Abstract
During lytic infections, the virion host shutoff (Vhs) protein (UL41) of herpes simplex virus destabilizes both host and viral mRNAs. By accelerating the decay of all mRNAs, it helps redirect the cell from host to viral gene expression and facilitates the sequential expression of different classes of viral genes. While it is clear that Vhs induces mRNA degradation, it is uncertain whether it is itself an RNase or somehow activates a cellular enzyme. This question was addressed by using a combination of genetic and biochemical approaches. The Vhs homologues of alphaherpesviruses share sequence similarities with a family of mammalian, yeast, bacterial, and phage nucleases. To test the functional significance of these similarities, Vhs was mutated to alter residues corresponding to amino acids known to be critical to the nuclease activity of cellular homologues. In every instance, mutations that inactivated the nuclease activity of cellular homologues also abolished Vhs activity. Recent experiments showed that Vhs interacts with the cellular translation initiation factor eIF4H. In this study, the coexpression of Vhs and a glutathione S-transferase (GST)-eIF4H fusion protein in bacteria resulted in the formation of a complex of the proteins. The wild-type Vhs/GST-eIF4H complex was isolated and shown to have RNase activity. In contrast, Vhs mutations that altered key residues in the nuclease motif abolished the nuclease activity of the recombinant Vhs/GST-eIF4H complex. The results provide genetic and biochemical evidence that Vhs is an RNase, either alone or as a complex with eIF4H.
During lytic herpes simplex virus (HSV) infections, viral and cellular gene expression is regulated through a complex set of transcriptional and posttranscriptional controls (57). Of the posttranscriptional mechanisms, one of the best characterized is the destabilization of host and viral mRNAs by the HSV virion host shutoff (Vhs) protein (UL41) (49). Soon after infection, copies of the Vhs polypeptide, which enter cells as components of infecting virions, accelerate the degradation of host mRNAs (20, 58, 66). This activity, together with the inhibition of pre-mRNA splicing by the immediate-early polypeptide ICP27 (24, 25), helps redirect the cell from the synthesis of host proteins to that of viral proteins. In addition, following the onset of viral transcription, the Vhs protein accelerates the turnover of viral mRNAs belonging to all kinetic classes (36, 45, 46, 66). In this role, it helps determine viral mRNA levels and facilitates the sequential expression of different classes of viral genes (46).
While not lethal, mutations that inactivate Vhs result in a severalfold reduction of virus growth in cell cultures (50, 51), and wild-type virus rapidly outgrows Vhs mutants in mixed infections (37). A number of studies suggest that Vhs plays a significant role in HSV pathogenesis (4, 38, 61, 63-65). Thus, the replication of Vhs mutants is markedly reduced in mouse cornea, trigeminal ganglion, and brain (38, 64), and Vhs is required for the efficient establishment of latency (63), although this latter phenotype probably reflects a requirement for Vhs for efficient virus replication in peripheral tissues (63). The mechanisms by which Vhs affects pathogenesis are unclear but may involve a role in reducing the expression of major histocompatibility complex class I (69) or suppressing cytokine production by infected cells (67).
Although it is clear that Vhs induces mRNA turnover, a central unanswered question is whether Vhs is itself an RNase or, instead, activates a cellular enzyme. Several studies demonstrated Vhs-dependent cleavage of target mRNAs in cytoplasmic extracts of infected cells (34, 62). Similarly, rabbit reticulocyte lysates containing in vitro-translated Vhs induced Vhs-dependent endonuclease cleavage of target mRNAs (15, 16, 75). In these in vitro systems, Vhs degraded mRNAs, while rRNAs were unaffected, an observation that parallels the specificity of Vhs for mRNAs that is seen in vivo (45, 46, 58, 66). In addition, recent studies suggested that, in vivo and in rabbit reticulocyte lysates, Vhs does not cleave mRNAs at random sites but rather appears to initiate degradation near regions of translation initiation. Thus, the degradation of mRNAs, which are translated by cap-dependent scanning, appears to be initiated near the 5′ end (15, 32), and in vitro-translated Vhs preferentially induces cleavage at sites downstream from a picornavirus internal ribosome entry site (16, 41). Extracts of partially purified virions contain an RNase activity that is blocked by Vhs-specific antisera and is absent from the virions of Vhs mutants (75). However, in several respects, this RNase activity lacks the selectivity of the Vhs activity in vivo. First, it is not restricted to mRNAs. Second, it cleaves target RNAs at multiple locations throughout the molecule without showing an apparent preference for sites near the 5′ end. These observations suggest that targeting of the Vhs activity may require one or more cellular factors that are present in reticulocyte lysates but absent from virion preparations. While all of these studies are consistent with Vhs being a nuclease, in each case the preparations of Vhs protein probably contained cellular polypeptides, making it impossible to exclude the possibility that Vhs activates a cellular RNase.
To investigate this question more fully, we used a combination of genetic and biochemical approaches. Vhs has been reported to share sequence similarities with a number of cellular nucleases (12, 17). These studies were extended by using hidden Markov modeling, with the result that the Vhs homologues of alphaherpesviruses were found to share more extensive similarities with a larger family of human, yeast, bacterial, and phage nucleases. In particular, nine charged or hydrophilic residues are highly conserved between Vhs and the cellular nucleases. For several of the nucleases, the conserved amino acids are located in the active site and are critical to nuclease activity. Alteration of the corresponding residues of Vhs by site-directed mutagenesis was found to abolish the ability of Vhs to degrade mRNAs. In other experiments, Vhs was expressed in bacteria along with a fusion protein of glutathione S-transferase (GST) and the mammalian translation initiation factor eIF4H. Vhs and eIF4H recently were shown to interact in mammalian cells (19), and the two proteins formed a complex when coexpressed in bacteria. A complex of GST-eIF4H and wild-type Vhs was isolated and shown to have RNase activity, while complexes containing either of two mutant forms of Vhs did not have such activity. These results provide genetic and biochemical evidence that Vhs is an RNase, either alone or as part of a complex with eIF4H.
MATERIALS AND METHODS
Cells.
Vero cells were purchased from the American Type Culture Collection and maintained in Eagle's minimum essential medium (GIBCO) supplemented with 10% (vol/vol) calf serum and antibiotics as described previously (17, 46, 51).
Plasmids.
The plasmid pKOSamp contains the Vhs (UL41) open reading frame from HSV type 1 (HSV-1) strain KOS cloned into the vector pcDNA1.1amp (Invitrogen) downstream from the cytomegalovirus immediate-early promoter as well as a promoter for T7 RNA polymerase (18). It was the parent plasmid for constructing all site-directed mutations in Vhs.
Homology searches and alignments.
To search for Vhs homologues, the UL41 open reading frame from HSV-1 (KOS) was compared to other known protein-encoding sequences by using the BLAST search program (2), and the results were refined by hidden Markov modeling (3, 13, 21, 35). The initial identification of cellular and phage nucleases with similarities to Vhs was achieved through a BLAST search with just the central region of UL41, including amino acids 165 through 265. Subsequent comparison of the sequences of the nucleases with the entire UL41 sequence revealed similarities outside of this central region of Vhs.
Vhs homologues from the alphaherpesviruses were aligned in a multiple alignment by using the ClustalW alignment algorithm of MacVector, version 6.0 (Oxford Molecular, Campbell, Calif.). Alignment of the Vhs proteins with the cellular and phage nucleases was done in several steps. First, the nucleases were divided into four groups (RAD2 DNA repair nucleases, xeroderma pigmentosum [XPG] proteins, flap endonucleases [FEN-1], and DNA polymerases) based on their relative homologies to each other. The ClustalW algorithm was then used to align the proteins in each group, after which the different groups were aligned with each other by performing a multiple alignment to achieve the best alignment of all of the proteins. Finally, the alignment of some proteins was adjusted by visual inspection, since the computer algorithm did not recognize some conserved motifs that were readily apparent by eye.
