RGD peptide, would function as linkers for any new ligand. These linkers would extend the ligand away from the knob to minimize potential steric interference during receptor binding. Additionally, the flexibility of the linkers would allow the ligand to assume its functional configuration within the fiber molecule. To this end, RGD-encoding sequences in all pXK.FHINNRGD plasmids were deleted and replaced with an oligonucleotide duplex containing a recognition sequence for the restriction endonuclease BaeI. BaeI was chosen due to its rare capacity to excise its recognition sequence from the target DNA molecule and thus form unique, noncomplementary sticky ends on the vector, which are suitable for efficient directional cloning of the ligand-encoding sequences. The sticky ends complementary to those of the vectors could then be generated on the DNA insert either by the sePCR technique or, for short sequences, by assembling them with oligonucleotides.
In this way, we generated a set of pHI.PBNN fiber shuttle vectors (pHI.PB10 through pHI.PB80). The design of the pHI.PBNN plasmids is such that upon cleavage with BaeI, they all have identical sticky ends and may thus be used for cloning of the same ligand-encoding sequence without additional manipulations (Fig. 5). Thus, a set of eight shuttle vectors could easily be derived, thereby allowing incorporation of an insert between the linkers of various lengths. This design facilitates the optimization of the ligand-linker structure in a time- and labor-efficient manner.
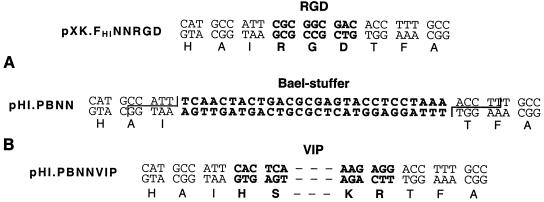
FIG. 5.
Cloning strategy employed to replace the RGD tripeptide within the modified fiber proteins with VIP. (A) Codons corresponding to the RGD peptide within the modified fiber genes of pXK.FHINNRGD vectors were replaced with a DNA stuffer containing the recognition sequence for the restriction endonuclease BaeI. Digestion of pHI.PBNN plasmids with BaeI results in excision of this stuffer and generation of unique, noncomplementary sticky ends on these vectors, thereby allowing for a seamless incorporation of the ligand-encoding sequence into the gene. (B) Schematic view of the VIP-coding sequence incorporated into the modified fiber between the flanking sequences derived from the penton base gene of Ad. Shown in bold are DNA sequences corresponding to the RGD tripeptide, BaeI recognition site, and VIP. They are shown in the context of adjacent sequences of the Ad5 penton base gene. Zigzags on the pHI.PBNN sequence show unique sticky ends formed upon BaeI cleavage.
VIP can be incorporated into the knob domains of modified fiber proteins.
To test whether the fibers modified with the penton base-derived linkers could be used to present a ligand, we chose to use these constructs to derive an Ad vector targeted to VIP receptors. This idea was supported by a body of data describing elevated levels of VIP receptors in a variety of diseased tissues (34), thereby suggesting that a VIP-targeted Ad could be used as a gene delivery vehicle to treat relevant illnesses. Additionally, both the small size (31 aa) and relatively simple structure of VIP implied that this polypeptide might function as a targeting ligand upon incorporation into the Ad fiber.
To test this concept, we used plasmids pHI.PB10, pHI.PB20, pHI.PB70, and pHI.PB80 to assemble the genes encoding fibers which incorporated VIP flanked with the penton base-derived linker sequences of 5, 10, 35, and 40 aa, respectively. Of note, the fiber protein, whose gene is contained in the pHI.80VIP vector, incorporated within the HI loop an insert of 111 aa. Ad5 vectors carrying these modified fiber genes were then derived using the pVK700 rescue vector as described in Materials and Methods. The presence of the VIP-encoding sequence in the genomes of the resultant vectors, Ad5Luc.10VIP, Ad5Luc.20VIP, Ad5Luc.70VIP, and Ad5Luc.80VIP, was confirmed by PCR, while the incorporation of VIP into the fibers of these viruses was shown by a Western blot analysis of purified virions (data not shown). The particle-to-infectious unit ratios for these viruses calculated from their titers, which had been determined in a spot assay on 293 cells, were 3- to 5.5-fold lower than that for Ad5Luc1. Similarly to what had been previously observed for the Ad5Luc.NNRGD vectors, these ratios for the Ad5Luc.NNVIP viruses inversely correlated with the overall length of the insert within the fiber.
