Abstract
The nucleocapsid protein VP30 of Ebola virus (EBOV), a member of the Filovirus family, is known to act as a transcription activator. By using a reconstituted minigenome system, the role of VP30 during transcription was investigated. We could show that VP30-mediated transcription activation is dependent on formation of a stem-loop structure at the first gene start site. Destruction of this secondary structure led to VP30-independent transcription. Analysis of the transcription products of bicistronic minigenomes with and without the ability to form the secondary structure at the first transcription start signal revealed that transcription initiation at the first gene start site is a prerequisite for transcription of the second gene, independent of the presence of VP30. When the transcription start signal of the second gene was exchanged with the transcription start signal of the first gene, transcription of the second gene also was regulated by VP30, indicating that the stem-loop structure of the first transcription start site acts autonomously and independently of its localization on the RNA genome. Our results suggest that VP30 regulates a very early step of EBOV transcription, most likely by inhibiting pausing of the transcription complex at the RNA structure of the first transcription start site.
Ebola virus (EBOV) and the closely related Marburg virus (MBGV) are the only members of the family Filoviridae, which belongs to the order Mononegavirales. A common feature of all Mononegavirales viruses is the possession of a nonsegmented negative-sense single-stranded RNA genome. Filoviruses are endemic in Africa and cause a severe hemorrhagic fever in humans and nonhuman primates with fatality rates up to 90% (33).
The RNA genome of EBOV is 19 kb in length and has a coding capacity for eight proteins encoded by seven genes (8). Located at the 3′ and 5′ ends of the genome are short nontranscribed leader and trailer regions containing the signals for replication, transcription initiation, and encapsidation (30). Each gene is flanked by highly conserved transcriptional start and stop signals, which determine the 5′ and 3′ ends of the nascent mRNA chains. Interestingly, the transcription start signals are predicted to form stable stem-loop structures (29, 35). Since the genome organizations of the different nonsegmented negative-sense RNA viruses are quite similar, it is presumed that the transcription process follows a general mechanism. The current model proposes a single binding site for the transcription complex within the leader region (6). Once bound to the genome, the transcription complex scans the viral RNA for the conserved transcription start and stop signals, where transcription initiation and termination of the individual genes occur. The genes are sequentially transcribed, leading to a gradient of mRNA abundance from the first gene toward the last gene (11, 19). Both processes, transcription and replication, are driven by the viral nucleocapsid proteins.
In contrast to most of the other members of the order Mononegavirales (except the pneumoviruses), MBGV and EBOV genomes encode four instead of three nucleocapsid proteins: NP, VP35, L, and VP30 (1, 5). These are the nucleoprotein NP encoded by the first gene (36, 37); the polymerase cofactor P, or VP35, encoded by the second gene (30); the viral protein VP30 encoded by the fifth gene (26); and the major component of the polymerase complex L encoded by the seventh gene (28, 40). The nucleocapsid proteins have a dual function in the viral replication cycle: they are involved in virus morphogenesis as structural components (22), and they catalyze replication and transcription of the RNA genome.
In a reconstituted EBOV minigenome system, NP, VP35, and L were essential and sufficient to support replication of EBOV-specific minigenomes. Transcription, however, was strongly enhanced by VP30 (30). In addition, VP30 was required to rescue a full-length recombinant EBOV (41). Thus, EBOV VP30 is assumed to be a transcription activation factor, which is essential for the viral replication cycle.
Here, we present evidence that EBOV VP30 regulates a very early step of transcription. Furthermore, we could show that secondary structure formation of the first transcription start signal is a prerequisite for VP30-regulated transcription. Transcription of the genes located downstream takes place independently of VP30.
MATERIALS AND METHODS
Cells and viruses.
HeLa and Vero E6 cells were maintained in Dulbecco's modified Eagle's medium (DMEM)-10% fetal calf serum (FCS). Infected cells were maintained in DMEM-2% FCS. EBOV strain Mayinga, subtype Zaire (35), was grown in Vero E6 cells and harvested as described elsewhere (28). Experiments with EBOV were performed in a high-containment laboratory according to the federal rules. Recombinant vaccinia virus MVA-T7 containing the T7 RNA polymerase gene (10) was propagated in chicken embryo fibroblasts.
Construction of EBOV-specific minigenomes.
Minigenome 3E-5EΔ250 was created by deleting nucleotides (nt) 251 to 472 of the 3′ region of minigenome 3E-5E (30). For construction of the bicistronic minigenome E-bici-1,2 (Fig. 1), first, a PCR fragment consisting of the first 300 nt of the chloramphenicol acetyltransferase (CAT) gene (ΔCAT) was amplified. The primers used for PCR were flanked either by an NdeI restriction site or by BglII and AseI restriction sites. The resulting fragment was digested with NdeI and AseI and ligated into the NdeI restriction site of clone 3E-5E. Thereafter, 401 nt of the intergenic region between the NP and VP35 gene of EBOV (nt 2728 to 3128; nucleotide numbers refer to the EBOV Zaire genome sequence, GenBank accession no. AF086833) were amplified by reverse transcription-PCR (RT-PCR). The primer used for generation of cDNA was flanked by a BglII site; the second primer was flanked by a BamHI restriction site. Finally, the PCR fragment was inserted into the BglII site of clone 3E-5E-ΔCAT.