Site-directed mutagenesis.
The Vhs-expressing plasmid pKOSamp was the parent plasmid used for all site-directed mutagenesis procedures. The wild-type Vhs allele was mutagenized by using a Chameleon double-stranded, site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions but with the modifications described previously (18). Mutagenic primers were designed to create not only the desired change in the Vhs coding sequence but also a new restriction enzyme site to be used in screening the mutant alleles. Mutagenized plasmids were screened by restriction enzyme analysis, sequenced to confirm the nucleotide changes, and analyzed by in vitro transcription and translation with a TnT T7 quick coupled transcription-translation system (Promega Corp., Madison, Wis.) to confirm that they encoded proteins of the expected molecular masses (18). Mutants were named for the number of the residue that is altered, preceded by the wild-type amino acid and followed by the amino acid to which it is changed. For example, in D34N, aspartic acid at residue 34 is changed to asparagine.
DNA isolation and sequencing.
Plasmids for transfection and sequencing were prepared from bacterial lysates by using MidiPrep and MaxiPrep systems as recommended by the manufacturer (Qiagen Inc., Chatsworth, Calif.). Sequencing of the Vhs alleles was performed with an Applied Biosystems model 377 DNA sequencer at the Molecular Biology Core Facility of the University of Missouri—Kansas City (18).
Transient expression assay of Vhs activity.
Vhs activity was measured by determining the ability of a transfected UL41 allele to inhibit the expression of a cotransfected reporter plasmid containing the Escherichia coli lacZ gene under the control of the simian virus 40 early promoter and enhancer (17, 18, 47). Transfection was performed by using a Profection mammalian transfection system (Promega) according to the manufacturer's instructions. Vero cells were plated on the day before transfection in 60-mm-diameter petri dishes at a density of 2.5 × 104 cells/cm2. Cultures were transfected with 0.6-ml aliquots containing calcium phosphate coprecipitates of 3 μg of the reporter plasmid pSV-β-galactosidase (Promega) and 0.73 pmol of either a UL41-containing effector plasmid or the expression vector pcDNA1.1amp. The precipitates also contained enough salmon sperm carrier DNA to bring the total amount of DNA to 12 μg.
Cell extracts were prepared 40 to 48 h after transfection and assayed for reporter gene expression by using a β-galactosidase enzyme assay system (Promega) and a Thermo Max microplate reader (Molecular Devices, Sunnyvale, Calif.) as described previously (17, 18, 47). For each transfection involving a UL41-containing effector plasmid, the amount of β-galactosidase activity was expressed as a fraction of that observed in a transfection involving 0.73 pmol of the empty expression vector pcDNA1.1amp.
Western blotting.
The expression of the Vhs polypeptide in extracts of transfected cells was confirmed by Western blotting with a polyclonal rabbit antiserum raised against a Vhs-LacZ fusion protein as described previously (51).
Expression of recombinant Vhs and GST-eIF4H.
Recently, the Vhs protein was shown to interact with the eukaryotic translation initiation factor eIF4H in vitro in yeast and mammalian cells (19). In the present study, efforts were undertaken to coexpress Vhs and a fusion protein of eIF4H and GST in E. coli. To this end, the Vhs gene was cloned into the NcoI site of pET19b (Novagen) to yield a plasmid encoding an untagged Vhs protein. pGST-4H was described previously (19) and encodes a fusion protein of eIF4H and GST. For the coexpression studies, an EcoRV-SalI fragment was excised from pGST-4H and inserted between the same sites of pLysS (Novagen) to yield a plasmid encoding GST-eIF4H under the control of an isopropyl-β-d-galactopyranoside (IPTG)-inducible tac promoter.
E. coli BL21(DE3) was transformed with both the Vhs- and the GST-eIF4H-expressing plasmids and selected for resistance to both carbenicillin and chloramphenicol. The cells were grown to mid-log phase and induced for 3 to 4 h with 1 mM IPTG. Induced cells were harvested and lysed, and protein complexes that bound glutathione-Sepharose 4B were isolated by using a bulk GST purification module from Amersham-Pharmacia Biotech. Proteins were eluted from glutathione-Sepharose, dialyzed overnight against 50 mM sodium phosphate (pH 7.0), and applied to a 1-ml HiTrap SP Sepharose HP column (Amersham-Pharmacia Biotech) by using a Waters 650E advanced protein purification system. Proteins were eluted with a nonlinear 0 to1 M NaCl gradient. Fractions were concentrated by using Centricon YM-10 filters (Amicon) and analyzed for protein content and RNase activity.
RNase activity.
Target RNAs encoding HSV thymidine kinase were synthesized by in vitro transcription of linearized pBK2 with SP6 RNA polymerase in the presence of a 5′ cap analogue (Stratagene) as described previously (32). The resulting 1,340-nucleotide transcripts contained a 35-nucleotide poly(A) tail encoded by the plasmid.
RNase activity was measured by using the reaction conditions of Zelus and coworkers (75). Reactions were initiated by mixing 5 μl of concentrated gradient fractions with 45 μl of degradation buffer (80 mM potassium acetate, 1.5 mM magnesium diacetate, 2 mM dithiothreitol, 0.1 mM EDTA, 10 U of RNasin [Promega], 25 mM Tris-HCl [pH 7.5]) containing 0.4 μg of target RNA. After incubation at 37°C for various intervals, the RNAs were extracted with phenol-chloroform, precipitated from ethanol, and analyzed by electrophoresis through 1.2% agarose gels (34).
RESULTS
Similarities between Vhs and cellular nucleases.
A comparison of the Vhs (UL41) homologues of HSV-1 and the other alphaherpesviruses reveals that they contain three regions of high sequence homology (5, 17, 31). These consist of amino-terminal and carboxyl-terminal regions of approximately 100 amino acids and a conserved internal region (Fig. 1, first line). Two less conserved regions that vary in length depending upon the Vhs homologue separate the conserved regions. Mutations that affect Vhs activity generally fall within the conserved regions, while those in the less conserved regions often do not affect activity (17, 18, 31, 47).
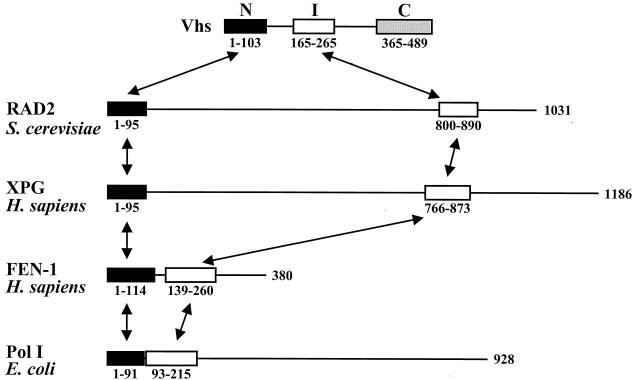
FIG. 1.