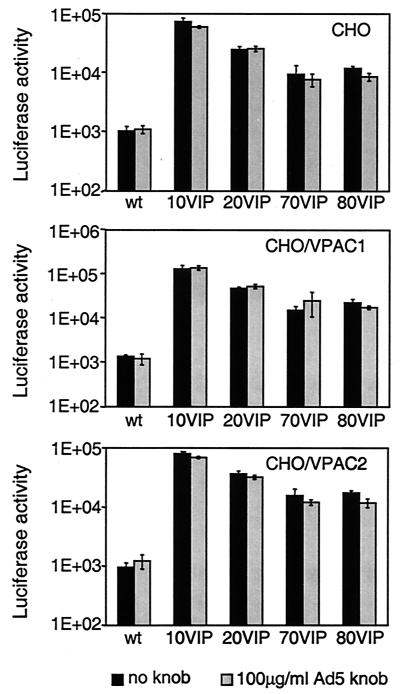
The ability of VIP to direct Ad5Luc.NNVIP virions to VIP receptors was assessed in a gene transfer experiment employing CAR-deficient CHO cells and their derivatives CHO/VPAC1 and CHO/VPAC2, which stably express one of the two VIP receptors, VPAC1 and VPAC2, respectively. As expected, incorporation of VIP into the Ad capsid led to a significant increase in the gene delivery capacity of the vectors: compared to unmodified control Ad, Ad5Luc.NNVIP vectors transduced VPAC-expressing CHO cells 10- to 100-fold more efficiently (Fig. 6). However, contrary to our expectations, a similar augmentation in transduction efficiency was observed in VPAC-negative CHO cells, thus indicating that the VPAC status of a target cell is irrelevant to its accessibility to VIP-modified Ad vectors.
FIG. 6.
Ad-mediated gene transfer to CHO, CHO/VPAC1, and CHO/VPAC2 cells. CHO, CHO/VPAC1, and CHO/VPAC2 cells were preincubated for 10 min with either DMEM-F-12 medium or the same medium containing recombinant Ad5 knob protein at a concentration of 100 μg/ml prior to infection for 30 min with Ad5Luc.NNVIP viruses (MOI of 40 viral particles per cell). Luciferase activity was determined in cell lysates 20 h postinfection. Each data point corresponds to an average of three measurements. Numbers at the bottom indicate the length of the penton base-derived sequences flanking the site of VIP incorporation. “wt” corresponds to the Ad5Luc1 control virus bearing unmodified Ad5 fibers.
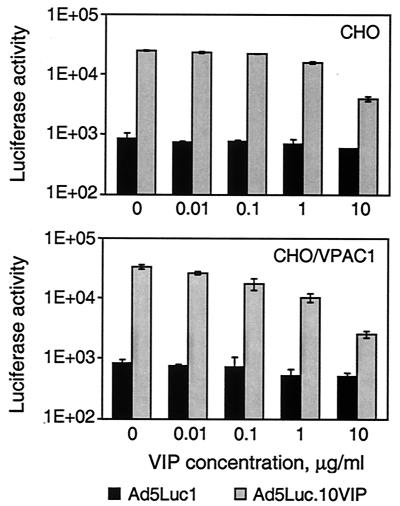
Gene transfer experiments employing synthetic VIP as a competitor demonstrated that cell transduction by the VIP-modified Ad vector is inhibited by VIP in a dose-dependent manner (Fig. 7). Whereas we observed some nonspecific inhibition of the gene transfer by VIP in the control samples, cell transduction by Ad5Luc.10VIP was blocked to a much greater extent, thereby indicating that the expanded tropism of this virus is indeed VIP mediated.