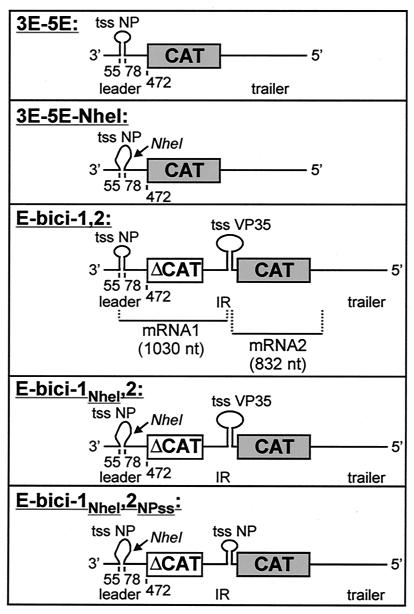
FIG. 1.
Schematic representations of the EBOV minigenomes. Construction of the negative-sense minigenome 3E-5E consisting of 472 nt of the EBOV leader, the CAT gene, and 731 nt of the EBOV trailer is described elsewhere (30). The conserved transcription start signal of the NP gene (nt 55 to 67) and 11 nt downstream of the NP transcription start signal are involved in formation of a hairpin loop as schematically depicted. In minigenome 3E-5E-NheI, 7 nt in the region between nt 69 and 78 were mutated, thus creating an NheI restriction site. These mutations prevented secondary structure formation. The negative-sense minigenome E-bici-1,2 contains 472 nt of the EBOV leader; the first 300 nt of the CAT gene (ΔCAT) as the first reporter gene; 401 nt of the intergenic region between the EBOV NP and VP35 genes, including the transcription stop signal of the NP gene and the transcription start signal of the VP35 gene; the full-length CAT gene as the second reporter gene; and 731 nt of the EBOV trailer. In minigenome E-bici-1NheI,2, secondary structure formation of the NP gene start signal was prevented as described for minigenome 3E-5E-NheI. Additionally, in minigenome E-bici-1NheI,2NPss, the transcription start signal of VP35 gene, including the nucleotides located downstream necessary for secondary structure formation, was exchanged with the respective nucleotides forming the NP gene hairpin loop. Nucleotide numbers refer to the EBOV Zaire genome. Restriction sites at the DNA level are indicated by arrows. tss NP, transcription start signal of the NP gene; tss VP35, transcription start signal of the VP35 gene; IR, intergenic region. [ ], secondary structure of the NP gene start region; [
], secondary structure of the NP gene start region; [ ], destroyed secondary structure of the NP gene start region; [
], destroyed secondary structure of the NP gene start region; [ ], secondary structure of the VP35 gene start region.
], secondary structure of the VP35 gene start region.
In vitro mutagenesis of minigenomes 3E-5E and E-bici-1,2 was performed with the QuikChange site-directed mutagenesis kit (Stratagene) according to the supplier's instructions. For generation of mutant 3E-5E-NheI, the nt 69, 71 to 73, and 75 to 77 within the EBOV-specific leader region were substituted, thus creating an NheI restriction site (Fig. 1). For generation of mutant E-bici-1NheI,2, the NdeI-NotI restriction fragment of 3E-5E-NheI was exchanged with the NdeI-NotI restriction fragment of E-bici-1,2 (Fig. 1). Mutant E-bici-1NheI,2NPss was constructed by in vitro mutagenesis of plasmid E-bici-1NheI,2. Here, the transcription start signal of the VP35 gene, including the nucleotides located downstream, which are involved in secondary structure formation (29), were exchanged with the transcription start region of the NP gene by substitution of nt 3034, 3044, 3046, 3047, and 3050 to 3055 (nucleotide numbers refer to the EBOV genome). Minigenomes 3E-5E(+) and 3E-5E-NheI(+) are the positive-sense counterparts of minigenomes 3E-5E and 3E-5E-NheI, respectively.
RNA isolation.
RNA isolation from MVA-T7-infected and transfected HeLa cells was performed with either an RNeasy kit (Qiagen) or with TRIZOL reagent (Gibco BRL) as recommended by the manufacturers. Polyadenylated RNA species were precipitated by using oligo(dT) cellulose (27). For isolation of prematurely terminated mRNA products, a biotinylated primer coupled to immobilized streptavidin (Roche) was used. The biotinylated primer binds to nt 28 to 53 of the EBOV NP mRNA. Binding of RNA to the coupled primer was performed with streptavidin binding buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 200 mM NaCl), and elution was performed with streptavidin elution buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA). Northern blot analysis performed with digoxigenin-labeled riboprobes is described elsewhere (27).
Transfection of MVA-T7-infected HeLa cells.
HeLa cells were infected with MVA-T7 at a multiplicity of infection of 5 PFU per cell and incubated for 1 h at 37°C. Subsequently, cells were transfected by the Lipofectin (GIBCO-BRL) precipitation technique. For transfection of 106 infected HeLa cells in a 7-cm2 well, 0.5 μg of pT/NPEBO, 0.5 μg of pT/VP35EBO, 1 μg of pT/LEBO, 0.1 μg of pT/VP30EBO, and 2 μg of the respective EBOV-specific minigenomic plasmid were used (30). After transfection, cells were incubated for 48 h at 33°C. RNA transfection was carried out as described previously (27).
Chemical probing of in vitro-transcribed RNA with DMS.
Dimethyl sulfate (DMS) modification was performed as described elsewhere with 1 μg of in vitro-transcribed RNA (42). After probing, RNA was resuspended in H2O to a final concentration of 0.1 μg/μl.
Chemical probing of in vitro-transcribed RNA with CMCT.