Primary structures of Vhs and selected human, yeast, and bacterial nucleases. The primary structure of the Vhs polypeptides is depicted in the first line, with the three homology regions shared by the UL41 homologues from various alphaherpesviruses depicted by the black, white, and grey rectangles labeled N, I (internal), and C. The coordinates of the residues encompassing each of the homology regions differ slightly among the UL41 homologues, as do the distances separating them in the primary sequence. The coordinates shown below the first line refer to the Vhs polypeptide of HSV-1. The primary structures of the RAD2 protein from S. cerevisiae, the human XPG and FEN-1 polypeptides, and DNA polymerase I from E. coli are diagrammed in the second through fifth lines (26, 68). Each of these polypeptides has an amino-terminal domain (depicted by a black rectangle) that is similar to the amino-terminal domain of Vhs and an internal domain (depicted by a white rectangle) that is similar to the internal domain of Vhs. The amino acids that comprise the domains of each of the proteins are shown below the corresponding rectangles, and the overall length of each polypeptide is shown at the right. The distances separating the amino-terminal and internal domains of each polypeptide differ considerably from nuclease to nuclease. Arrows connect homologous domains in the different proteins.
Previous studies revealed sequence similarities between regions of Vhs and several cellular nucleic acid binding proteins and nucleases (12, 17). To extend these studies, the Vhs sequence of HSV-1 was used to perform a BLAST search (2), and the results were refined by hidden Markov modeling (3, 13, 14, 35). The results revealed similarities between the amino-terminal and internal regions of Vhs proteins and a larger family of mammalian, yeast, bacterial, and phage nucleases, including members of the RAD2 DNA repair nucleases, XPG proteins, flap endonucleases (FEN-1), and DNA polymerases (Table 1). The primary structures of the Vhs proteins and representative members of the cellular nucleases are compared in Fig. 1. The nucleases contain two regions of homology: a conserved amino-terminal region and an internal region (10, 26, 39, 68). These, in turn, are similar to the conserved amino-terminal and internal regions of the Vhs proteins. The nucleases differ in overall size and in the length of the primary sequence separating the conserved amino-terminal and internal regions. No obvious homology was observed between the cellular nucleases and the conserved carboxyl-terminal region of the Vhs proteins.
TABLE 1.
Nuclease family
| Category | Proteina | Organismb | Abbreviation | Entrez accession no. |
|---|---|---|---|---|
| DNA polymerase associated proteins | DNA polymerase I | B. burgdorferi | Bbu POLI | 2688462 |
| DNA polymerase I | B. caldotenax | Bca POLI | 416913 | |
| DNA polymerase I | Bacillus stearothermophilus | Bst POLI | 3041672 | |
| DNA polymerase I | Escherichia coli | Eco POLI | 118825 | |
| DNA polymerase I | Helicobacter pylori | Hpy POLI | 2494177 | |
| DNA polymerase I | Streptococcus pneumoniae | Spn POLI | 118827 | |
| DNA polymerase | Thermus aquaticus | Taq POLI | 1942938 | |
| Exonuclease | E. coli | Eco EXO | 2507020 | |
| Exonuclease | Mycoplasma pneumoniae | Mpn EX53 | 2494184 | |
| Exonuclease | Bacillus subtilis | Bsu YPCP | 1730895 | |
| YY30 | Mycobacterium tuberculosis | Mtu YY30 | 1731347 | |
| Exodeoxyribonuclease | Bacteriophage T5 | T5 EXO5 | 119684 | |
| RNase H | Coliphage T4 | T4 RNH | 133162 | |
| Exodeoxyribonuclease | Bacteriophage T3 | T3 EXRN | 119705 | |
| IXPG proteins | XPG | C. elegans | Cel XPG | 2773206 |
| XPG | Xenopus laevis | Xle XPG | 267421 | |
| XPG | Homo sapiens | Hsa XPG | 267420 | |
| XPG | Mus musculus | Mmu XPG | 549454 | |
| Flap endonucleases | Flap endonuclease 1 | X. laevis | Xle FEN1 | 2674207 |
| Flap endonuclease 1 | M. musculus | Mmu FEN1 | 729476 | |
| Flap endonuclease 1 | H. sapiens | Hsa FEN1 | 729475 | |
| RAD2 DNA repair proteins | YEN1 | Saccharomyces cerevisiae | Sce YEN1 | 731457 |
| RAD2 | C. elegans | Cel RAD2 | 529362 | |
| Exonuclease I | Schizosaccharomyces pombe | Spo EXOI | 1706728 | |
| DIN7 | S. cerevisiae | Sce DIN7 | 2501673 | |
| Exonuclease I | S. cerevisiae | Sce EXOI | 1706421 | |
| TOSCA | Drosophila melanogaster | Dme TOSCA | 1419489 | |
| RAD2 | S. cerevisiae | Sce RAD2 | 131811 | |
| RAD13 | S. pombe | Spo RAD13 | 131777 | |
| RA27 | S. cerevisiae | Sce RA27 | 140964 | |
| RAD2 | S. pombe | Spo RAD2 | 730469 | |
| YA31 | S. pombe | Spo YA31 | 1175380 | |
| RAD2 | A. julgidus | Afu RAD2 | 2650376 | |
| RAD2 | Methanobacterium thermoautotrophicum | Mth RAD2 | 2622760 | |
| RAD2 | M. jannaschii | Mja RAD2 | 2127857 |
Nucleases with which Vhs shares homology.
Organisms from which the Vhs homologues are obtained.
Figure 2 shows a sequence comparison of the amino-terminal and internal regions of the Vhs proteins and representative members of the cellular nucleases. The Vhs polypeptides of the alphaherpesviruses are highly homologous, with a large number of identical or conserved residues (Fig. 2, alleles below the double line). A smaller, but significant, amount of similarity is observed in both the amino-terminal and the internal regions of the Vhs proteins and the cellular nucleases. The similarity is most striking for eight charged residues and one hydrophilic residue (highlighted in black in Fig. 2) that are conserved across all of the aligned proteins. These include three residues (D34, D82, and K96) in the amino-terminal region and six amino acids (E192, D194, T211, D213, D215, and D261) in the internal region.
FIG. 2.
Sequences of the amino-terminal and internal domains of Vhs and selected cellular nucleases. The amino acid sequences of the amino-terminal and internal domains of the Vhs polypeptides from seven alphaherpesviruses are shown below the double line; the sequences of the corresponding domains of selected cellular and phage nucleases (listed in Table 1) are shown above the double line. The amino acid coordinates of the Vhs polypeptide from HSV-1 strain KOS are shown at the bottom of each panel. Amino acids that are identical to the residues in HSV-1 strain KOS are shown in white lettering on a dark grey background. Amino acids that represent a conservative change from the residues in HSV-1 strain KOS are shown in black lettering on a light grey background. The names of cellular and phage nucleases for which structural and/or genetic data have identified key residues that are in the active site are highlighted by a light grey background in the leftmost column. The active-site residues from these nucleases that are conserved in other cellular and phage nucleases and in the Vhs polypeptides are shown in white lettering on a black background. These residues were the focus of site-directed mutagenesis of the Vhs polypeptide (see Fig. 3 and 4). VZV, varicella-zoster virus; PRV, pseudorabies virus; BHV, bovine herpesvirus; Galid HV-2, Gallid herpesvirus 2; EHV, equine herpesvirus.
Within the amino-terminal region, an invariant aspartate is present at position 34 of the HSV-1 polypeptide and the corresponding residues of the other Vhs proteins as well as all of the cellular nucleases listed in Table 1 and Fig. 2. Aspartate is found at position 82 of the HSV-1 polypeptide and the corresponding residues of all but two of the other viral and cellular proteins, in which it is glutamate. A third conserved residue is a basic amino acid at position 96 of the HSV-1 sequence. For all of the viral and cellular homologues, this residue is either lysine or arginine.