FIG. 7.
Inhibition of Ad5Luc.10VIP-mediated gene transfer to CHO and CHO/VPAC1 cells with synthetic VIP. CHO and CHO/VPAC1 cells had been preincubated for 10 min with either DMEM-F-12 medium or the same medium containing VIP peptide at concentrations ranging from 0.01 to 10 μg/ml before they were infected for 30 min with Ad5Luc.10VIP virus (MOI of 40 viral particles per cell). Bars show luciferase activity detected in lysates of infected cells 20 h postinfection. Each data point corresponds to an average of three measurements.
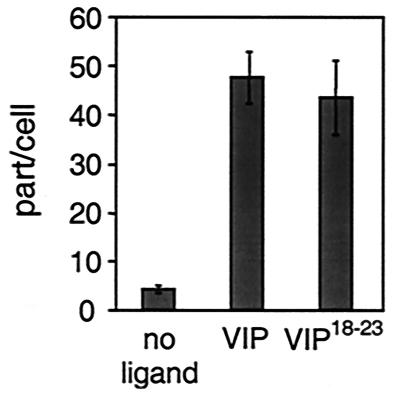
The observed enhancement of gene transfer by the Ad5Luc.NNVIP vectors and its VIP dependence, taken together with the lack of VPAC involvement in virus binding, all suggested that the incorporation of VIP into the Ad capsid promotes nonspecific, although rather efficient binding of these viruses to cell surface molecules other than known VPAC receptors. The fact that VIP is a highly basic polypeptide with an isoelectric point above 11 suggested that the augmentation of gene transfer by the VIP-modified Ad vectors may be due to a charge-based interaction of the virions with target cells. We chose to test this hypothesis by generating a new Ad vector containing a mutated VIP sequence in which the positions of residues Ala18 and Leu23 were exchanged. It has been reported previously that Leu23 plays a key role in VIP binding to both VPAC1 and VPAC2 and that its replacement with Ala residues dramatically decreases the affinity of VIP for these receptors (29, 30). Therefore, by switching Ala18 to Leu18 and Leu23 to Ala23 within VIP, we designed its derivative VIP18-23, whose ability to specifically bind native VIP receptors was impaired but whose molecular charge was not affected. An Ad vector incorporating this mutated VIP within the HI loop of the fiber, Ad5Luc.10VIP18-23, was designed essentially as described above for vectors of the Ad5Luc.NNVIP series. The virus was then radiolabeled with [3H]thymidine and used for a cell-binding assay side by side with radiolabeled Ad5Luc.10VIP and Ad5Luc1. As shown in Fig. 8, we observed similar binding efficiencies for both VIP-containing vectors on CHO/VPAC1 cells, which was approximately an order of magnitude higher than that seen for the control vector. Therefore, these findings showed that the enhancement of the gene transfer by Ad5Luc.NNVIP vectors was due to more efficient binding of these viruses to target cells and also that this binding, although VIP mediated, is not dependent on VPAC receptors and is most likely caused by charge-based interactions of VIP sequence, native or scrambled, with unidentified cell surface molecules.
FIG. 8.
Analysis of binding of Ad5Luc.10VIP and Ad5Luc.10VIP18-23 viruses to CHO/VPAC1 cells. [3H]thymidine-labeled viruses were incubated for 1 h with cells at an MOI of 3,500 viral particles per cell. Unbound virions were then removed by washing the cells with the virus-free medium, and the number of particles bound per cell was calculated based on the specific radioactivities of the virions. Each data point corresponds to an average of three measurements. Cell attachment of the labeled Ad5Luc1 vector, which was used as a control, is shown by the “no ligand” bar.