Fifty micrograms of in vitro-transcribed EBOV-specific minigenome RNA was incubated in a total volume of 25 μl containing 12.5 μl of 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT) buffer (50 mM potassium borate [pH 8.0], 10 mM MgCl2, and 100 mM NH4Cl) and 12.5 μl of CMCT stock solution (100 mM CMCT in CMCT buffer) for 10 min at 37°C. The reaction was terminated by addition of 2.5 volumes of cold ethanol. After precipitation, the pellet was resuspended in 200 μl of extraction buffer (0.3 M sodium acetate [pH 6.5], 0.5% sodium dodecyl sulfate, 5 mM EDTA [pH 8.0]), extracted with phenol-chloroform, and precipitated. Finally, modified RNA was resuspended in 400 μl of H2O.
Primer extension analysis.
Chemically modified RNA molecules (0.5 μg) were mixed with ∼2 × 105 cpm of radiolabeled oligonucleotides. Primer extension was performed at 37°C for 50 min with 100 U of Superscript II RNase H− reverse transcriptase (Gibco-BRL) under standard conditions as recommended by the manufacturer. The location of modification sites was determined by analyzing primer extension products electrophoretically on an 8% denaturing polyacrylamide gel containing 4 M urea. In parallel, the corresponding DNA sequences were determined by sequencing with the same radiolabeled primers.
RNase protection assay.
RNase protection assays were performed with the HybSpeed RPA kit (Ambion) according to the supplier's instructions. An in vitro-transcribed RNA labeled with [α-32P]UTP was used as a riboprobe. This riboprobe was 523 nt in length and contained the first 270 nt of the EBOV leader in negative-sense orientation.
Immunoprecipitation of EBOV mRNA.
A total of 107 HeLa cells were infected with EBOV at a multiplicity of infection of 1 PFU per cell. At 3 days postinfection (p.i.), total cellular RNA was isolated and resuspended in 50 μl of H2O. Ten micrograms of monoclonal antibody H20 (2) was coupled to 30 μl of preswollen protein A-Sepharose beads (Sigma) in 400 μl of phosphate-buffered saline at 4°C overnight. Then the beads were washed three times with IPP buffer (31) and resuspended in the same buffer. Ten microliters of purified RNA from EBOV-infected cells was incubated with the protein A-Sepharose-antibody conjugate for 1 h at 4°C. The beads were washed three times with IPP buffer, dried, and incubated with 2 μg of proteinase K for 1 h at 55°C. Purified RNA was detected by Northern blot analysis with a digoxigenin-labeled negative-sense riboprobe directed to VP40 mRNA.
RESULTS
VP30 does not act as a transcription elongation factor.
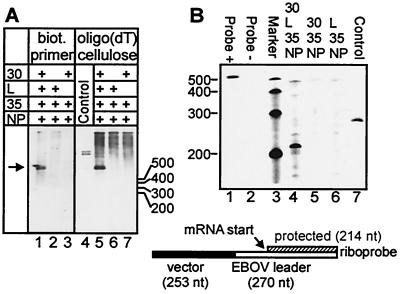
The effects of VP30 for EBOV transcription were examined by using a minigenome system in which replication and transcription are reconstituted from plasmid-supplied EBOV-specific minigenomic RNA and nucleocapsid proteins (30). It has been shown previously that replication of minigenomic RNA is unaffected by VP30, whereas transcription is strongly dependent on the presence of VP30 (30). If VP30 was involved in an early step of transcription (i.e., initiation or early antitermination), it would be assumed that in the absence of VP30, transcription activity would be suppressed, and, consequently, mRNA synthesis would not occur. The putative function of VP30 as an elongation factor would imply that in the absence of VP30, prematurely terminated mRNA species would be synthesized. In order to identify shortened mRNA species, RNA purification was performed with a biotinylated primer binding to the 5′ end of the nascent mRNA chains. Bound RNA was precipitated via immobilized streptavidin. In parallel, polyadenylated mRNA species were isolated by oligo(dT) precipitation. If VP30 prevented premature termination during transcription, one would expect that in the absence of VP30, a smear of incompletely synthesized and presumably nonpolyadenylated mRNAs would appear. These shortened, nonpolyadenylated RNAs should only be detected by using the biotinylated primer for purification and not by oligo(dT) precipitation. However, Fig. 2 shows that no prematurely terminated mRNA species were precipitated by using the biotinylated primer. Only the full-length polyadenylated mRNA product was detected in the presence of VP30 (Fig. 2A, lanes 1 and 5). In the absence of VP30, neither the full-length product nor shorter RNA species could be detected (Fig. 2A, lanes 2 and 6). The second positive-sense RNA species besides the mRNA is a replication intermediate, the mini-antigenome, which is complementary to the negative-sense minigenome. This RNA was also bound by the biotinylated primer, but due to its small quantity, to a much lesser extent. Since the mini-antigenome is synthesized in very small amounts relative to the mRNA, the corresponding RNA band could only be detected after a prolonged exposure time (data not shown).
FIG. 2.