Within the internal region, acidic amino acids are conserved at positions 192, 194, 213, 215, and 262 of the HSV-1 polypeptide. Aspartate is found at the residue corresponding to position 213 in every one of the proteins and at positions 215 and 262 of all but one of the other viral and cellular homologues. Similarly, glutamate is found at the residue corresponding to position 192 in all but one of the proteins, in which it is aspartate; the residue corresponding to position 194 is uniformly either aspartate or glutamate. The other highly conserved site within the internal region is a serine or threonine at the residue corresponding to position 211 of the HSV-1 polypeptide.
Functional significance of the conserved amino acids.
For a number of the nucleases, structural and genetic studies indicate that many of the conserved residues are located in the active site and are essential for catalytic activity (Tables 2 and 3). Crystal structures have been determined for several of the FEN-1 nucleases (28, 29) as well as T4 RNase H (44), T5 5′ exonuclease (11), and the 5′ exonuclease domain of TaqI DNA polymerase (33). Each of these nucleases binds two or more divalent metal ions, and the conserved residues play key roles in metal binding (10). For the FEN-1 nucleases, one Mg2+ binding site is formed by amino acids corresponding to residues D34, D82, E192, and D194 of Vhs, while a second Mg2+ site is formed by amino acids corresponding to Vhs residues D213, D215, and D261 (28) (Table 2). Alteration of any of these residues to alanine by site-directed mutagenesis abolishes the nuclease activity of human FEN-1, a finding further supporting the conclusion that these amino acids are important for catalysis (59, 60). Similar results were obtained for T4 RNase H. For this nuclease, aspartates homologous to Vhs residues D34, D82, D194, and D213 are important for the binding of one metal ion, while amino acids corresponding to Vhs residues D194, D215, and D261 form hydrogen bonds with the water molecules that are coordinated by a second metal ion (44) (Table 2). In addition, a serine homologous to threonine 211 of Vhs forms a hydrogen bond with the carboxylate of the aspartic acid corresponding to residue D34 of Vhs. Site-directed mutagenesis that alters the residue homologous to D213 of Vhs or any of the aspartates forming the first metal binding site abolishes or greatly reduces the nuclease activity of T4 RNase H, as does altering the serine homologous to threonine 211 of Vhs (6) (Table 3). Interestingly, a point mutation that changes threonine 214 of Vhs to isoleucine abolishes detectable Vhs activity. These and similar results for other nucleases (Tables 2 and 3) indicate that these conserved residues are components of a motif shared by a large group of nucleases.
TABLE 2.
Residues implicated in activity and metal binding by structural studies of nucleasesa
| Vhs residue | Corresponding residue in:
|
|||
|---|---|---|---|---|
| T5 5′ exonuclease (10, 11) | Taq I DNA polymerase (10, 33) | T4 RNase H (10, 44) | Human FEN-1 (10, 28, 29) | |
| D34 | D26 (Mn-1) | D18 (M-1) | D19 (Mg-1) | |
| D82 | D71 (Mg-1) | |||
| K96 | ||||
| E192 | E128 (Mn-1) | E117 (M-3) | ||
| D194 | D119 (M-1, M-3) | D132 (Mg-1, M-2) | ||
| D195 | E131 (Mn-1) | D120 (M-3) | ||
| T211 | S153↔D19 | |||
| D213 | D153 (Mn-1, Mn-2) | D142 (M-1, M-2) | D155 (Mg-1) | |
| D215 | D155 (Mn-2) | D144 (M-2) | D157 (Mg-2) | D181 (Mg-2) |
| D261 | D204 (Mn-2) | D200 (Mg-2) | ||
Key conserved residues of the Vhs polypeptide are shown in column 1. The corresponding residues of cellular and phage nucleases are shown in columns 2 through 5. T5 5′ exonuclease and T4 RNase H have been shown to bind two manganese (Mn-1 and Mn-2) and two magnesium (Mg-1 and Mg-2) ions, respectively. Human FEN-1 has been shown to bind two magnesium ions, while Taq I DNA polymerase binds three metal ions (M-1, M-2, and M-3). Residues that have been shown in structural studies to play key roles in metal binding are indicated for the different nucleases. S153 and D19 of T4 RNase H have been shown to interact by hydrogen bonding. Data are from the references listed at the top of each column.
TABLE 3.
Site-directed mutagenesis of key nuclease residuesa
| Vhs residue | Mutated residue in:
|
||||
|---|---|---|---|---|---|
| T4 RNase H (6) | Human FEN-1 (59, 60, 73) | Human XPG (73) | M. tuberculosis YY30 (43) | E. coli DNA polymerase I (74) | |
| D34 | D19N (nuclease −, sub. binding −) | D34A (nuclease −, sub. binding <<) | D21N (nuclease −) | D13N (nuclease −) | |
| D82 | D71N (nuclease −, sub. binding −) | D86A (nuclease −, sub. binding <) | D77A (reduced) | D73N (nuclease −) | D63A (nuclease −) |
| K96 | K87A (nuclease −, sub. binding −) | R103A (nuclease +) | |||
| E192 | E158A (nuclease −, sub. binding −) | E123Q (nuclease −) | E113A (nuclease −) | ||
| D194 | D132N (nuclease −, sub. binding −) | D125N (nuclease −) | D115A (nuclease −) | ||
| D195 | D126N (nuclease −) | D116A (nuclease −) | |||
| T211 | S153A (nuclease <, sub. binding <) | ||||
| D213 | D155N (nuclease −, sub. binding −) | D179A (nuclease +) | D148N (nuclease −) | D138N (nuclease −) | |
| D215 | D157N (nuclease <<, sub. binding <<) | D181A (nuclease −, sub. binding +) | D812A (nuclease −) | D150N (nuclease −) | D140N (nuclease −) |
| D261 | D200N (nuclease +, sub. binding +) | D233A (nuclease −, sub. binding −) | D202N (nuclease −) | D188A (nuclease −) | |
Key conserved residues of the Vhs polypeptide are shown in column 1. Mutations that have been constructed in the corresponding residues of cellular and phage nucleases are shown in columns 2 through 6, along with their effects upon nuclease activity and substrate (sub.) binding. −, no activity; <<, greatly decreased activity; <, decreased activity; +, active. Data are from the references listed at the top of each column.
Site-directed mutagenesis of Vhs.
If one believes that the sequence similarities shared by Vhs and the cellular and phage nucleases indicate that Vhs is a nuclease, then mutations that alter key conserved residues of the nuclease motif should abolish Vhs activity, just as they do for the nucleases discussed above. To test this prediction, Vhs was altered by site-directed mutagenesis to change eight acidic residues (D34, D82, E192, D194, D195, D213, D215, and D261) as well as threonine 211. The acidic amino acids were changed to their basic counterparts, aspartate to asparagine and glutamate to glutamine. Threonine 211 was changed to serine in one mutant and alanine in another. The activities of the mutant Vhs alleles were assessed by using a transient expression assay of Vhs activity in which Vero cells were transfected with plasmids encoding a lacZ reporter gene and mutant or wild-type Vhs alleles. Vhs activity was determined by the ability of a transfected Vhs allele to inhibit lacZ activity. This assay was previously used to compare the mRNA degradation activities of various mutant Vhs alleles as well as the Vhs polypeptides expressed by HSV-1 and HSV-2 strains (17, 18, 23, 47).