DISCUSSION
Previous work on the genetic alteration of Ad vectors' tropism has primarily been focused on the modification of Ad fiber using short peptides as targeting ligands. Whereas those efforts have shown the feasibility of such a targeting approach, they have also identified a number of shortcomings of this strategy. One drawback is related to the size limitations imposed on the ligand by the structural constrains of the fiber knob domain, where the ligands are typically incorporated. For instance, it has been shown that ligands whose size exceeds a limit of about 25 to 30 aa residues cannot be configured into the carboxy terminus of the fiber, as they appear to destabilize the fiber structure (13, 44). These early findings narrowed the range of potential ligands for Ad targeting to short peptides and thus a priori limited the ranges of prospective target receptors and cell types.
While the structural properties of the HI loop of the Ad fiber, which was recently identified as an alternative locale for incorporation of targeting ligands (5, 16), appears to favor the insertion of larger ligands, thereby expanding the repertoire of targeting moieties and the versatility of the strategy overall, this potential advantage of the loop has never been tested. Therefore, the purpose of this study was to assess in a systematic manner the capacity of the HI loop to accommodate ligands.
To reach this end, we chose to incrementally increase the size of the HI loop by incorporating into it a series of ligand-mimicking polypeptide sequences and test the ability of the resultant fibers to maintain their functional trimeric structure, be incorporated into an Ad particle, and mediate receptor binding. Further, we chose to approach this goal by transferring fragments of the integrin-binding loop of the Ad5 penton base protein into the HI loop of the fiber. Several considerations supported this choice. First, the loop in the penton base does not have a rigid structure. Thus, fragments of this loop incorporated into the fiber were not expected to cause any significant structural conflict with the carrier protein. Second, being a component of an Ad5 capsid protein, this loop does not require any posttranslational modification, which would not be normally available for the Ad fiber. Third, the loop contains an apical RGD motif, which possesses a receptor-binding capacity and upon transfer to the fiber could be expected to alter the tropism of the virus. Fourth, the length of the loop appeared to be sufficient to derive ligand mimics large enough to destroy the structure of the fiber knob domain upon insertion, thereby defining the size limit for similar ligands. Finally, should the viruses containing the fibers modified with the penton base-derived sequences be rescued and the integrin-binding ability of the RGD peptide in the context of these chimeric proteins be retained, it would then be possible to use these fibers as frameworks for incorporation of novel ligands in place of the RGD. In this scenario, the fragments of the penton base protein flanking the site of the ligand insertion would be expected to function as flexible linkers by extending the ligand away from the knob and facilitating its correct folding and receptor binding.
Realization of this strategy led to the generation of eight Ad5Luc.NNRGD vectors incorporating within the HI loop of the fiber inserts ranging in size from 13 to 83 aa residues. According to our data on the productivity and infectivity of Ad5Luc.NNRGD viruses, these significant increases of the size of the knob domain (up to 46%) did not cause any dramatic reduction in these key properties of the virions. The maximum decrease in the yields and infectivities was a two- to threefold reduction, which was observed for the vectors incorporating the largest inserts (63 to 83 aa). These data suggest that, as expected based on the structure of the HI loop and its location within the knob relative to the CAR-binding site, rather significant enlargements of its size via incorporation of targeting sequences can be achieved without compromising the productivity or infectivity of the vectors. However, the gradual decrease in both parameters seen in the vectors bearing longer insertions implies that enlargements of the loop significantly exceeding those described here would most likely cause more serious damage to the fiber structure and function and should thus be avoided.
