Abstract
Epstein-Barr virus (EBV) is a lymphotrophic herpesvirus infecting most of the world's population. It is associated with a number of human lymphoid and epithelial tumors and lymphoproliferative diseases in immunocompromised patients. Recent studies have shown an in vitro and in vivo interaction between the EBV nuclear antigen 3C (EBNA3C) and the metastatic suppressor Nm23-H1, known to be downregulated in human invasive breast carcinoma. In this study, we have identified the domain of EBNA3C that specifically binds to Nm23-H1. This domain lies within the region comprising amino acids 637 to 675 of EBNA3C flanked by the proline- and glutamine-rich domains. Furthermore, we show that Nm23-H1 activates transcription when fused to the Gal4 DNA-binding domain and is coexpressed with a luciferase reporter construct containing the Gal4 binding sites upstream of a basal promoter. Gal4-Nm23-H1, when tethered to the promoter by binding to the Gal4 DNA binding sequences, consistently activated transcription. The level of activation increased when increasing amounts of Gal4-Nm23-H1 were introduced into the system. Moreover, EBNA3C when cotransfected with Gal4-Nm23-H1 enhanced the transcriptional activity. These results suggest that Nm23-H1 may have intrinsic transcription activities in EBV-infected cells and that this activity can be modulated in the presence of the essential latent antigen EBNA3C.
Epstein-Barr virus (EBV) is a human oncogenic gammaherpesvirus predominantly infecting the oropharyngeal epithelium and B lymphocytes (17, 33). It is the etiological agent of infectious mononucleosis and is associated with a wide variety of human malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's lymphoma, AIDS-associated lymphoma, and other lymphoproliferative diseases (17, 33). Recent studies have also shown the presence of EBV DNA and latent EBV nuclear antigen 1 (EBNA1) in invasive human breast carcinomas (7; A. A. Brink, A. J. van Den Brule, P. van Diest, and C. J. Meijer, Letter, J. Natl. Cancer Inst. 92:655-656, 2000). In vitro infection of primary B lymphocytes with EBV gives rise to lymphoblastoid cell lines (LCLs). In immortalized LCLs, EBV persists as an episome from which characteristic repertoires of latent genes are expressed. This type of latency is referred to as type III latency (17, 33). There are at least 11 genes expressed during latent infection, and these include the six EBNAs, three latent membrane proteins (LMPs), and two EBV early RNAs (EBERs) (17, 33, 44). Genetic recombination studies have revealed that EBNA1, -LP, -2, -3A, and -3C and LMP1 are critical for growth transformation of primary B lymphocytes, whereas EBNA3B, LMP2A, LMP2B, and EBERs are dispensable for the immortalization of primary B cells (9, 16, 17, 45). EBNA1 protein binds to viral DNA and is involved in the maintenance of the EBV genome as a circular episome in the infected cell (17, 32). The additional EBNAs function primarily as regulators of transcription in association with other cellular transcription factors (17, 33).
Three distinct genes that are similar in structure and are tandemly arranged in the EBV genome encode the EBNA3 family of proteins (17, 34). The EBNA3 proteins all share limited homology in a region near their amino terminus (46). A conserved domain within this amino terminus mediates binding to the transcriptional regulator RBP-Jκ (36, 46). This results in downregulation of transcription from the major EBV latent promoters. EBNA3 proteins compete with EBNA2 for binding to RBP-Jκ and prevent binding of RBP-Jκ to the cognate DNA element (34).
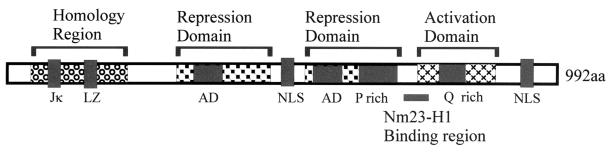
The basic structure of EBNA3C protein shows a large polypeptide of 992 amino acids (aa) (Fig. 1), which contains a potential leucine zipper motif near the amino terminus, acidic domains, a glutamine- and proline-rich domain that functions as a transactivation domain in gene fusion assays in its carboxy terminus, and several arginine and lysine residues that are potentially important for nuclear translocation (17, 34, 40). EBNA3C acts both as an activator and as a repressor of transcription in transient reporter assays. EBNA3C acts as a repressor of transcription when tethered to DNA as a fusion protein with the Gal4 DNA-binding domain (DBD) (6). Furthermore, EBNA3C can interact with the cellular transcription factor RBP-Jκ/CBF1 (35). Expression of EBNA3C was shown to derepress another essential EBV latent antigen, LMP1, in cells arrested in the G1 phase of the cell cycle (3). Moreover, EBNA3C can function as an immortalizing oncoprotein, similar to the E7 and E1A of the small DNA viruses (28). EBNA3C also binds to a transcriptional repression complex that includes histone deacetylase 1, which is targeted to Cp by the DNA binding protein RBP-Jκ (30, 31). EBNA3C contributes to the negative autoregulatory control loop of the major latent promoters (28). EBNA3C also activates the LMP1 promoter in conjunction with EBNA2 through the Spi-1/Spi-B binding site (47).
FIG. 1.
EBNA3C protein and its functional domains. Genetic recombination studies indicated that the introduction of a stop codon at aa 365 renders the gene incapable of growth transformation (45). The glutamine- and proline-rich transactivation domain towards the carboxy-terminal region, the Jκ binding site, the leucine zipper (LZ) towards the amino-terminal region, putative nuclear localization signals (NLS), and the acidic domains (AD) are indicated. E3C, EBNA3C.