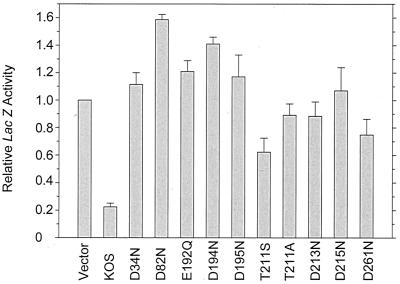
In these experiments, the wild-type Vhs allele reduced lacZ expression to a level 15 to 20% that seen in cells transfected with the reporter gene plus the empty expression vector lacking a Vhs allele (Fig. 3). In contrast, seven of the mutants containing alterations in acidic residues (D34N, D82N, E192Q, D194N, D195N, D213N, and D215N) exhibited reporter gene expression that was at least as great as that seen with the empty expression vector, indicating that they had no detectable Vhs activity in this assay. The same was true for one of the mutants containing an alteration in amino acid 211, T211A. Two of the mutants, T211S and D261N, showed slight inhibition of reporter gene activity, indicating that they had some activity. However, the more active of the two, T211S, reduced reporter gene expression to only 60% that seen in transfections with the empty expression vector (Fig. 3).
FIG. 3.
Ability of wild-type Vhs and site-directed Vhs mutants to inhibit the expression of a cotransfected reporter gene. Triplicate cultures of Vero cells were transfected with 3 μg of the reporter plasmid pSV-β-galactosidase plus 0.73 pmol of pcDNA1.1amp (Vector) or UL41-containing effector plasmids encoding wild-type Vhs or the indicated mutant forms of Vhs. Transfection mixtures also contained enough salmon sperm carrier DNA to bring the total amount of DNA to 12 μg. Cell extracts were prepared 40 to 48 h after transfection and assayed for β-galactosidase activity as described in the text. For each transfection, the amount of β-galactosidase activity was expressed as a fraction of that observed in the transfection involving the vector pcDNA1.1amp. Errors bars indicate the standard error of the mean.
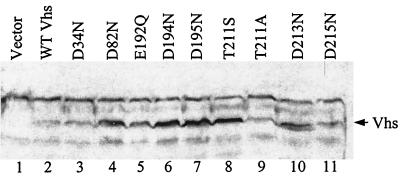
To ensure that the lack of activity of the Vhs mutants was not simply due to reduced levels of the Vhs polypeptide, lysates were prepared from cells transfected with wild-type or mutant Vhs alleles and were analyzed by Western blotting for the Vhs polypeptide. The Vhs protein was detected in all of the lysates from cells transfected with a Vhs allele but not in those from control cells transfected with the empty expression vector (Fig. 4). Significantly, cells transfected with each of the mutant alleles contained at least as much Vhs polypeptide as did cells transfected with the wild-type allele (Fig. 4, compare lane 2 with lanes 3 to 11). Interestingly, cells transfected with some of the mutant alleles contained significantly more Vhs protein than did cells transfected with the wild-type allele (for example; D82N, D194N, D195N, and T211S). This finding was reported previously for other mutants lacking Vhs activity and is presumably due to the fact that an active Vhs protein degrades its own mRNA, thereby reducing the amount of the protein that is produced (47). In any event, the lack of activity that was observed for the mutant alleles clearly was not due to a lower level of expression of the Vhs polypeptide.
FIG. 4.
Expression of mutant and wild-type Vhs polypeptides in transfected cells. Vero cells were transfected with 0.73 pmol of pcDNA1.1amp (Vector) or plasmids encoding wild-type Vhs (WT Vhs) or the indicated mutant forms of Vhs. Whole-cell lysates were prepared 48 h after transfection and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with polyclonal rabbit antiserum raised against a Vhs-LacZ fusion protein. The arrow to the right of lane 11 indicates the position of the Vhs (UL41) polypeptide.
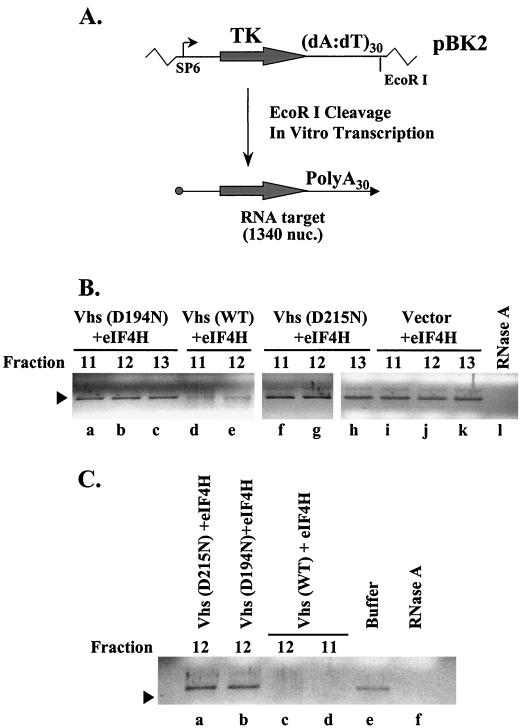
Preparations containing recombinant Vhs and GST-eIF4H have RNase activity.
The above experiments demonstrated that the conserved residues of the nuclease motif are important for Vhs activity and provided strong genetic data that Vhs is itself a nuclease. Nevertheless, to date, the Vhs protein has not been purified and shown to have nuclease activity. To this end, we attempted to purify recombinant Vhs expressed in E. coli. Initial efforts to express His-tagged or GST-tagged Vhs resulted in a protein that was insoluble, except in buffers containing high concentrations of guanidine hydrochloride, urea, or Sarkosyl (data not shown). Attempts to refold the Vhs protein by dialysis or rapid dilution into buffers that lacked a denaturant proved unsuccessful.
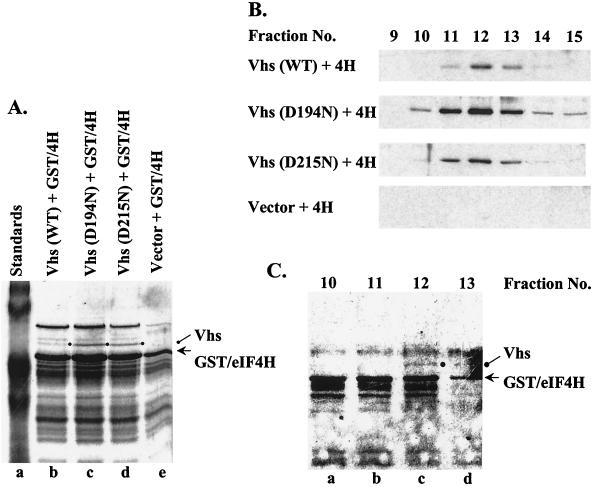
As an alternative approach, we took advantage of our recent observation that, in mammalian cells, Vhs interacts with the cellular translation initiation factor eIF4H. We tried coexpressing untagged Vhs and a GST-eIF4H fusion protein, with the hope that a complex of the two proteins would be more soluble than Vhs alone. E. coli was transformed with plasmids encoding GST-eIF4H and one of three Vhs alleles: wild type, D194N, or D215N. As shown above, the latter two mutants express proteins which have alterations of key residues in the nuclease motif and which do not inhibit reporter gene expression in the transient expression assay. Nevertheless, both mutant proteins have been shown to still bind eIF4H (19). Control bacteria were transformed with the plasmid encoding GST-eIF4H and the vector lacking Vhs. Following induction with IPTG, the bacteria were lysed and the soluble fraction was applied to a column of glutathione-Sepharose to isolate complexes containing GST-eIF4H. GST-eIF4H was the most prominent protein in the bound fraction from each of the four types of bacteria (Fig. 5A). In addition, a 58-kDa polypeptide was observed in bacteria expressing wild-type Vhs or either of the two mutant forms of Vhs (Fig. 5A, lanes b to d) but not in cells expressing just GST-eIF4H (Fig. 5A, lane e). This polypeptide comigrated with Vhs from infected Vero cells and was verified as Vhs by Western blotting (data not shown).