We have also shown that the RGD tripeptide contained in the fibers of all modified virions retains its natural ability to bind integrins. Thus, the position of the RGD sequence within the modified HI loop favors its successful interaction with integrins, thereby implying that it may also be optimal for other targeting ligands. Interestingly, gene transfer by various RGD-containing vectors to CAR-negative U118MG cells clearly showed that vectors of the Ad5Luc.NNRGD series are just as efficient as the previously designed vector Ad5LucFHIRGD (5) in transducing these cells. Of note, Ad5LucFHIRGD contains the RGD peptide in a configuration which is considered optimal for binding integrins. The RGD-4C peptide incorporated into the fiber of Ad5LucFHIRGD contains two pairs of cysteine residues (CDCRGDCFC), introduced into the sequence to stabilize it via formation of disulfide bonds between the cysteines. In contrast, none of the penton base-derived polypeptides incorporated into the Ad5Luc.NNRGD vectors contains such constraining cysteines. Equal efficiencies of gene transfer observed in our study for all RGD-containing vectors, regardless of the presence of cysteine residues, show that Ad-targeting peptides do not have to be constrained to be functional. This conclusion is supported by the successful targeting of Ad vectors with peptide ligands that lack constraining cysteines (28, 45). Additionally, the formation of disulfide bonds requires the involvement of disulfide isomerases, enzymes that are not found in the cytoplasm or nucleoplasm, the cellular compartments where trafficking of the newly synthesized Ad proteins and subsequent virus assembly take place. These theoretical considerations, taken together with our comparison of the infectivities of Ad5LucFHIRGD and Ad5Luc.NNRGD vectors, imply that for the purpose of Ad targeting, the cysteine-constrained peptides do not have any advantage over their linear counterparts.
Our findings on the productivity, infectivity, and expanded tropism of Ad5Luc.NNRGD viruses supported subsequent attempts to use these viruses as prototypes for the derivation of Ad vectors targeted with known polypeptide ligands to receptors, which would be useful for gene therapy. To facilitate this task, we first derived a series of the fiber shuttle vectors, in which the genes encoding the modified fibers were redesigned to accommodate ligand-encoding inserts in place of the RGD codons. These vectors of the pHI.PBNN series allow for the simple and efficient cloning of the desired sequences within the extended HI loops of all eight fiber constructs derived herein, thereby providing flexibility in the selection of linkers flanking the targeting moiety within the fiber. We then reasoned that VIP would be a rational choice as a targeting ligand for Ad. Elevated expression of the VIP receptors VPAC1 and VPAC2 in a number of malignant tissues (34) suggested that VIP targeting of Ad may result in a vector suitable for a wide range of therapeutic applications. The relatively simple structure of VIP and its moderate size provided additional reasons for the insertion of this molecule into the Ad fiber. Several considerations, however, complicated this otherwise rational choice. First, it is known that VIP is normally produced as a precursor protein which has to undergo a multistep processing before the final, mature configuration of VIP is achieved. It is further known that one of the final steps of VIP maturation involves amidation of its carboxy-terminal residue. Obviously, this step could not take place in a VIP which has been incorporated into the HI loop of the fiber, as its carboxy terminus is fused with the sequence of the loop. The way around this problem was to use a configuration of VIP corresponding to its incompletely processed form. Specifically, it has been shown that 31-aa-long VIP (VIP31), which differs from the fully processed 28-aa-long polypeptide by a carboxy-terminal Gly-Lys-Arg sequence, possesses specific VPAC-binding activity characteristic of the fully processed and amidated VIP (8, 38). Led by these considerations, we incorporated VIP31 into extended HI loops of the fibers containing 5-, 10-, 35-, and 40-aa-long flanking sequences derived from the Ad5 penton base.
Experiments employing the resultant Ad vectors for gene transfer to CHO-derived cells stably expressing VPAC1 or VPAC2 gave unexpected results. While we observed very significant (10- to 100-fold) augmentation of gene delivery to these cells by the VIP-modified Ad vectors, similar levels of transduction were also demonstrated using VPAC-negative parental CHO cells. Therefore, while modification of the Ad5 fiber with VIP did result in tropism expansion of the vector, this infectivity enhancement was not dependent on the VPAC status of the target cells. Importantly, the use of synthetic VIP as a competitor of binding resulted in an efficient, dose-dependent inhibition of Ad5Luc.NNVIP-mediated gene delivery, thereby further implying that it was incorporation of VIP into the capsids of these vectors that caused the alteration of viral tropism. A comparison of the cell-binding capacity of radiolabeled Ad5Luc.10VIP to that of the control vector demonstrated that the augmentation of infectivity of Ad5Luc.10VIP was due to its increased binding to target cells. Taken together, these results suggested that a mechanism of VIP-mediated cell binding other than the VIP-VPAC interaction was responsible for the observed increase of transduction efficiency.