Previous genetic studies indicated that introduction of an amber stop codon after aa 365 in the EBNA3C open reading frame resulted in EBV recombinants incapable of growth transformation of primary B lymphocytes (45). These results clearly indicated that interactions downstream of aa 365 might play an important role in EBV immortalization. One of the proteins interacting downstream of aa 365 is a cellular protein, Nm23-H1, functionally associated with suppression of metastasis (42). Interestingly, EBNA3C reverses the ability of Nm23-H1 to suppress the migration of Burkitt's lymphoma cells and breast carcinoma cells in vitro (42).
The nm23 gene family has nucleoside dinucleotide phosphate kinase enzyme activity and is highly conserved among a wide variety of eukaryotic species (19). Additionally, the nm23 homologs are also shown to be involved in various cellular processes, such as stimulation of transcription, cell differentiation, and cell proliferation (11). Eight distinctly different genes (nm23-H1 to -H8) have been identified in humans (19). The proteins Nm23-H1 and -H2 have similar amino acid sequences, but differ in function and cellular localization (18, 29, 41). Nm23-H1 is known to function as a metastatic suppressor, whereas Nm23-H2 binds DNA and activates transcription of c-myc (41). Nm23-H1 and Nm23-H2 proteins are localized to the cytoplasm; however, studies carried out with isoform-specific antibody showed that Nm23-H2 is predominantly located in the nucleus (18, 29). Functional studies demonstrated that suppression of metastasis was observed in several tumor cell lines transfected with Nm23-H1 (20, 22, 23, 39).
Although the exact role of Nm23-H1 in metastasis is not clearly understood, the experimental data accumulated thus far implicate Nm23-H1 as playing a role in regulating the metastasis of human cancer. Additionally, EBV is associated with a number of lymphoid and epithelial cancers. However, there has been no previous study linking a known etiologic agent and the metastatic suppressor protein. Nevertheless, a recent study has shown that the EBV antigen EBNA3C can interact with Nm23-H1 (42). Now, we show that the region of EBNA3C that binds to NM23-H1 is located within the proline- and glutamine-rich domains associated with the transcriptional activation function of EBNA3C.
MATERIALS AND METHODS
Cell lines and constructs.
Cells from the human embryonic kidney (HEK) 293T cell line used in transient transfection experiments were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 25 U of penicillin-streptomycin per ml, and 20 μg of Gentamicin per ml.
Glutathione S-transferase (GST)-Nm23-H1 was prepared as described previously (42). PGEX vector and EBNA3C truncations were cut with EcoRI and SmaI for the GST-EBNA3C aa 582 to 792 fusion construct (EBNA3C 582-792). The other two GST fusion constructs, EBNA3C 637-742 and 629-910, were prepared as in-frame GST fusions as described previously (10).
EBNA3C expression constructs were prepared as in-frame myc fusion proteins cloned into pA3M vector (4). For the preparation of the pA3M-EBNA3C 366-623 clone, the PCR-amplified product was cut with BamHI and NotI and ligated into the BamHI and NotI sites of pA3M in frame with the myc epitope. Similarly, the pA3M-EBNA3C 621-992 construct was prepared by amplification by PCR and digestion of the amplified product with EcoRI and NotI, followed by ligation of these sites in frame with the myc epitope on the pA3M vector. The pA3M-EBNA3C 621-675 and 675-714 constructs were PCR amplified as described before and cloned into the EcoRI and EcoRV sites of pA3M.
The pSG5 clones of EBNA3C were prepared from the pBS KS+ clones. Fragments containing the specific EBNA3C domains were excised from the pBS construct with SmaI, and the overhanging ends were filled with the Klenow fragment of DNA polymerase. The pSG5 vector was prepared by digesting the vector with BglII and end filling with Klenow DNA polymerase. The pBS clones were prepared as described previously (43).
In vitro binding and Western blot assays.
GST-Nm23-H1 and the GST-EBNA3C mutants used in the binding assay were prepared as previously described (42). Briefly, cells were induced and then pelleted by centrifugation. Pellets were sonicated, and the cell debris was removed. The supernatant collected was rotated with glutathione Sepharose beads washed with NETN buffer (0.5% NP-40, 20 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA) supplemented with protease inhibitors (100 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg of pepstatin per ml, 1 μg of aprotinin per ml) and stored at 4°C before use (42).
The EBNA3C full-length protein and the truncated proteins, as well as the Nm23-H1 proteins cloned into the pA3M or pSG5 expression vector, were transcribed and translated in vitro with [35S]Met/Cys translabel and the T7 TNT system (Promega, Inc.). The in vitro-translated proteins were precleared first with glutathione Sepharose beads in the binding buffer (1× phosphate-buffered saline, 0.1% NP-40, 0.5 mM dithiothreitol [DTT], 10% glycerol, 1 mM PMSF, 2 mg of aprotinin per ml) for 30 min. The beads were removed by centrifugation, and the proteins were incubated with glutathione Sepharose-bound fusion proteins for 2 h at 4°C. The beads were then collected and washed with the binding buffer. Proteins bound to the GST and GST fusion proteins were denatured with sodium dodecyl sulfate (SDS)-β-mercaptoethanol lysis buffer and were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE [8% or 15% polyacrylamide]). The dried gel was analyzed with a PhosphorImager and quantified with the ImageQuant program (Molecular Dynamics).