Prematurely terminated mRNA was detected neither in the presence nor in the absence of VP30. (A) Northern blot analysis of EBOV-specific RNA-species. HeLa cells were infected with MVA-T7 and subsequently transfected with pT/NPEBO, pT/VP35EBO, pT/LEBO, pT/VP30EBO, and 3E-5EΔ250 as indicated. Forty-eight hours p.i., total RNA was isolated with TRIZOL reagent and purified either by using a biotinylated (biot.) primer coupled to immobilized streptavidin, which binds to the 5′ end of the minigenomic mRNA species (lanes 1 to 3), or by using oligo(dT) cellulose (lanes 5 to 7). Precipitated RNA was transferred onto nylon membranes and finally probed with the negative-sense digoxigenin-labeled riboprobe DIG-BS/CAT (27). The arrow indicates the position of the full-length mRNA species, which is 921 nt in length [without poly(A) tail]. (B) RNase protection assay. HeLa cells were infected and transfected as described above. Total RNA was isolated by using TRIZOL reagent. The radiolabeled negative-sense riboprobe comprised nt 1 to 270 of the EBOV leader (open bar) and 253 nt of upstream vector sequences (solid bar). Due to binding of the riboprobe to the 5′ end of the minigenomic mRNA (striped bar), a 214-nt segment of the probe was protected against RNase digestion. As a control, in vitro-transcribed positive-sense minigenome 3E-5E(+) was used for both experiments. Numbers indicate the length of the respective RNA marker bands. Probe −, riboprobe without RNase; Probe +, riboprobe with RNase.
The finding that no truncated mRNAs were generated in the absence of VP30 was confirmed by RNase protection assays (Fig. 2B). Here, an RNA probe was used that bound to the first 214 nt of the nascent mRNA chains. In the absence of VP30, the riboprobe was not protected, indicating that prematurely terminated mRNA was not synthesized (Fig. 2B, lane 6). Taken together, these results demonstrate that EBOV VP30 does not act as an elongation factor. Since mRNA synthesis was strongly dependent on the presence of VP30, it is assumed that VP30 is involved in an early step of transcription.
VP30-regulated transcription is dependent on secondary structure formation of the NP gene start sequence.
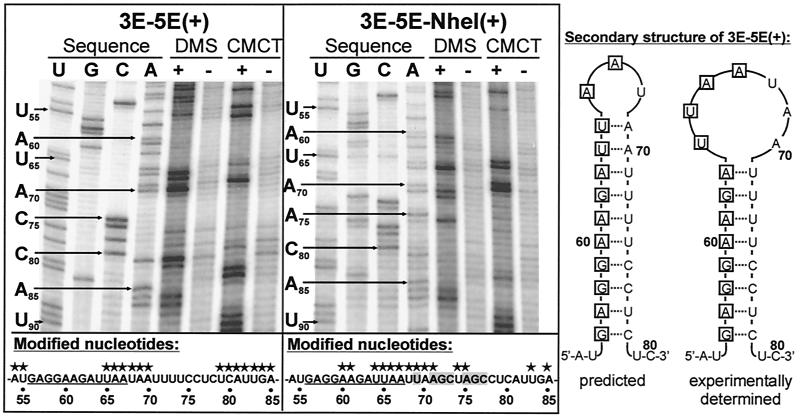
To further determine the role of VP30, we analyzed whether VP30-dependent transcription activation was influenced by the transcription start signals preceding the EBOV genes. As mentioned above, all filoviral transcription start signals are predicted to be involved in formation of stable stem-loop structures. Since the sequences downstream of the highly conserved transcription start signals are not conserved, the structure of the predicted stem-loops varies from gene to gene (29). First, secondary structure formation was experimentally determined. Therefore, an in vitro-transcribed minigenome containing the EBOV leader sequence and the 3′ nontranslated region of the NP gene was used to perform chemical modification assays. As modifying agents, DMS and CMCT were used. DMS specifically modifies unpaired A and C residues, and CMCT is specific for unpaired U and G residues (18). The sites of chemical modification were identified by primer extension analysis, whereby progress of the reverse transcriptase was inhibited by the modified nucleotides. In parallel, the corresponding DNA sequence was determined by sequencing with the primer also used for primer extension. On the left-hand side of Fig. 3, the autoradiograph of the sequence analysis of minigenome 3E-5E(+) between nt 50 and 90 and the corresponding primer extension reactions are shown. Since RT terminates 1 nt 3′ of the position of the modified nucleotide, the primer extension products are displaced by 1 nt relative to the DNA sequencing lanes. Unpaired nucleotides that have reacted with either DMS or CMCT resulted in clear bands in the primer extension analysis (e.g., nt 65 to 70). Paired nucleotides that were not attacked by the chemicals did not lead to transcription termination, and, consequently, no specific bands could be seen at the positions of these nucleotides (e.g., nt 71 to 78). The data obtained by chemical modification assay revealed that, indeed, the NP gene start signal is involved in formation of a stem-loop structure. The experimentally determined structure differs from the computer-predicted secondary structure in that the loop is extended by an additional 4 nt (Fig. 3).
FIG. 3.
The NP gene start region is involved in formation of a stable secondary structure. In vitro-transcribed RNAs of positive-sense minigenomes 3E-5E(+) (left-hand side) and 3E-5E-NheI(+) (right-hand side), respectively, were subjected to chemical modification assay with the drugs DMS (A and C specific) and CMCT (U and G specific). In order to visualize minor RNase contaminations, samples were also incubated in the respective buffer without addition of DMS or CMCT, respectively. Modified RNA was purified and analyzed by the primer extension method. In parallel, the corresponding DNA sequence was determined by sequencing with the same radiolabeled primer. Samples were separated on a denaturing 8% polyacrylamide gel, and the gel was processed for autoradiography. Note that the cDNA fragments synthesized from modified RNA are displaced by 1 nt relative to the sequencing lanes. The sequence of the NP gene start region is shown below the autoradiographs. The conserved NP transcription start signal is underlined; modified nucleotides are marked by asterisks. Mutated nucleotides in the sequence of minigenome 3E-5E-NheI(+) are highlighted by gray boxes. The predicted and experimentally determined secondary structures of minigenome 3E-5E(+) are shown schematically on the right-hand side. Nucleotides belonging to the conserved transcription start signal are boxed. Nucleotide numbers refer to the genome sequence of EBOV Zaire.