FIG. 5.
Expression of recombinant Vhs and GST-eIF4H. (A) Coomassie blue-stained gel of recombinant proteins bound to glutathione-Sepharose and eluted with 10 mM glutathione. Proteins are from E. coli expressing GST-eIF4H (GST/4H) (lanes b to e) and wild-type (WT) Vhs (lane b), D194N (lane c), or D215N (lane d). Material in lane e is from bacteria expressing GST-eIF4H and no Vhs. Vhs is indicated by a closed circle to the right of lanes b to d, and GST-eIF4H is indicated by an arrow. (B) Material that eluted from glutathione-Sepharose was applied to a column of HiTrap SP Sepharose and eluted with a gradient of 0 to 1 M salt. Fractions were analyzed for Vhs by Western blotting. 4H, eIF4H. (C) Coomassie blue-stained gel of HiTrap SP Sepharose fractions 10 to 13 from cells cells expressing wild-type Vhs and GST-eIF4H. Vhs is indicated by a closed circle, and GST-eIF4H is indicated by an arrow.
Material that eluted from glutathione-Sepharose was applied to a HiTrap SP Sepharose HP cation-exchange column and eluted with a gradient of 0 to 1 M salt. Vhs-containing fractions were identified by Western blotting. Peak amounts of wild-type Vhs and mutant Vhs eluted in fraction 12 of the gradient (Fig. 5B). As expected, Western blotting detected no Vhs protein in fractions from bacteria expressing just GST-eIF4H. Figure 5C shows a Coomassie blue-stained gel of the material in fractions 10 through 13 from bacteria expressing wild-type Vhs. A prominent band corresponding to GST-eIF4H was observed in all four fractions, while the 58-kDa Vhs polypeptide was observed predominantly in fraction 12 and to a lesser extent in fraction 13. Several other proteins were observed in the peak fractions. Whether they were breakdown products of Vhs or of GST-eIF4H or bacterial proteins that copurified with them is unknown.
To test for RNase activity, peak fractions containing GST-eIF4H and each of the three forms of Vhs were incubated for 3 or 16 h with a capped, polyadenylated target RNA produced by in vitro transcription. Three hours of incubation with partially purified GST-eIF4H and wild-type Vhs caused degradation of most of the target RNA (Fig. 6B, lanes d and e), while incubation for 16 h resulted in complete degradation of the target RNA to products that were no longer visible on the gel (Fig. 6C, lanes c and d). In contrast, GST-eIF4H alone or preparations containing GST-eIF4H and either D194N or D215N failed to induce any detectable degradation (Fig. 6B, lanes a to c and f to k, and Fig. 6C, lanes a and b), even after 16 h of incubation, indicating that the mutant polypeptides lacked nuclease activity and that no nonspecific bacterial nuclease copurified with Vhs and GST-eIF4H. Although the peak fractions did not contain a purified complex of Vhs and GST-eIF4H, they were highly enriched for the two proteins. Significantly, Vhs and GST-eIF4H were the only eukaryotic or viral proteins present. Taken together, the results indicate that partially purified preparations of recombinant Vhs and GST-eIF4H have RNase activity.
FIG. 6.
RNase activity of peak fractions containing Vhs and GST-eIF4H. (A) EcoRI cleavage of pBK2 followed by in vitro transcription with SP6 RNA polymerase in the presence of a 5′ cap analogue resulted in the production of a capped, 1,340 nucleotide (nuc.) target RNA with a 35-nucleotide poly(A) tail encoded by the plasmid. TK, thymidine kinase. (B and C) Target RNAs were incubated for 3 h (B) or 16 h (C) with degradation buffer (C, lane e), with RNase A (B, lane l, and C, lane f), or with the indicated HiTrap SP Sepharose fractions from bacteria expressing just GST-eIF4H (B, lanes i to k), GST-eIF4H plus Vhs D194N (B, lanes a to c, and C, lane b), GST-eIF4H plus Vhs D215N (B, lanes f to h, and C, lane A), or GST-eIF4H plus wild-type (WT) Vhs (B, lanes d and e, and C, lanes c and d). RNAs were extracted with phenol-chloroform, precipitated from ethanol, electrophoresed through 1.2% agarose gels, and visualized by staining with ethidium bromide.
DISCUSSION
The Vhs protein is known to accelerate the decay of host and viral mRNAs (49). However, despite considerable study, it has remained unclear whether Vhs is itself an RNase or somehow activates a cellular enzyme. The present study demonstrates that bacterial fractions which are highly enriched for recombinant Vhs and GST-eIF4H have RNase activity. In addition, Vhs shares significant sequence similarities with a number of cellular and phage nucleases, and mutations that alter key conserved residues in Vhs and the nucleases abolish the activities of both. Taken together, the results provide strong evidence that Vhs is an mRNA RNase (mRNase), either alone or as part of a complex with eIF4H.
At present, it is unclear whether recombinant Vhs/GST-eIF4H is an endonuclease, an exonuclease, or both. Data suggesting that it is an endonuclease come from the observation of Elgadi and coworkers that rabbit reticulocyte lysates containing in vitro-translated Vhs contain a Vhs-dependent endonuclease (15). In the present study, recombinant Vhs/GST-eIF4H did not produce discrete degradation intermediates but instead degraded target mRNAs to products that were not observed on the gel. If Vhs is an endonuclease, then such a result could be explained if it cleaved each RNA molecule many times or cleaved each molecule at one or a few sites but at sites that differed from molecule to molecule. In either scenario, the isolated Vhs nuclease would lack the specificity that it demonstrates in vivo, where it shows a strong preference for mRNAs and appears to initiate mRNA degradation at sites of translation initiation (16, 32, 46). This information suggests that one or more additional factors may be required for the targeting of Vhs that is observed in vivo.
In this regard, our observation that Vhs binds the translation factor eIF4H suggests a mechanism for targeting the Vhs nuclease (19). Evidence that this interaction is biologically important is provided by the observation that several Vhs point mutations which abolish its ability to bind eIF4H also abolish its ability to degrade mRNAs in vivo (19). eIF4H shares sequence homologies with eIF4B and appears to be functionally similar in that both stimulate the RNA helicase activity of eIF4A (52-56). eIF4A, in turn, is a component, along with eIF4E and eIF4G, of the tripartite cap binding complex eIF4F (22). Thus, the available data suggest that eIF4H acts at an early stage of translation initiation to help unwind mRNA secondary structures and facilitate ribosome scanning (27).
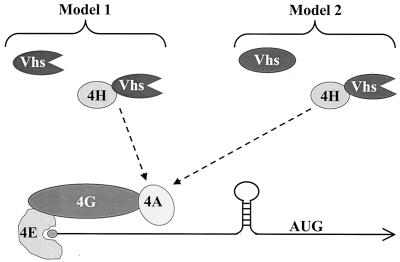
Our data are consistent with two alternative models of Vhs activity (Fig. 7). In model 1, Vhs is itself a nuclease. It requires no other factors to have basal, relatively nonselective endonuclease activity. Binding to eIF4H targets Vhs to mRNAs, as opposed to non-mRNAs, and to regions of translation initiation. In model 2, Vhs is an essential component of an mRNase, perhaps the component that contains the active site. However, by itself the Vhs protein lacks activity. Binding of Vhs to eIF4H serves two purposes. First, it activates the nuclease activity of Vhs, and second, it targets Vhs to mRNAs and regions of translation initiation. Recently, Lu and coworkers published data that would appear to favor model 2 (40). Unfractionated lysates from yeast cells that expressed the Vhs polypeptide lacked RNase activity until after they were supplemented with unfractionated rabbit reticulocyte lysates. This observation is consistent with a model in which one or more mammalian factors are required to activate the nuclease activity of Vhs. However, alternative explanations are possible. An important test of these models will be to purify Vhs to homogeneity and determine whether it has nuclease activity in the absence of eIF4H.