It remains unclear why VIP incorporated into the Ad5 fiber failed to bind its cognate receptors while being able to mediate Ad attachment to another cell surface molecule(s). It should be noted in this regard that the generation of functional genetic fusions of VIP with other proteins has not been reported previously. Therefore, it is largely unknown whether VIP can retain its receptor binding specificity when it is included in such fusions. Perhaps binding of VIP to its native receptors requires that either one or both termini of the polypeptide should not be engaged in covalent bonding with other protein sequence.
While specific targeting to VPAC receptors has not been achieved in this work, the Ad5Luc.NNVIP series of viruses described herein may find utility as gene delivery vehicles to cells which, similar to CHO cells, are refractory to transduction by regular Ad5 vectors while supporting infection by VIP-modified Ad. The rationale for using VIP-modified vectors in this manner is the same as that underlying the use of Ad vectors, whose tropism was expanded by directing them to either broadly expressed types of cell surface molecules, such as integrins (5, 44) and heparan sulfate-containing receptors (43, 44), or to the unidentified native receptors for alternative serotypes of Ad, whose elevated or preferential expression in the studied cells or tissues supports Ad targeting (18, 37, 39).
In the aggregate, the findings described here show the feasibility of incorporation of relatively large targeting sequences into the HI loop of the fiber knob domain and thus expand the range of potential ligands and target receptors and tissues. However, the results with respect to the yields and infectivities of the Ad vectors containing extended HI loops suggest that ligands significantly larger than those used in our work would likely have a detrimental effect on Ad viability. While expanding the practical utility of Ad targeting via the modification of the fiber knob domain, our work also provides a rationale for further development of Ad tropism modification strategies based on fiber replacement (15, 21, 42). The approaches that have emerged most recently solve the problem of structural and functional compatibility between the ligand and Ad capsid components by disengaging the folding of the ligand and the ligand-presenting protein. Finally, as these Ad targeting strategies appear to have advantages and disadvantages, their strengths may be further increased by designing mosaic Ad virions incorporating both modified full-size fibers and fiber-replacing protein chimeras.
Acknowledgments
We thank Jeffrey Engler and Glen Nemerow for providing antifiber and anti-penton base antibodies to us. Mark Laburthe is thanked for making CHO, CHO/VPAC1, and CHO/VPAC2 cells available for this work. We are grateful to Joanne T. Douglas, Alexander Pereboev, and Nikolay Korokhov for critical reading of the manuscript.
This work was supported by the following grants: R01 CA86881, R01 CA83821, P50 CA89019, and DAMD17-02-1-0002.
REFERENCES
- 1.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 2.Bewig, B., and W. E. Schmidt. 2000. Accelerated titering of adenoviruses. BioTechniques 28: 870-873. [DOI] [PubMed] [Google Scholar]
- 3.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chroboczek, J., R. W. Ruigrok, and S. Cusack. 1995. Adenovirus fiber. Curr. Top. Microbiol. Immunol. 199:163-200. [DOI] [PubMed] [Google Scholar]
- 5.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Einfeld, D. A., D. E. Brough, P. W. Roelvink, I. Kovesdi, and T. J. Wickham. 1999. Construction of a pseudoreceptor that mediates transduction by adenoviruses expressing a ligand in fiber or penton base. J. Virol. 73:9130-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freimuth, P. 1996. A human cell line selected for resistance to adenovirus infection has reduced levels of the virus receptor. J. Virol. 70:4081-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gafvelin, G., M. Andersson, R. Dimaline, H. Jornvall, and V. Mutt. 1988. Isolation and characterization of a variant form of vasoactive intestinal polypeptide. Peptides 9:469-474. [DOI] [PubMed] [Google Scholar]