Cell lysates from the reporter assay were used for Western blot analysis of Nm23-H1 and EBNA3C. SDS-lysis buffer was added to the lysed cells, and the proteins were fractionated by SDS-PAGE with either 15% polyacrylamide (Nm23-H1) or 8% polyacrylamide (EBNA3C). The fractionated proteins were transferred to a 0.45-μm-pore-diameter nitrocellulose membrane. The membranes were blocked with 5% milk and then incubated with either mouse Nm23-H1 antibody for detection of Nm23-H1 (Santa Cruz, Inc.) or A10 monoclonal antibody for detection of EBNA3C (37). The membranes were washed and then incubated with horseradish peroxidase (HRP)-conjugated antimouse secondary antibody, and the proteins were visualized by chemilluminescence against X-ray film.
Transfections and reporter assay.
HEK 293T cell lines were transiently transfected with the myc- or Gal4-tagged Nm23-H1, myc- or Gal4-tagged full-length EBNA3C, or EBNA3C 634-992. The EBNA3C clones in the eukaryotic expression vector were transfected either alone or with pM-nm23-H1. The pFr reporter construct (Stratagene, Inc.), which contains five Gal4 binding sites and a TATA box, was added to all transfections. The total DNAs were balanced with parental vector for Nm23-H1 and EBNA3C DNA, and the transfections were normalized for efficiency of transfection by counting green fluorescent protein (GFP)-positive cells with an IX70 inverted Olympus fluorescence microscope. Cells were harvested for transfections at approximately 70 to 80% confluence by trypsinization. Ten million cells were transiently transfected by electroporation at 210 V and 975 μF with a Bio-Rad Gene Pulser II. At 24 h posttransfection, cells were collected, washed with 1× phosphate-buffered saline (Invitrogen-Gibco), and lysed with 200 μl of reporter lysis buffer (Promega, Inc.). Luciferase activity was measured with an Opticomp Luminometer (MGM Instruments, Inc.). All transfections were done in triplicate two times, and the mean was plotted.
RESULTS
The metastatic suppressor protein Nm23-H1 interacts with the carboxy terminus of EBNA3C.
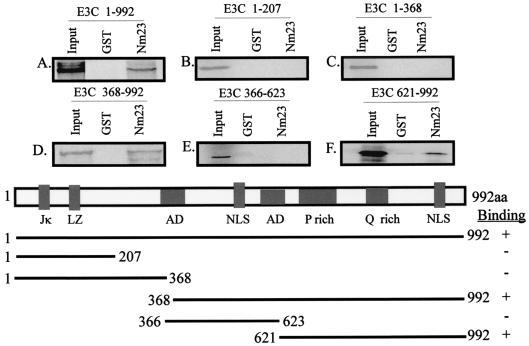
Genetic studies in which a stop codon was introduced at the SpeI site of EBNA3C showed that the region of EBNA3C from aa 366 to aa 992 is essential for growth transformation (45). Therefore, we wanted to determine the interacting domain of EBNA3C that bound Nm23-H1. Truncated EBNA3C clones were in vitro translated and labeled with [35S]Met/Cys Translabel (ICN). The labeled EBNA3C proteins were then bound to either GST alone or GST-Nm23-H1. The results presented in Fig. 2 show that the full-length EBNA3C protein bound to GST-Nm23-H1 as expected and that binding was also seen with other fusion constructs within the carboxy-terminal 627-aa region of EBNA3C (Fig. 2). The efficiency of binding calculated for GST and GST-Nm23-H1 with full-length EBNA3C indicated that the level of GST-Nm23-H1 binding is 375 times greater than that seen with GST alone (compare lanes 1 and 3 in Fig. 2A). Quantitation was done with the Molecular Dynamics ImageQuant program. Moreover, arbitrary counts obtained for EBNA3C aa 368 to 992 indicated that the efficiency of binding was about 10% compared to the input level of the truncated protein (compare lanes 1 and 3 in Fig. 2D and F). It should be noted that little or no binding was seen with the amino-terminal region of EBNA3C (1 to 368 aa), thereby indicating that the amino-terminal region does not interact with Nm23-H1. These results corroborate previous yeast data indicating that a region in the carboxy-terminal region of EBNA3C is required for mediating the interaction with Nm23-H1 (42).
FIG. 2.
Carboxy-terminal region of EBNA3C interacts with NM23-H1 in vitro. Full-length EBNA3C (E3C) and EBNA3C clones that have truncations at the carboxy-terminal and amino-terminal regions were in vitro translated and transcribed. The 35S-labeled proteins were incubated with GST as well as GST-Nm23-H1. The beads were then washed with the binding buffer and then fractionated on SDS-PAGE gel (12% polyacrylamide), dried, and visualized with a PhosphorImager. In all of the binding experiments, a 10% input control was used, and the bands were quantified with the ImageQuant program. The schematic giving the amino acid positions of the different clones used is given at the bottom of the figure. LZ, leucine zipper; NLS, nuclear localization signal; AD, acidic domains.
In order to further determine the region of EBNA3C interacting with Nm23-H1 in the carboxy-terminal 627 aa, truncated EBNA3C clones representing aa 366 to 623 and 621 to 992 were tested for their ability to bind Nm23-H1. Truncated EBNA3C proteins were in vitro translated, labeled with [35S]Met/Cys translabel, and bound to glutathione Sepharose beads and GST-Nm23-H1 beads. Bound proteins were fractionated by SDS-PAGE (12% polyacrylamide) and dried. The signal from the 35S-labeled protein quantified with the Molecular Dynamics ImageQuant program indicated that aa 621 to 992 of EBNA3C are required for the binding with Nm23-H1 and that negligible binding was observed with the region comprising aa 366 to 623 (Fig. 2, compare panels E and F). These data suggest that the binding domain lies in the region adjacent to the proline- and glutamine-rich region at the carboxy terminus of EBNA3C.