To elucidate the function of the determined secondary structure for transcription, a monocistronic minigenome was constructed in which the sequences downstream of the highly conserved transcription start signal of the NP gene, which are involved in secondary structure formation, were mutated by insertion of an NheI restriction site (Fig. 1, 3E-5E-NheI). After it was confirmed that formation of the secondary structure was impaired (Fig. 3, right-hand panel), minigenome 3E-5E-NheI was used as a template for transcription in the reconstituted minigenome assay. Surprisingly, 3E-5E-NheI was not only efficiently transcribed in the presence of VP30, but was also efficiently transcribed in the absence of VP30 (Fig. 4A, lanes 1 and 3), indicating that proper secondary structure formation of the NP gene start signal is a prerequisite for VP30-dependent transcription activation.
FIG. 4.
Destruction of the secondary structure formed by the NP gene start signal leads to VP30-independent transcription initiation. (A) Northern blot analysis of transcribed mRNA species. HeLa cells were infected with MVA-T7 and subsequently transfected with pT/NPEBO, pT/VP35EBO, pT/LEBO, pT/VP30EBO, and either 3E-5E or 3E-5E-NheI as indicated. Forty-eight hours p.i., total RNA was isolated and precipitated with oligo(dT) cellulose. Purified RNA was transferred onto nylon membranes and probed with the negative-sense digoxigenin-labeled riboprobe DIG-BS/CAT. Arrows indicate the positions of mRNA bands. (B) Prevention of secondary structure formation does not impede translation of the minigenomic mRNA. HeLa cells were infected with MVA-T7; transfected with pT/NPEBO, pT/VP35EBO, pT/LEBO, and pT/VP30EBO as indicated; and subsequently transfected with 5 μg of RNA 3E-5E or RNA 3E-5E-NheI, respectively. Forty-eight hours p.i., cells were lysed. CAT activity was determined, and acetylated products were separated by thin-layer chromatography.
To analyze whether translation of the reporter gene was also influenced by destruction of the secondary structure formed by the NP gene start signal, expression of the CAT gene from 3E-5E and 3E-5E-NheI, respectively, was checked. As shown in Fig. 4B, the CAT gene was expressed by both minigenomes in the presence of VP30. In the absence of VP30, CAT activity was only observed in cells transfected with minigenome 3E-5E-NheI. Thus, it was concluded that prevention of secondary structure formation did not interfere with translation.
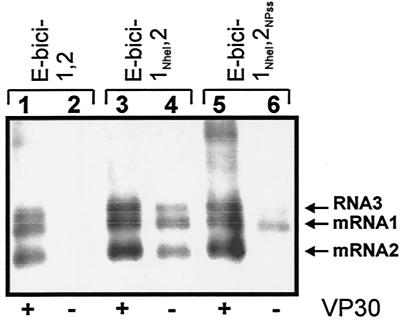
The NP gene start signal is the first transcription start site downstream of the transcription promoter. Since the transcription start signals of all internally located EBOV genes are predicted to form stem-loop structures as well, it was of interest to examine whether VP30 was needed to reinitiate transcription at the gene boundaries. To this end, bicistronic minigenomes were constructed, comprising two reporter genes separated by the intergenic region of the first and the second EBOV genes. While minigenome E-bici-1,2 contained the authentic NP and VP35 gene start signals upstream of the first and second reporter genes, respectively, the NP gene start sequence of minigenome E-bici-1NheI,2 was mutated by insertion of an NheI restriction site comparable to minigenome 3E-5E-NheI (Fig. 1). The effect of VP30 on transcription of E-bici-1,2 and E-bici-1NheI,2 was then assayed with the reconstituted minigenome system. In the presence of VP30, both E-bici-1NheI,2-specific mRNAs were synthesized as efficiently as the mRNAs derived from the wild-type bicistronic minigenome E-bici-1,2 (Fig. 5, lanes 1 and 3). Besides the two expected monocistronic mRNA bands (Fig. 5, mRNA1 and mRNA2), a third RNA band was detected (RNA3), which was about 1.2 kb in length. To elucidate the origin of this RNA, the first reporter gene of minigenome E-bici-1,2, the ΔCAT gene, was exchanged with the green fluorescent protein (GFP) gene. When the mRNA species transcribed from this minigenome were probed with the CAT gene-specific riboprobe, neither mRNA1 nor RNA3 hybridized to the probe, indicating that mRNA3 did not contain sequences of the second gene (data not shown). According to the size of RNA3, it is presumed that the promoter-proximal mRNA1 was attached to the leader RNA, resulting in synthesis of RNA3.
FIG. 5.
VP30-mediated transcription of bicistronic minigenomes is dependent on a proper secondary structure formation of the NP gene start region. HeLa cells were infected with MVA-T7 and subsequently transfected with pT/NPEBO, pT/VP35EBO, pT/LEBO, or pT/VP30EBO and E-bici-1,2, E-bici-1NheI,2, or E-bici-1NheI,2NPss, respectively as indicated. Forty-eight hours p.i., total RNA was isolated and precipitated with oligo(dT) cellulose. Northern blot analyses were performed as described in the legend to Fig. 2 to detect specific mRNAs in the presence (+) or absence (−) of VP30. The position of the RNA bands is indicated by arrows.