FIG. 7.
Models for Vhs targeting. In model 1, Vhs is itself an RNase. It is targeted to mRNAs and to regions of translation initiation through its interaction with eIF4H. The functional, and perhaps physical, interaction of eIF4H with eIF4A is depicted by the broken arrow. In model 2, Vhs is an essential component of an RNase. However, it does not become active until after binding to eIF4H. The active nuclease is targeted by virtue of its eIF4H component.
The Vhs homologues of the alphaherpesviruses share three blocks of homology (Fig. 1), only two of which, the amino-terminal and internal domains, are related to regions of the cellular and phage nucleases. This result raises the question as to the function of the carboxyl-terminal domain of Vhs. One possibility is that the carboxyl-terminal domain contains sequences required for the interaction of Vhs with eIF4H. In apparent support of this model, a point mutation that changes arginine 435 to histidine abolishes the interaction, as does shortening Vhs from the carboxyl terminus to 454 amino acids (19). However, the interaction is also abolished by several point mutations within the internal domain, as well as by the removal of the first 88 residues from the amino terminus of the protein (19). This finding may indicate that the interaction domain of Vhs is formed by the folding together of residues from different parts of the protein. Alternatively, it is possible that the interaction domain consists of contiguous residues in the primary sequence, but mutations in other parts of the molecule inhibit the interaction with eIF4H by disrupting the folding of the protein. Clearly, additional experiments are required to define the domain of Vhs required for its interaction with eIF4H.
In view of the role of Vhs in mRNA degradation, it is interesting that many of the eukaryotic, bacterial, and phage nucleases with which it shares similarities are involved in DNA repair and replication. One possible explanation is that the conserved residues in Vhs and the cellular nucleases are part of a motif shared by a number of DNases and RNases and that residues responsible for making Vhs an RNase lie elsewhere.
The observation that Vhs binds a helicase accessory factor is particularly interesting in view of studies showing that, in bacteria and yeasts, nucleases involved in mRNA decay also interact with RNA helicases. In bacteria, the degradation of many mRNAs is catalyzed by the degradosome, a multiprotein complex containing endo- and exonucleases (RNase E and polynucleotide phosphorylase), as well as enolase and RhlB, a member of the DEAD-box family of ATP-dependent RNA helicases (8, 9, 48, 72). The purified degradosome has an ATP-dependent activity that facilitates its ability to degrade structured RNAs, and antibody against RhlB inhibits this activity (48). These results suggest that the unwinding of RNA secondary structures by RhlB plays an important role in the degradosome-mediated degradation of mRNAs. Similarly, the exosome is a large complex of proteins that mediates a number of RNA-processing reactions in yeast cells and perhaps mammalian cells (1, 42, 70, 71). Among these, at least in yeast cells, is the 3′-to-5′ degradation of mRNAs (30, 71). The yeast exosome contains at least 10 core proteins, several of them exonucleases, as well as a number of associated factors, including the Ski2p RNA helicase (1, 7, 71). Genetic studies indicate that Ski2p facilitates the 3′-to-5′ degradation of mRNAs, suggesting that the helicase may be important for mRNA decay via disruption of RNA secondary structures or RNA-protein interactions or, perhaps, by targeting of RNA to the exosome (30, 71). Whether the eIF4H-stimulated helicase activity of eIF4A facilitates or in some way modulates the nuclease activity of Vhs remains an important unanswered question.
The above models of Vhs activity hypothesize that the interaction with eIF4H is important for targeting Vhs to mRNAs and regions of translation initiation. However, in this study, preparations containing recombinant Vhs and GST-eIF4H lacked the specificity that is observed for Vhs in vivo. This result suggests that the interaction with eIF4H may be necessary for the targeting of Vhs but not sufficient for it. The interaction between Vhs and eIF4H may be just one in a chain of protein-protein or protein-RNA interactions that are important for normal Vhs activity. Other important interactions may include those between eIF4H and eIF4A, between eIF4A and eIF4G, between eIF4G and eIF4E, or between eIF4G and internal ribosome entry site elements. These and other possibilities are under investigation.
Acknowledgments
We thank Lindsey Hutt-Fletcher for helpful discussion about many aspects of this work. We are indebted to Kelley Thomas and Krys Morris at the University of Missouri—Kansas City (UMKC) Molecular Biology Core Facility for sequencing mutant Vhs alleles. Finally, we thank Marino Martinez-Carrion for inspirational leadership of the UMKC School of Biological Sciences.
This work was supported by grant AI21501 from the National Institute of Allergy and Infectious Diseases and by a grant from the University of Missouri Research Board.
REFERENCES
- 1.Allmang, C., E. Petfalski, A. Podtelejnikov, M. Mann, D. Tollervey, and P. Mitchell. 1999. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 13:2148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldi, P., Y. Chauvin, T. Hunkapiller, and M. A. McClure. 1994. Hidden Markov models of biological primary sequence information. Proc. Natl. Acad. Sci. USA 91:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, Y., E. Tavor, Y. Asher, C. Berkowiltz, and M. Moyal. 1993. Effect of herpes simplex virus type-1 UL41 gene on the stability of mRNA from the cellular genes: beta-actin, fibronectin, glucose transporter-1, and docking protein, and on virus intraperitoneal pathogenicity of newborn mice. Virus Genes 7:133-143. [DOI] [PubMed] [Google Scholar]
- 5.Berthomme, H., B. Jacquemont, and A. Epstein. 1993. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology 193:1028-1032. [DOI] [PubMed] [Google Scholar]
- 6.Bhagwat, M., D. Meara, and N. G. Nossal. 1997. Identification of residues of T4 RNase H required for catalysis and DNA binding. J. Biol. Chem. 272:28531-28538. [DOI] [PubMed] [Google Scholar]
- 7.Brouwer, R., C. Allmang, R. Raijmakers, Y. van Aarssen, W. V. Egberts, E. Petfalski, W. J. van Venrooij, D. Tollervey, and G. J. Pruijn. 2001. Three novel components of the human exosome. J. Biol. Chem. 276:6177-6184. [DOI] [PubMed] [Google Scholar]
- 8.Carpousis, A. J., G. VanHouwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889-900. [DOI] [PubMed] [Google Scholar]
- 9.Carpousis, A. J., N. F. Vanzo, and L. C. Raynal. 1999. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 15:24-28. [DOI] [PubMed] [Google Scholar]
- 10.Ceska, T. A., and J. R. Sayers. 1998. Structure-specific DNA cleavage by 5′ nucleases. Trends Biochem. Sci. 23:331-336. [DOI] [PubMed] [Google Scholar]
- 11.Ceska, T. A., J. R. Sayers, G. Stier, and D. Suck. 1996. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature 382:90-93. [DOI] [PubMed] [Google Scholar]
- 12.Doherty, A. J., L. C. Serpell, and C. P. Pointing. 1996. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 24:2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy, S. R. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6:361-365. [DOI] [PubMed] [Google Scholar]
- 14.Eddy, S. R. 1998. Profile hidden Markov models. Comput. Appl. Biosci. 14:755-763. [DOI] [PubMed] [Google Scholar]
- 15.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus Vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elgadi, M. M., and J. R. Smiley. 1999. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J. Virol. 73:9222-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everly, D. N., Jr., and G. S. Read. 1997. Mutational analysis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): characterization of HSV type 1 (HSV-1)/HSV-2 chimeras. J. Virol. 71:7157-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everly, D. N., Jr., and G. S. Read. 1999. Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles. J. Virol. 73:9117-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, P., D. N. Everly, Jr., and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenwick, M. L., and M. M. McMenamin. 1984. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J. Gen. Virol. 65:1225-1228. [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara, Y., M. Asogawa, and A. Konagaya. 1994. Stochastic motif extraction using hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:121-129. [PubMed] [Google Scholar]