- 9.Graham, F. L., and L. Prevec. 1991. Manipulation of adenovirus vectors. Methods Mol. Biol. 7:109-128. [DOI] [PubMed] [Google Scholar]
- 10.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 11.Haviv, Y. S., and D. T. Curiel. 2001. Conditional gene targeting for cancer gene therapy. Adv. Drug Deliv. Rev. 53:135-154. [DOI] [PubMed] [Google Scholar]
- 12.Hemmi, S., R. Geertsen, A. Mezzacasa, I. Peter, and R. Dummer. 1998. The presence of human coxsackievirus and adenovirus receptor is associated with efficient adenovirus-mediated transgene expression in human melanoma cell cultures. Hum. Gene Ther. 9:2363-2373. [DOI] [PubMed] [Google Scholar]
- 13.Hong, J. S., and J. A. Engler. 1991. The amino terminus of the adenovirus fiber protein encodes the nuclear localization signal. Virology 185:758-767. [DOI] [PubMed] [Google Scholar]
- 14.Hong, J. S., and J. A. Engler. 1996. Domains required for assembly of adenovirus type 2 fiber trimers. J. Virol. 70:7071-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krasnykh, V., N. Belousova, N. Korokhov, G. Mikheeva, and D. T. Curiel. 2001. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J. Virol. 75:4176-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krasnykh, V., I. Dmitriev, G. Mikheeva, C. R. Miller, N. Belousova, and D. T. Curiel. 1998. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J. Virol. 72:1844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krasnykh, V. N., J. T. Douglas, and V. W. van Beusechem. 2000. Genetic targeting of adenoviral vectors. Mol. Ther. 1:391-405. [DOI] [PubMed] [Google Scholar]
- 18.Krasnykh, V. N., G. V. Mikheeva, J. T. Douglas, and D. T. Curiel. 1996. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J. Virol. 70:6839-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon, R. P., T. Hedlund, S. J. Meech, S. Li, J. Schaack, S. P. Hunger, R. C. Duke, and J. DeGregori. 1998. Adenoviral-mediated gene transfer in lymphocytes. Proc. Natl. Acad. Sci. USA 95:13159-13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y., R. C. Pong, J. M. Bergelson, M. C. Hall, A. I. Sagalowsky, C. P. Tseng, Z. Wang, and J. T. Hsieh. 1999. Loss of adenoviral receptor expression in human bladder cancer cells: a potential impact on the efficacy of gene therapy. Cancer Res. 59:325-330. [PubMed] [Google Scholar]
- 21.Magnusson, M. K., S. S. Hong, P. Boulanger, and L. Lindholm. 2001. Genetic retargeting of adenovirus: novel strategy employing “deknobbing” of the fiber. J. Virol. 75:7280-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115-125. [DOI] [PubMed] [Google Scholar]
- 23.Michael, S. I., J. S. Hong, D. T. Curiel, and J. A. Engler. 1995. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 2:660-668. [PubMed] [Google Scholar]
- 24.Miller, C. R., D. J. Buchsbaum, P. N. Reynolds, J. T. Douglas, G. Y. Gillespie, M. S. Mayo, D. Raben, and D. T. Curiel. 1998. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res. 58:5738-5748. [PubMed] [Google Scholar]
- 25.Mittal, S. K., M. R. McDermott, D. C. Johnson, L. Prevec, and F. L. Graham. 1993. Monitoring foreign gene expression by a human adenovirus-based vector using the firefly luciferase gene as a reporter. Virus Res. 28:67-90. [DOI] [PubMed] [Google Scholar]
- 26.Mizuguchi, H., N. Koizumi, T. Hosono, N. Utoguchi, Y. Watanabe, M. A. Kay, and T. Hayakawa. 2001. A simplified system for constructing recombinant adenoviral vectors containing heterologous peptides in the HI loop of their fiber knob. Gene Ther. 8:730-735. [DOI] [PubMed] [Google Scholar]