In the absence of VP30, only minigenome E-bici-1NheI,2, which lacks the secondary structure of the first gene, was used as a template for transcription (Fig. 5, lanes 2 and 4). Interestingly, both mRNA species could be detected, although the amount of mRNA2 was lower than the level of transcription in the presence of VP30 (Fig. 5, lanes 3 and 4). These results clearly indicate that transcription reinitiation at internally located gene start signals with intact stem-loop structures is not a VP30-regulated process. Transcription activation at the first promoter-proximal gene start signal, however, is strongly regulated by VP30. This regulation is dependent on a proper secondary structure formation of the transcription start site.
The secondary structure formed by the NP gene start site acts as an autonomous signal.
To further investigate whether the position of the NP gene start signal adjacent to the transcription promoter is responsible for VP30-dependent transcription activation or the sequence itself, a mutant of minigenome E-bici-1NheI,2 was constructed in which the sequence forming the VP35 gene stem-loop upstream of the second gene was replaced by the sequence forming the NP gene stem-loop structure (Fig. 1, E-bici-1NheI,2NPss). In the presence of VP30, E-bici-1NheI,2NPss was transcribed like E-bici-1,2 and E-bici-1NheI,2, respectively. Thus, both mRNA species were synthesized (Fig. 5, lane 5). In the absence of VP30, however, only mRNA1 was visible, whereas mRNA2 could not be detected (Fig. 5, lane 6), suggesting that synthesis of mRNA2 now was regulated by VP30. This result clearly showed that the secondary structure of the NP gene start signal acts as an autonomous regulatory signal independently of its localization on the RNA genome.
The EBOV-specific mRNA species are capped.
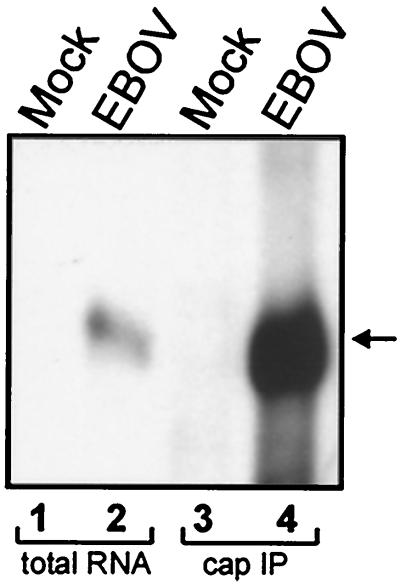
Since transcription reinitiation at the EBOV gene boundaries was shown to be a VP30-independent process, the question arose of whether the predicted secondary structures formed by the gene start signals might be involved in processes other than transcription initiation. RNA structures were found to be involved in cap-independent translation initiation, where they serve as internal ribosomal entry sites (IRES) (20, 32). It is assumed that if the predicted secondary structures of EBOV genes functioned in an IRES-like manner, the mRNAs would not be capped. To analyze whether the EBOV-specific mRNAs were capped, total RNA was isolated from EBOV-infected HeLa cells and subjected to immunoprecipitation with the monoclonal antibody H20 directed to the m7G-cap (2). Precipitated RNA species were then analyzed by Northern hybridization. As a probe, a negative-sense RNA directed to the VP40 mRNA, which is the most abundant EBOV-specific mRNA species, was used. Figure 6, lane 4, shows that VP40 mRNA could be precipitated with the anticap antibody, indicating that the EBOV mRNAs are capped. Hence, it is not likely that the secondary structures at the gene start signals are involved in cap-independent translation.
FIG. 6.
EBOV-specific mRNAs are capped. HeLa cells were either infected with EBOV Zaire or mock infected, and total cellular RNA was isolated at 3 days p.i. Isolated RNA was immunoprecipitated with anti-m7G-cap monoclonal antibody H20. Finally, precipitated RNA species were purified and subjected to Northern blot analysis (lanes 3 and 4) together with total RNA isolated from EBOV- and mock-infected cells (lanes 1 and 2). As a probe, a negative-sense digoxigenin-labeled VP40 gene-specific RNA was used. The position of VP40 mRNA is indicated by an arrow.
DISCUSSION
In a reconstituted plasmid-based minigenome system, the fourth EBOV nucleocapsid protein, VP30, has been shown to efficiently enhance transcription. The only other members of the order Mononegavirales possessing a fourth nucleocapsid protein besides filoviruses are the pneumoviruses. The fourth nucleocapsid protein of human respiratory syncytial virus (hRSV), M2-1, is known to function as an intra- and intergenic transcription antitermination factor by promoting chain elongation and by increasing synthesis of readthrough mRNA species (4, 7, 13, 15). Besides their role as transcription transactivators, VP30 and M2-1 also show some structural similarities: both proteins are part of the nucleocapsid, both are phosphorylated, and both contain a putative Zn binding domain located in their amino-terminal regions (3, 5, 14, 23, 26, 38). Due to these striking similarities, it was first hypothesized that VP30 might function in an M2-1-similar manner. However, this is not the case. Analysis of the transcription products derived from mono- and bicistronic EBOV-specific minigenomes in the absence and presence of VP30 clearly indicated that VP30 acts neither as an elongation factor nor as an intergenic antitermination factor. When bicistronic minigenomes were used as templates for transcription, an additional polyadenylated RNA species was detected besides the two expected monocistronic mRNAs (Fig. 5, RNA3). This band most likely was due to synthesis of the promoter-proximal mRNA containing the attached leader sequence. A similar finding has also been described for transcription of RSV-specific bicistronic minigenomes (7). However, RNA3 did not represent a real readthrough mRNA product, because, first, the migration velocity of RNA3 in the agarose gels did not fit to the expected size of a readthrough mRNA and, second, RNA3 did not contain the sequences of the second gene.