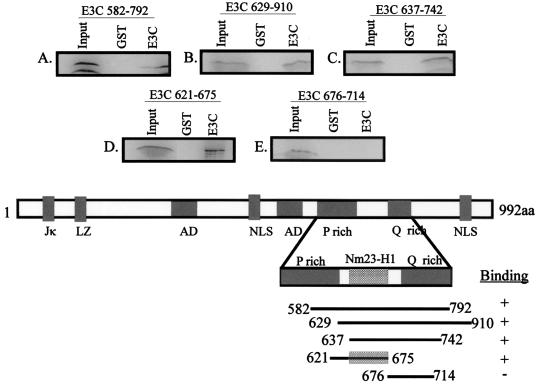
Nm23-H1 specifically interacts with a region of EBNA3C located between the proline- and glutamine-rich domains.
In vitro-translated Nm23-H1 protein was 35S labeled and tested with a number of GST-EBNA3C constructs from the carboxy-terminal aa 628 to 992 to map the binding region. The results of these assays indicate that the region of EBNA3C that interacts with Nm23-H1 lies between aa 629 and 792, which encompasses the proline- and glutamine-rich region of EBNA3C (Fig. 3). Additionally, the data indicated that the amino-terminal region comprising aa 1 to 628 of EBNA3C is not involved in binding, because there is little or no significant interaction seen for the GST-EBNA3C fusion proteins with the amino-terminal 628 aa when compared to the carboxy terminus of EBNA3C (compare Fig. 2 and 3). In vitro-translated Nm23-H1 bound GST-EBNA3C fusion proteins that include aa 582 to 792 and 629 to 910 with similar levels of interaction (Fig. 3A and B). A smaller GST-EBNA3C fusion that includes aa 637 to 742 also had similar levels of binding (Fig. 3C), indicating that the interaction domain lies somewhere within the domain of EBNA3C containing the glutamine-rich activation domain, but not to the adjacent proline-rich domain upstream of the glutamine-rich domain (Fig. 3A and B). Two additional GST-EBNA3C constructs that separate the glutamine-rich domain from the adjacent domain located between the proline- and glutamine-rich domains were further tested. The results showed that the interaction domain of Nm23-H1 with EBNA3C is not at the glutamine- or proline-rich domain, but is located within a 38-aa stretch between the two domains (Fig. 3D and E). GST-EBNA3C 675-714, which includes the glutamine-rich activation domain, does not bind Nm23-H1 (Fig. 3E). However, GST-EBNA3C aa 621 to 675 indicated a specific interaction with an efficiency of about 13% compared to 10% of the input according to arbitrary counts with the Molecular Dynamics ImageQuant program (Fig. 3D). These binding studies using the truncated EBNA3C constructs show that the region comprising aa 637 to 675 of EBNA3C is critical for the interaction of Nm23-H1.
FIG. 3.
The region between the proline- and glutamine-rich domain of EBNA3C binds to the metastatic suppressor NM23-H1. The Nm23-H1 protein was in vitro translated with the TNT-coupled reticulocyte lysate system. The in vitro-translated protein was first incubated with GST beads, and the supernatant was then bound to GST-EBNA3C mutants. After 2 h, the bound proteins were washed with the binding buffer and fractionated by SDS-PAGE (15% polyacrylamide). The bands were then visualized with the PhosphorImager and quantified by ImageQuant analysis. LZ, leucine zipper; NLS, nuclear localization signal; AD, acidic domains.
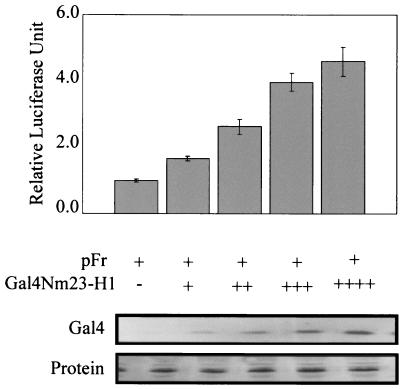
Nm23-H1 can activate transcription of a Gal4 promoter when fused to the Gal4 DBD.
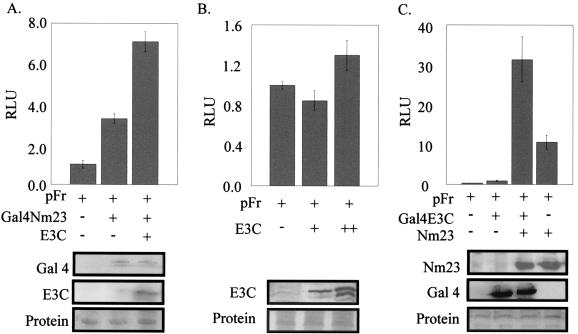
In an effort to address the strong interaction of EBNA3C with Nm23-H1, we wanted to determine if Nm23-H1 itself is associated with transcriptional activity, because EBNA3C has been shown to have both activation as well as repression activities. Nm23-H1 was therefore fused to the Gal4 DBD and tested for transcriptional activity on a Gal4-responsive promoter. Gal4-Nm23-H1 was transfected into 293T cells with pFr, a reporter plasmid that contains the Gal4-responsive sites. The transfections were balanced with vector DNA, and the transfection efficiency was determined by cotransfecting pEGFP (Clontech, Inc.) and manually counting the number of green-fluorescing cells. The results from our analyses indicate that Nm23-H1 is capable of activating transcription when tethered to responsive promoters. As the levels of Gal4-Nm23-H1 increased from 5 μg to 20 μg, the levels of activation increased consistently (Fig. 4). Similar levels of protein loading showed that the levels of activation were due to the changes in levels of Gal4-Nm23-H1 (Fig. 4). Although the levels of activation were not high, it was clear from multiple assays that Nm23-H1 was able to consistently activate transcription when targeted to the promoters and that the activity was dependent on the levels of Nm23-H1 expressed as the levels of activation continued to increase with increasing amounts of Gal4-Nm23-H1 (Fig. 4).