- 27.Nalbantoglu, J., G. Pari, G. Karpati, and P. C. Holland. 1999. Expression of the primary coxsackie and adenovirus receptor is downregulated during skeletal muscle maturation and limits the efficacy of adenovirus-mediated gene delivery to muscle cells. Hum Gene Ther. 10:1009-1019. [DOI] [PubMed] [Google Scholar]
- 28.Nicklin, S. A., D. J. Von Seggern, L. M. Work, D. C. Pek, A. F. Dominiczak, G. R. Nemerow, and A. H. Baker. 2001. Ablating adenovirus type 5 fiber-CAR binding and HI loop insertion of the SIGYPLP peptide generate an endothelial cell-selective adenovirus. Mol. Ther. 4:534-542. [DOI] [PubMed] [Google Scholar]
- 29.Nicole, P., L. Lins, C. Rouyer-Fessard, C. Drouot, P. Fulcrand, A. Thomas, A. Couvineau, J. Martinez, R. Brasseur, and M. Laburthe. 2000. Identification of key residues for interaction of vasoactive intestinal peptide with human VPAC1 and VPAC2 receptors and development of a highly selective VPAC1 receptor agonist. Alanine scanning and molecular modeling of the peptide. J. Biol. Chem. 275:24003-24012. [DOI] [PubMed] [Google Scholar]
- 30.O'Donnell, M., R. J. Garippa, N. C. O'Neill, D. R. Bolin, and J. M. Cottrell. 1991. Structure-activity studies of vasoactive intestinal polypeptide. J. Biol. Chem. 266:6389-6392. [PubMed] [Google Scholar]
- 31.Okegawa, T., Y. Li, R. C. Pong, J. M. Bergelson, J. Zhou, and J. T. Hsieh. 2000. The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res. 60:5031-5036. [PubMed] [Google Scholar]
- 32.Okegawa, T., R. C. Pong, Y. Li, J. M. Bergelson, A. I. Sagalowsky, and J. T. Hsieh. 2001. The mechanism of the growth-inhibitory effect of coxsackie and adenovirus receptor (CAR) on human bladder cancer: a functional analysis of car protein structure. Cancer Res. 61:6592-6600. [PubMed] [Google Scholar]
- 33.Pickles, R. J., D. McCarty, H. Matsui, P. J. Hart, S. H. Randell, and R. C. Boucher. 1998. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J. Virol. 72:6014-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reubi, J. C., U. Laderach, B. Waser, J. O. Gebbers, P. Robberecht, and J. A. Laissue. 2000. Vasoactive intestinal peptide/pituitary adenylate cyclase-activating peptide receptor subtypes in human tumors and their tissues of origin. Cancer Res. 60:3105-3112. [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. Russell. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simoncsits, A., M. L. Tjornhammar, M. Kalman, I. Cserpan, G. Gafvelin, and T. Bartfai. 1988. Synthesis, cloning and expression in Escherichia coli of artificial genes coding for biologically active elongated precursors of the vasoactive intestinal polypeptide. Eur. J. Biochem. 178:343-350. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson, S. C., M. Rollence, J. Marshall-Neff, and A. McClelland. 1997. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J. Virol. 71:4782-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart, P. L., C. Y. Chiu, S. Huang, T. Muir, Y. Zhao, B. Chait, P. Mathias, and G. R. Nemerow. 1997. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 16:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Beusechem, V. W., A. L. van Rijswijk, H. H. van Es, H. J. Haisma, H. M. Pinedo, and W. R. Gerritsen. 2000. Recombinant adenovirus vectors with knobless fibers for targeted gene transfer. Gene Ther. 7:1940-1946. [DOI] [PubMed] [Google Scholar]
- 43.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 44.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia, H., B. Anderson, Q. Mao, and B. L. Davidson. 2000. Recombinant human adenovirus: targeting to the human transferrin receptor improves gene transfer to brain microcapillary endothelium. J. Virol. 74:11359-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zabner, J., P. Freimuth, A. Puga, A. Fabrega, and M. J. Welsh. 1997. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J. Clin. Investig. 100:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng, G. 1998. Sticky-end PCR: new method for subcloning. BioTechniques 25:206-208. [DOI] [PubMed] [Google Scholar]