Since it was not possible to detect any prematurely terminated mRNA species in the absence of VP30, it was concluded that VP30 is rather involved in a very early step of transcription: possibly initiation or early antitermination. This assumption was consistent with the observation that VP30-dependent transcription was regulated by an RNA structure formed by the first transcription start signal, which is located immediately 5′ of the leader region. However, when the RNA structure was destroyed, transcription was initiated independently of VP30, indicating that the first step of transcription, namely assembly of the transcription complex at the promoter, is not regulated by VP30. We suggest a model whereby the EBOV transcription complex initially binds to a single promoter site within the leader region. In the absence of VP30, secondary structure formation of the first transcription start signal might hamper movement of the polymerase along the RNA template. In the presence of VP30, the RNA structure would not interfere with transcription initiation. A possible explanation could be that VP30 might resolve or cover the secondary structure, either by RNA binding or by directing an additional cofactor to the folded RNA. So far, an RNA binding activity for VP30 has not been described. Since the EBOV genome is tightly enwrapped by the nucleocapsid proteins, it is hard to imagine how secondary structure formation might occur. The only naked RNA species are the positive-sense mRNAs. Therefore, we checked positive-sense minigenomic RNAs for their ability to form the predicted hairpin loops, and, indeed, secondary structure formation was observed. Thus, it could be the case that secondary structure formation takes place on the mRNA level. Since the first 23 nt of the nascent NP mRNA are involved in stem-loop structure formation, one can consider that RNA folding immediately after transcription initiation might interfere with progression of transcription. Consequently, transcription should proceed when secondary structure formation is inhibited, even in the absence of VP30. A similar mechanism has been described for antiterminator proteins that recognize control signals (i.e., RNA structures) near the promoter and prevent transcriptional termination. An example of an early antiterminator is the DNA binding protein Q of phage λ, which prevents pausing of the RNA polymerase complex only 16 nt downstream of the transcription initiation site (12). Antitermination controlled by simply organized RNA structures within the nascent RNA chain has been reported for the N protein of phage λ (34).
RNA folding also plays an essential role in initiation and elongation of human immunodeficiency virus type 1 transcription (16, 24). An early step of elongation is regulated by the transactivating protein Tat through binding to virus-encoded folded RNA called the “transactivation response element” (TAR) (39). In the absence of Tat, transcription complexes assemble at the viral promoter leading to synthesis of short (≤200 nt) RNA transcripts. Interaction of Tat with TAR renders the RNA polymerase II complex competent for elongation (21). The TAR element is not only essential for elongation, but also has been shown to influence transcription initiation (16). However, there are striking differences from VP30-regulated transcription activation. First, it was not possible to detect RNA species shorter than 200 nt in the absence of VP30 as shown by RNase protection assays. Second, the transactivating capacity of Tat mediated through binding to the TAR stem-loop structure is a prerequisite for transcription (9), whereas VP30 is dispensable for EBOV transcription when secondary structure formation is impeded.
Among the other members of the order Mononegavirales, a comparable process has not yet been described. So far, predicted or experimentally determined RNA structures play no discernible role in replication and transcription (17, 24). For the closely related MBGV, it was shown previously that transcriptional activity was not enhanced by addition of VP30 (27). Interestingly, the predicted RNA structure formed by the MBGV-specific NP gene start region is completely different from the EBOV NP hairpin loop (29). The differences in secondary structure formation are due to several nucleotide exchanges downstream of the conserved transcription start signals. So far, it is not known whether the altered RNA structure is responsible for MBGV-specific VP30-independent transcription.
For VP30-dependent EBOV transcription activation, only RNA folding of the first transcription start site seems to be important. Thus, the function of secondary structure formation of the other EBOV genes remains unclear. Since viral mRNA isolated from EBOV-infected cells was shown to be capped, it is not very likely that the RNA secondary structures formed by each gene start region act in an IRES-like manner. Furthermore, the known IRES are highly structured (25), whereas the EBOV secondary structures are far more simply organized.
Although the precise mechanism of EBOV-specific transcription regulation is not yet known, this study demonstrates that VP30 is a key regulator of EBOV transcription and that this regulatory process is modulated by an RNA structure.
Acknowledgments
We thank Angelika Lander for excellent technical assistance. We are grateful to S. E. Behrens and H. Yu for introduction to the CMA technique. We thank R. Lührmann for supplying the monoclonal antibody H20.
This work was supported by the Boehringer Ingelheim Fonds (to M. Weik), by the FAZIT Stiftung (to J. Modrof), and by the Deutsche Forschungsgemeinschaft (SFB 535 and 286).
REFERENCES
- 1.Becker, S., C. Rinne, U. Hofsäss, H.-D. Klenk, and E. Mühlberger. 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249:406-417. [DOI] [PubMed] [Google Scholar]
- 2.Bringmann, P., J. Rinke, B. Appel, R. Reuter, and R. Lührmann. 1983. Purification of snRNPs U1, U2, U4, U5 and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1)RNP antisera. EMBO J. 2:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartee, T. L., and G. W. Wertz. 2001. Respiratory syncytial virus M2-1 protein requires phosphorylation for efficient function and binds viral RNA during infection. J. Virol. 75:12188-12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, P. L., M. G. Hill, J. Cristina, and H. Grosfeld. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott, L. H., M. P. Kiley, and J. B. McCormick. 1985. Descriptive analysis of Ebola virus proteins. Virology 147:169-176. [DOI] [PubMed] [Google Scholar]
- 6.Emerson, S. U. 1982. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell 31:635-642. [DOI] [PubMed] [Google Scholar]
- 7.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann, H., and M. P. Kiley. 1999. Classification, structure, and replication of filoviruses. Curr. Top. Microbiol. Immunol. 235:1-21. [DOI] [PubMed] [Google Scholar]
- 9.Feng, S., and E. C. Holland. 1988. HIV-1 tat trans-activation requires the loop sequence within tar. Nature 334:165-167. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galinski, M. S., and S. L. Wechsler. 1991. The molecular biology of the paramyxovirus genus, p. 41-82. In D. W. Kingsbury (ed.), The paramyxoviruses. Plenum Press, New York, N.Y.