FIG. 4.
Nm23-H1 activates transcription. HEK 293T cells were transfected with the vector containing the Gal4 DBD, Nm23-H1 fused in frame with the Gal4 DBD, and the luciferase reporter plasmid pFr. Increasing amounts of Nm23-H1 were transfected along with the reporter plasmid. Approximately 10 million cells were transfected by electroporation, and 24 h after transfection, the cells were collected, washed, and lysed with lysis buffer, and luciferase activity was measured. The activity of the control vector was taken as 1, and the activities of the others were calculated based on that value. All experiments were done at least twice in duplicate, and the average values are presented. A portion of the lysed cells were run by SDS-PAGE (15% polyacrylamide), transferred onto a 0.45-μm-pore-diameter nitrocellulose membrane, and Western blotted with mouse Nm23-H1 monoclonal antibody (Santa Cruz). Transfection efficiencies were determined on the basis of GFP-positive cells manually counted on slides.
EBNA3C enhances the transcriptional activity of Nm23-H1 in vitro.
To examine the effect of EBNA3C on the transcriptional activity of Nm23-H1, EBNA3C was cotransfected with Gal4-Nm23-H1 in 293T cells along with the Gal4-responsive reporter plasmid. All transfections were balanced for total DNA, and the assays were normalized for transfection efficiency by cotransfecting pEGFP and counting the green-fluorescing cells as before. In addition, EBNA3C alone was transfected along with the reporter construct pFr to determine if EBNA3C has any effect on the reporter plasmid. The data from these assays indicate that EBNA3C gradually increased the transcriptional activity in the presence of Gal4-Nm23-H1 (Fig. 5A). This activity continued to increase with increasing amounts of EBNA3C from 5 μg to 20 μg (data not shown). We were curious as to whether the levels seen were due to the additive effects of EBNA3C and Gal4-Nm23-H1 on the Gal4 reporter plasmids. We tested separately the effects of EBNA3C on the Gal4-responsive promoter, and the results show that there was little or no significant change in activity with increases in the concentration of EBNA3C (Fig. 5B).
FIG. 5.
EBNA3C modulates the transcriptional activity of Nm23-H1. To determine the role of EBNA3C (E3C) in altering the transcriptional activity of Nm23-H1, we transfected into 293T cells various concentrations of EBNA3C (full length) alone or along with pA3M-Nm23-H1 and pFr. Increasing amounts of EBNA3C (5 to 20 μg) were transfected alone to determine if EBNA3C had some activation function in the absence of Nm23-H1. The basic expression vector minus insert was transfected as a control. The cells were collected after 24 h, washed, and then lysed. The luciferase activity was measured (relative luciferase units [RLU]), and the value was calculated as described above. The experiments were repeated twice in duplicate, and the average values are presented. The lysed cells were run on SDS-PAGE (8% polyacrylamide), transferred, and Western blotted for EBNA3C with A10 mouse monoclonal antibody (26).
To determine if the increased activation seen depended on whether EBNA3C or NM23-H1 was tethered, we fused EBNA3C to the Gal4 DBD and tested it in our reporter assay. As expected, the level of activation was increased when Nm23-H1 was expressed in the presence of Gal4-EBNA3C. Moreover, the effect was at least threefold over that of Nm23-H1 or Gal4-EBNA3C alone. These results strongly suggest that Nm23-H1 and EBNA3C can influence transcriptional activity through activation of promoters when coexpressed in the same cell. Additionally, the degree of activation is dependent on the amount of Nm23-H1 or EBNA3C expressed. In our assay, the level of activation was clearly greater when EBNA3C was fused to Gal4. However, when Nm23-H1 was fused to Gal4, the level of protein expressed was much lower—approximately 10-fold when comparing the Western blots for Gal4-Nm23-H1 and pA3M-Nm23-H1. This is a result of the difference in the expression from the simian virus 40 (SV40) promoter compared to the cytomegalovirus (CMV) immediate-early (IE) promoter (compare Gal4 Western blot I in panels A and C, Fig. 5). Nonetheless, the level of transcription activation was clearly synergized when EBNA3C was coexpressed with Nm23-H1 in our in vitro reporter assay. The levels of proteins were determined by Western blotting with specific antibody to Nm23-H1 and EBNA3C fusion proteins.
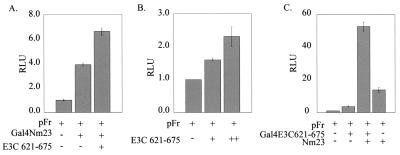
The EBNA3C region comprising aa 621 to 675 activates transcription in the presence of Nm23-H1.