- 22.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 23.Hamouda, T., R. McPhee, S. C. Hsia, G. S. Read, T. C. Holland, and S. R. King. 1997. Inhibition of human immunodeficiency virus replication by the herpes simplex virus virion host shutoff protein. J. Virol. 71:5521-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardwicke, M. A., and R. M. Sandri-Goldin. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and the regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington, J. J., and M. R. Lieber. 1994. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for excision repair. Genes Dev. 8:1344-1355. [DOI] [PubMed] [Google Scholar]
- 27.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-89. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Hosfield, D. J., C. D. Mol, B. H. Shen, and J. A. Tainer. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 95:135-146. [DOI] [PubMed] [Google Scholar]
- 29.Hwang, K. Y., K. Baek, H. Y. Kim, and Y. Cho. 1998. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 5:707-713. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs Anderson, J. S., and R. Parker. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, F. E., C. A. Smibert, and J. R. Smiley. 1995. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that Vhs functions in the absence of other viral proteins. J. Virol. 69:4863-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karr, B. M., and G. S. Read. 1999. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology 264:195-204. [DOI] [PubMed] [Google Scholar]
- 33.Kim, Y., S. H. Eom, J. Wang, D. S. Lee, S. W. Suh, and T. A. Steitz. 1995. Crystal structure of Thermus aquaticus DNA polymerase. Nature 376:612-616. [DOI] [PubMed] [Google Scholar]
- 34.Krikorian, C. R., and G. S. Read. 1991. An in vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J. Virol. 65:112-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogh, A., M. Brown, I. S. Mian, K. Sjolander, and D. Haussler. 1994. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235:1501-1531. [DOI] [PubMed] [Google Scholar]
- 36.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieber, M. R. 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19:240.. [DOI] [PubMed] [Google Scholar]
- 40.Lu, P., F. E. Jones, H. A. Saffran, and J. R. Smiley. 2001. Herpes simplex virus virion host shutoff protein requires a mammalian factor for efficient in vitro endoribonuclease activity. J. Virol. 75:1172-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu, P., H. A. Saffran, and J. R. Smiley. 2001. The Vhs1 mutant form of herpes simplex virus virion host shutoff protein retains significant internal ribosome entry site-directed RNA cleavage activity. J. Virol. 75:1072-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91:457-466. [DOI] [PubMed] [Google Scholar]
- 43.Mizrahi, V., and P. Huberts. 1996. Deoxy- and dideoxynucleotide discrimination and identification of critical 5′ nuclease domain residues of the DNA polymerase I from Mycobacterium tuberculosis. Nucleic Acids Res. 24:4845-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueser, T. C., N. G. Nossal, and C. C. Hyde. 1996. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell 85:1101-1112. [DOI] [PubMed] [Google Scholar]
- 45.Oroskar, A. A., and G. S. Read. 1987. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J. Virol. 61:604-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pak, A. S., D. N. Everly, K. Knight, and G. S. Read. 1995. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology 211:491-506. [DOI] [PubMed] [Google Scholar]
- 48.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 49.Read, G. S. 1997. Control of mRNA stability during herpes simplex virus infections, p. 311-321. In J. B. Harford and D. R. Morris (ed.), mRNA metabolism and post-transcriptional gene regulation. Wiley-Liss, Inc., New York, N.Y.
- 50.Read, G. S., and N. Frenkel. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate-early) polypeptides. J. Virol. 46:498-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richter, N. J., G. W. Rogers, J. O. HEnsold, and W. C. Merrick. 1999. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 274:35415-35424. [DOI] [PubMed] [Google Scholar]
- 53.Richter-Cook, N. J., T. E. Dever, J. O. HEnsold, and W. C. Merrick. 1998. Purification and characterization of a new eukaryotic protein translation factor-eukaryotic initiation factor 4H. J. Biol. Chem. 273:7579-7587. [DOI] [PubMed] [Google Scholar]
- 54.Rogers, G. W., W. F. Lima, and W. C. Merrick. 2001. Further Characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 276:12598-12608. [DOI] [PubMed] [Google Scholar]
- 55.Rogers, G. W., Jr., N. J. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914-30922. [DOI] [PubMed] [Google Scholar]
- 56.Rogers, G. W., Jr., N. J. Richter, and W. C. Merrick. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274:12236-12244. [DOI] [PubMed] [Google Scholar]
- 57.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Williams, Philadelphia, Pa.
- 58.Schek, N., and S. L. Bachenheimer. 1985. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J. Virol. 55:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen, B. H., J. P. Nolan, L. A. Sklar, and M. S. Park. 1996. Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J. Biol. Chem. 271:9173-9176. [DOI] [PubMed] [Google Scholar]
- 60.Shen, B. H., J. P. Nolan, L. A. Sklar, and M. S. Park. 1997. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 25:3332-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, T. J., C. E. Ackland-Berglund, and D. A. Leib. 2000. Herpes simplex virus virion host shutoff (Vhs) activity alters periocular disease in mice. J. Virol. 74:3598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sorenson, C. M., P. A. Hart, and J. Ross. 1991. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 19:4459-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strelow, L., T. Smith, and D. A. Leib. 1997. The virion host shutoff function of herpes simplex virus type 1 plays a role in corneal invasion and functions independently of the cell cycle. Virology 231:28-34. [DOI] [PubMed] [Google Scholar]
- 64.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (Vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strelow, L. I., and D. A. Leib. 1996. Analysis of conserved domains of UL41 of herpes simplex virus type 1 in virion host shutoff and pathogenesis. J. Virol. 70:5665-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strom, T., and N. Frenkel. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 61:2198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyama, and M. Azuma. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 81:1763-1771. [DOI] [PubMed] [Google Scholar]
- 68.Szankasi, P., and G. R. Smith. 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267:1166-1169. [DOI] [PubMed] [Google Scholar]
- 69.Tigges, M. A., S. Leng, D. C. Johnson, and R. L. Burke. 1996. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J. Immunol. 156:3901-3910. [PubMed] [Google Scholar]
- 70.Tollervey, D., and J. F. Caceres. 2000. RNA processing marches on. Cell 103:703-709. [DOI] [PubMed] [Google Scholar]
- 71.Van Hoof, A., and R. Parker. 1999. The exosome: a proteasome for RNA? Cell 99:347-350. [DOI] [PubMed] [Google Scholar]
- 72.Vanzo, N. F., Y. S. Li, B. Py, E. Blum, C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wakasugi, M., J. T. Reardon, and A. Sancar. 1997. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J. Biol. Chem. 272:16030-16034. [DOI] [PubMed] [Google Scholar]
- 74.Xu, Y., V. Derbyshire, K. Ng, X. C. Sun, N. D. Grindley, and C. M. Joyce. 1997. Biochemical and mutational studies of the 5′-3′ exonuclease of DNA polymerase I of Escherichia coli. J. Mol. Biol. 268:284-302. [DOI] [PubMed] [Google Scholar]
- 75.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]