- 12.Greenblatt, J., J. R. Nodwell, and S. W. Mason. 1993. Transcriptional antitermination. Nature 364:401-406. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy, R. W., and G. W. Wertz. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy, R. W., and G. W. Wertz. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrich, D., C. Ulich, and R. B. Gaynor. 1996. A critical role for the TAR element in promoting efficient human immunodeficiency virus type 1 reverse transcription. J. Virol. 70:4017-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman, M. A., and A. K. Banerjee. 2000. Analysis of RNA secondary structure in replication of human parainfluenza virus type 3. Virology 272:151-158. [DOI] [PubMed] [Google Scholar]
- 18.Inoue, T., and T. R. Cech. 1985. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl. Acad. Sci. USA 82:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iverson, L. E., and J. K. Rose. 1982. Sequential synthesis of 5′-proximal vesicular stomatitis virus mRNA sequences. J. Virol. 44:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang, S. K., H.-G. Krausslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 22.Kolesnikova, L., E. Mühlberger, E. Ryabchikova, and S. Becker. 2000. Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J. Virol. 74:3899-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert, D. M., J. Hambor, M. Diebold, and B. Galinski. 1988. Kinetics of synthesis and phosphorylation of respiratory syncytial virus polypeptides. J. Gen. Virol. 69:313-323. [DOI] [PubMed] [Google Scholar]
- 24.Marquet, R., C. Isel, C. Ehresmann, and B. Ehresmann. 1995. tRNAs as primer of reverse transcriptases. Biochimie 77:113-124. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Salas, E., R. Ramos, E. Lafuente, and S. Lopez de Quinto. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82:973-984. [DOI] [PubMed] [Google Scholar]
- 26.Modrof, J., C. Moritz, L. Kolesnikova, T. Konakova, B. Hartlieb, A. Randolf, E. Mühlberger, and S. Becker. 2001. Phosphorylation of Marburg virus VP30 at serines 40 and 42 is critical for its interaction with NP inclusions. Virology 287:171-182. [DOI] [PubMed] [Google Scholar]
- 27.Mühlberger, E., B. Lotfering, H.-D. Klenk, and S. Becker. 1998. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 72:8756-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mühlberger, E., A. Sanchez, A. Randolf, C. Will, M. P. Kiley, H.-D. Klenk, and H. Feldmann. 1992. The nucleotide sequence of the L gene of Marburg virus, a filovirus: homologies with paramyxoviruses and rhabdoviruses. Virology 187:534-547. [DOI] [PubMed] [Google Scholar]
- 29.Mühlberger, E., S. Trommer, C. Funke, V. Volchkov, H.-D. Klenk, and S. Becker. 1996. Termini of all mRNA species of Marburg virus: sequence and secondary structure. Virology 223:376-380. [DOI] [PubMed] [Google Scholar]
- 30.Mühlberger, E., M. Weik, V. E. Volchkov, H.-D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nottrott, S., K. Hartmuth, P. Fabrizio, H. Urlaub, I. Vidovic, R. Ficner, and R. Lührmann. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 8:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters, C. J., and A. S. Khan. 1999. Filovirus diseases. Curr. Top. Microbiol. Immunol. 235:85-95. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, J. W. 1988. Phage lambda and the regulation of transcription termination. Cell 52:5-6. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez, A., M. P. Kiley, B. P. Holloway, and D. D. Auperin. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29:215-240. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, A., M. P. Kiley, B. P. Holloway, J. B. McCormick, and D. D. Auperin. 1989. The nucleoprotein gene of Ebola virus: cloning, sequencing, and in vitro expression. Virology 170:81-91. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez, A., M. P. Kiley, H.-D. Klenk, and H. Feldmann. 1992. Sequence analysis of the Marburg virus nucleoprotein gene: comparison to Ebola virus and other non-segmented negative-strand RNA viruses. J. Gen. Virol. 73:347-357. [DOI] [PubMed] [Google Scholar]
- 38.Tang, R. S., N. Nguyen, X. Cheng, and H. Jin. 2001. Requirement of cysteines and length of the human respiratory syncytial virus M2-1 protein for protein function and virus viability. J. Virol. 75:11328-11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 40.Volchkov, V. E., V. A. Volchkova, A. A. Chepurnov, V. M. Blinov, O. Dolnik, S. V. Netesov, and H. Feldmann. 1999. Characterization of the L gene and 5′ trailer region of Ebola virus. J. Gen. Virol. 80:355-362. [DOI] [PubMed] [Google Scholar]
- 41.Volchkov, V. E., V. A. Volchkova, E. Mühlberger, L. V. Kolesnikova, M. Weik, O. Dolnik, and H.-D. Klenk. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965-1969. [DOI] [PubMed] [Google Scholar]
- 42.Yu, H., C. W. Grassmann, and S.-E. Behrens. 1999. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J. Virol. 73:3638-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]