Since Nm23-H1 interacts with the region of EBNA3C located between the proline- and glutamine-rich domains downstream of aa 637, we were curious to know if the residual transcriptional activity of EBNA3C region 621 to 675 when it is fused to Gal4 DBD is enhanced when Nm23-H1 is coexpressed, because this region interacts with Nm23-H1 and does not include the glutamine-rich region of EBNA3C known to activate transcription. The results of the reporter assay in which the Gal4-EBNA3C 621-675 fusion protein is cotransfected with the Gal4-responsive reporter pFr showed a small amount of transcriptional activity for this region of EBNA3C, as expected, due to the presence of only the predominant activation domain of EBNA3C (Fig. 6B and C). The level of activity was gradually increased with increasing amounts of Gal4-EBNA3C 621-675 fusion protein. It should be noted that both repression domains of full-length EBNA3C are absent in this construct. Hence, the level of activation was only about twofold when compared to activation with reporter alone. This activity, however, was further enhanced by the presence of Nm23-H1 (Fig. 6C). In this context, the results suggest that the region downstream of aa 634 of EBNA3C, which includes the Nm23-H1 binding site, can act as a potent activator of transcription in in vitro assays when coexpressed with Nm23-H1. The activity seen when Nm23-H1 was fused to Gal4 and coexpressed with an expression construct of the polypeptide of EBNA3C 621-675 indicated some level of increased transcriptional activity (Fig. 6A). The levels of protein expression for the Gal4 fusion proteins and expression protein were determined by Western blotting against Gal4 and Nm23-H1 with specific antibodies (data not shown).
FIG. 6.
Nm23-H1 synergistically activates the transcription of Gal4-EBNA3C 621-675. Reporter assays were carried out by transfecting into 293T cells the Gal4DBD-EBNA3C (E3C) 621-675 fusion protein along with the responsive reporter construct pFr and the expression construct for Nm23-H1. As a control, vector alone and the Gal4 fusion protein alone were transfected. Twenty-four hours after transfection, cells were collected and tested for luciferase activity (relative luciferase units [RLU]). As before, cells were lysed 24 h posttransfection, and luciferase activity was determined.
DISCUSSION
EBV, an oncogenic gammaherpesvirus, transforms human primary B cells into continuously proliferating LCLs in vitro (17, 33). This process requires the expression of a number of latent genes; of which only five are essential for growth transformation (17, 33). Genetic analysis with recombinant virus revealed that EBNA3C is essential for the efficient immortalization of primary B cells (45). EBNA3C is a large multifunctional protein expressed in EBV-transformed LCLs. The steady-state level of EBNA3C produced is low, but remarkably constant in the LCLs (1, 2).
The carboxy-terminal portion of EBNA3C (aa 365 to 992) critical for the growth transformation of B cells interacts with a number of cellular targets, including ProTα (10) and Nm23-H1. The interaction of ProTα with EBNA3C is likely to result in regulation of cell cycle events, because ProTα has been shown to be associated with cell proliferation (12, 14, 15, 24). Additionally, EBNA3C was shown to interact with p300, a cellular acetyltransferase (10). The interaction of EBNA3C with both ProTα and p300 provided new evidence implicating EBNA3C in modulating the acetylation of cellular factors, including histones (10).
The specific interaction observed between the metastatic suppressor Nm23-H1 and the viral oncoprotein EBNA3C is interesting, because it is the first demonstration of the interaction of a viral oncoprotein with a cellular molecule associated with suppression of metastasis (42). Immunoprecipitation studies and cotransfection studies demonstrated the association of these proteins in vivo in EBV-transformed LCLs and in Burkitt's lymphoma cells expressing EBV latent proteins (42). EBNA3C was also shown to be capable of reversing the ability of Nm23-H1 to suppress the migration of MDA-MB435 cancer cell lines and in Burkitt's cell lines when Nm23-H1 is overexpressed in vitro (42). These results suggested a potential role for EBV in metastasis of EBV-associated human malignancies and prompted us to further investigate in detail the interaction of EBNA3C and Nm23-H1.
In this study, we further delineate the interaction of Nm23-H1 with the specific domain of EBNA3C. The results of the binding assay carried out with in vitro-translated 35S-labeled EBNA3C fusion proteins from the carboxy-terminal and amino-terminal portions indicated that the carboxy-terminal region of EBNA3C essential for the growth transformation interacts strongly with Nm23-H1. There was little or no interaction seen for the amino-terminal region. Similar binding experiments were carried out with mutations at three functionally important potential domains of EBNA3C. These results indicated that the deletion of these functionally important domains does not alter the binding efficiency of Nm23-H1 with EBNA3C. The leucine motif found in DNA-binding transcription factors, a potential DNA binding basic region, and a stretch of amino acids that resembles the polyarginine motifs of a class of RNA-binding transactivator proteins are not essential for the binding of EBNA3C with the metastatic suppressor Nm23-H1. A more detailed analysis indicated that the binding site is located in a region between the proline- and glutamine-rich domains of EBNA3C. This is important, because studies have shown that the proline- and glutamine-rich domains located at the carboxy-terminal portion of EBNA3C can function as a transactivation domain (25). The binding site maps to aa 637 to 675 of EBNA3C and does not include the glutamine- or proline-rich region. These binding studies therefore suggested the involvement of Nm23-H1 in altering the functions of EBNA3C in terms of modulation of transcription.
The interaction of EBNA3C with Nm23-H1 suggests a potential role for Nm23-H1 in transcriptional activity. This role may allow Nm23-H1 to direct a broad spectrum of phenotypic changes through multiple target genes. Alternatively, its biological effect may be due to interaction with a series of cellular protein targets with specific enzymatic activities indirectly modifying the expression of other genes through association with factors that directly target the transcription factors. We wanted to determine the direct transcriptional role of Nm23-H1 and the effect of EBNA3C on any observed transcriptional activity, because this might provide an explanation for some of its biological properties. This study indicates that Nm23-H1 is capable of activating transcription when tethered to DNA. This is in contrast to previous studies with the same methods in which no transactivation potential was seen for either the full-length Nm23-H1 or for the amino-terminal region of Nm23-H1 (8). However, in the same study, the carboxy-terminal region of Nm23-H1 was found to have transactivation potential in a yeast system, suggesting that the activity may be cell type specific (8). After further analysis, we show that in 293T cells, Nm23-H1 can increase activation of a basal promoter when fused to the Gal4 DBD capable of targeting the responsive sites. Strikingly, EBNA3C increased this activation, showing a role for EBNA3C in regulating Nm23-H1 transcriptional activity. The fact that the Nm23-H1 binding domain had relatively little activation suggests a requirement for the glutamine-rich activation domain for EBNA3C activation, as expected. However, in the presence of Nm23-H1, this activity is greatly enhanced, suggesting that Nm23-H1 may have general transcription activities outside its interaction with EBNA3C and may require no DNA binding activities, but only interaction with other potential transcription factors that can target it to promoters in which it may have effects of transcription activation. Nevertheless, the effects of cellular Nm23-H1 may vary in different cell types and tissues.
A series of cellular events are involved in tumor progression and in the development of metastasis (38). The role of the metastatic suppressor protein Nm23-H1 in human neoplasia is most likely complex, and the precise mechanism by which Nm23-H1 interferes with the metastatic process is still somewhat unclear. Therefore, any change in the function of Nm23-H1 gene will have some effect in tumor progression and metastasis. In cell line model systems as well as in cohort studies of human breast, gastric, cervical, ovarian, and hepatocellular carcinomas, as well as melanoma, reduced expression of Nm23-H1 has been correlated with poor patient survival or incidence of high tumor metastatic potential (11). Interestingly in vivo reduction in tumor metastatic potential of melanoma and carcinoma cell lines was observed when transfected Nm23-H1 was overexpressed, providing strong evidence that this protein is involved in suppression of metastasis (5, 13, 20, 21, 27). It would be critical to more clearly define the role of EBNA3C in regulating the antimetastatic function of Nm23-H1. It is clear that more studies are needed to determine the specific effects of Nm23-H1 in cells. We are currently investigating this line of studies to determine the cellular targets of Nm23-H1 and how EBNA3C influences this effect on specific cellular promoters.
Acknowledgments
We are thankful to Elliott Kieff for the EBNA3C reagents. Our special thanks go to Patricia S. Steeg and Melanie T. Hartstough for the pET3C-nm23H1 and pCMV nm23-H1 constructs.
This work was supported by grants from the Leukemia and Lymphoma Society of America and the National Cancer Institute (CA072150-07), as well as the National Institutes of Dental and Craniofacial Research (DE14136-01 [to E.S.R.]). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America. C.S. is a postdoctoral fellow of the Lady Tata Memorial Trust.
REFERENCES
- 1.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. (Erratum, 185:946, 1991.) [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., D. H. Crawford, and B. E. Griffin. 1989. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J. Gen. Virol. 70:1755-1764. [DOI] [PubMed] [Google Scholar]
- 3.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J. Biol. Chem. 272:11336-11343. [DOI] [PubMed] [Google Scholar]
- 5.Baba, H., T. Urano, K. Okada, K. Furukawa, E. Nakayama, H. Tanaka, K. Iwasaki, and H. Shiku. 1995. Two isotypes of murine nm23/nucleoside diphosphate kinase, nm23-M1 and nm23-M2, are involved in metastatic suppression of a murine melanoma line. Cancer Res. 55:1977-1981. [PubMed] [Google Scholar]
- 6.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, M., J. M. Guinebretiere, E. Kremmer, V. Grunewald, E. Benhamou, G. Contesso, and I. Joab. 1999. Detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 91:1376-1381. [DOI] [PubMed] [Google Scholar]
- 8.Chae, S. K., N. S. Lee, K. J. Lee, and E. Kim. 1998. Transactivation potential of the C-terminus of human Nm23-H1. FEBS Lett. 423:235-238. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter, M. A., II, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Rosa, A., R. L. Williams, and P. S. Steeg. 1995. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. Bioessays 17:53-62. [DOI] [PubMed] [Google Scholar]
- 12.Eschenfeldt, W. H., R. E. Manrow, M. S. Krug, and S. L. Berger. 1989. Isolation and partial sequencing of the human prothymosin alpha gene family. Evidence against export of the gene products. J. Biol. Chem. 264:7546-7555. [PubMed] [Google Scholar]
- 13.Fukuda, M., A. Ishii, Y. Yasutomo, N. Shimada, N. Ishikawa, N. Hanai, N. Nagata, T. Irimura, G. L. Nicolson, and N. Kimura. 1996. Decreased expression of nucleoside diphosphate kinase alpha isoform, an nm23-H2 gene homolog, is associated with metastatic potential of rat mammary-adenocarcinoma cells. Int. J. Cancer 65:531-537. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Marquez, J., F. Segade, M. Dosil, J. G. Pichel, X. R. Bustelo, and M. Freire. 1989. The expression of prothymosin alpha gene in T lymphocytes and leukemic lymphoid cells is tied to lymphocyte proliferation. J. Biol. Chem. 264:8451-8454. [PubMed] [Google Scholar]
- 15.Haritos, A. A., G. J. Goodall, and B. L. Horecker. 1984. Prothymosin alpha: isolation and properties of the major immunoreactive form of thymosin alpha 1 in rat thymus. Proc. Natl. Acad. Sci. USA 81:1008-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieff, E. 1996. Epstein-Barr virus and its replication, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 18.Kraeft, S. K., F. Traincart, S. Mesnildrey, J. Bourdais, M. Veron, and L. B. Chen. 1996. Nuclear localization of nucleoside diphosphate kinase type B (nm23-H2) in cultured cells. Exp. Cell Res. 227:63-69. [DOI] [PubMed] [Google Scholar]
- 19.Lacombe, M.-L., L. Milon, A. Munier, J. G. Mehus, and D. O. Lambeth. 2000. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 32:247-258. [DOI] [PubMed] [Google Scholar]
- 20.Leone, A., U. Flatow, C. R. King, M. A. Sandeen, I. M. Margulies, L. A. Liotta, and P. S. Steeg. 1991. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 65:25-35. [DOI] [PubMed] [Google Scholar]
- 21.Leone, A., U. Flatow, K. VanHoutte, and P. S. Steeg. 1993. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene 8:2325-2333. [PubMed] [Google Scholar]
- 22.Leone, A., R. C. Seeger, C. M. Hong, Y. Y. Hu, M. J. Arboleda, G. M. Brodeur, D. Stram, D. J. Slamon, and P. S. Steeg. 1993. Evidence for nm23 RNA overexpression, DNA amplification and mutation in aggressive childhood neuroblastomas. Oncogene 8:855-865. [PubMed] [Google Scholar]
- 23.Lim, S., H. Y. Lee, and H. Lee. 1998. Inhibition of colonization and cell-matrix adhesion after nm23-H1 transfection of human prostate carcinoma cells. Cancer Lett. 133:143-149. [DOI] [PubMed] [Google Scholar]
- 24.Makarova, T., N. Grebenshikov, C. Egorov, A. Vartapetian, and A. Bogdanov. 1989. Prothymosin alpha is an evolutionary conserved protein covalently linked to a small RNA. FEBS Lett. 257:247-250. [DOI] [PubMed] [Google Scholar]
- 25.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maunders, M. J., L. Petti, and M. Rowe. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J. Gen. Virol. 75:769-778. [DOI] [PubMed] [Google Scholar]
- 27.Parhar, R. S., Y. Shi, M. Zou, N. R. Farid, P. Ernst, and S. T. al-Sedairy. 1995. Effects of cytokine-mediated modulation of nm23 expression on the invasion and metastatic behavior of B16F10 melanoma cells. Int. J. Cancer 60:204-210. [DOI] [PubMed] [Google Scholar]
- 28.Parker, G. A., T. Crook, M. Bain, E. A. Sara, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13:2541-2549. [PubMed] [Google Scholar]
- 29.Pinon, V. P., G. Millot, A. Munier, J. Vassy, G. Linares-Cruz, J. Capeau, F. Calvo, and M. L. Lacombe. 1999. Cytoskeletal association of the A and B nucleoside diphosphate kinases of interphasic but not mitotic human carcinoma cell lines: specific nuclear localization of the B subunit. Exp. Cell Res. 246:355-367. [DOI] [PubMed] [Google Scholar]
- 30.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 34.Robertson, E. S. 1997. The Epstein-Barr virus EBNA3 protein family as regulators of transcription. Epstein-Barr Virus Rep. 4:143-150. [Google Scholar]
- 35.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe, M., C. M. Rooney, A. B. Rickinson, G. M. Lenoir, H. Rupani, D. J. Moss, H. Stein, and M. A. Epstein. 1985. Distinctions between endemic and sporadic forms of Epstein-Barr virus-positive Burkitt's lymphoma. Int. J. Cancer 35:435-441. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz, P., and U. Gunthert. 1996. The cellular basis of metastasis. World J. Urol. 14:141-150. [DOI] [PubMed] [Google Scholar]
- 39.Russell, R. L., A. N. Pedersen, J. Kantor, K. Geisinger, R. Long, N. Zbieranski, A. Townsend, B. Shelton, N. Brunner, and T. E. Kute. 1998. Relationship of nm23 to proteolytic factors, proliferation and motility in breast cancer tissues and cell lines. Br. J. Cancer 78:710-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sample, C., and B. Parker. 1994. Biochemical characterization of Epstein-Barr virus nuclear antigen 3A and 3C proteins. Virology 205:534-539. [DOI] [PubMed] [Google Scholar]
- 41.Steeg, P. S., G. Bevilacqua, R. Pozzatti, L. A. Liotta, and M. E. Sobel. 1988. Altered expression of NM23, a gene associated with low tumor metastatic potential, during adenovirus 2 Ela inhibition of experimental metastasis. Cancer Res. 48:6550-6554. [PubMed] [Google Scholar]
- 42.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7:350-355. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian, C., S. Hazan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. The Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugden, B., and N. Warren. 1989. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 63:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]