Abstract
Maturation of vaccinia virus (VV) core proteins is required for the production of infectious virions. The VV G1L and I7L gene products are the leading candidates for the viral core protein proteinase (vCPP). Using transient-expression assays, data were obtained to demonstrate that the I7L gene product and its encoded cysteine proteinase activity are responsible for vCPP activity.
Traditional antiviral compounds have focused on viral nucleic acid-synthesizing enzymes, but since viruses are obligate intracellular parasites which utilize many of the host cell enzymes during their replication, it has proved difficult to identify compounds that specifically block viral enzymes. Fortunately, the emerging realization that most viruses use proteolysis catalyzed by virus-encoded proteinases as a key step in their developmental cycle has opened up a new class of targets for antiviral-drug development. Recently, proteinase inhibitors have been developed that specifically target human immunodeficiency virus, rhinovirus, and influenza virus enzymes and have proven very effective at preventing disease in the human host. Based on the fact that conditional lethal mutants and metabolic inhibitors of late protein synthesis such as α-amanitin result in the assembly of immature particles but no proteolytic maturation and no infectivity, it appears that proteolytic maturation of orthopoxvirus core proteins is required for infectious progeny to be produced (8).
There are two types of proteolytic processing that occur during viral replication, formative and morphogenic (7), both of which are used by poxviruses such as vaccinia virus (VV). Obligatory morphogenic cleavage has been demonstrated for three of the major structural proteins found in the mature VV virion, 4a, 4b, and 25K (15), thereby providing a viable target, the orthopoxvirus core protein, for poxvirus antiviral-drug development. The goal of the experiments reported here was to identify the VV gene that encodes the viral core protein proteinase (vCPP). Currently, there are two putative VV proteinases, the products of the G1L and I7L open reading frames (ORFs).
The VV G1L ORF encodes a 67-kDa late protein suspected to be a metalloproteinase by virtue of its homology to the insulin-degrading enzyme family of metalloproteinases. In common with this family, the G1L protein contains both the inverted H-X-X-E-H active site and a downstream E-N-E metal-binding site. Furthermore, the G1L protein was previously demonstrated to direct the in vivo endoproteolytic cleavage of the VV P25K core protein precursor, albeit at a cryptic cleavage motif. This cleavage activity was inhibited if either the active site or metal-binding domain were mutated, suggesting that G1L-mediated catalysis was required (17).
The VV I7L ORF encodes an approximately 47-kDa late protein believed to be a cysteine proteinase due to its homology to the African swine fever virus proteinase and the adenovirus proteinase, both of which are known to process viral core proteins in their respective systems. The I7L protein, like these other enzymes, contains putative catalytic dyad residues, histidine and cysteine, embedded in a conserved region containing an aspartic acid. These enzymes, including the I7L protein, also contain an invariant glutamine (Q) residue just upstream of the cysteine residue, which is predicted to form the oxyanion hole in the active site (10). The I7L gene is known to be essential for viral replication because a conditional lethal mutant, ts16, has been mapped to this locus (5). Interestingly, at the nonpermissive temperature, ts16 displays a defective late phenotype in which immature particles are assembled containing uncleaved core protein precursors, which is consistent with I7L having a role in core protein processing.
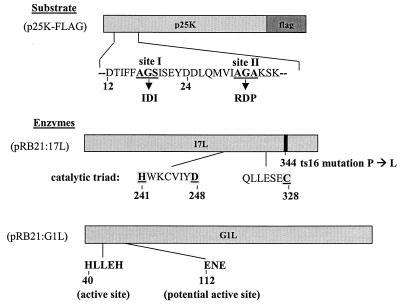
In order to determine whether G1L or I7L is the vCPP, it was necessary to develop an in vivo trans-processing assay because all previous attempts to demonstrate vCPP activity in cell extracts have failed. Earlier studies conducted in our lab have shown that the vCPP substrates include P4a, P4b, P25K, and P17K (12, 13, 15, 17), which are all proteolytically processed during viral assembly. Alignment of the cleavage sites in these precursors revealed a conserved AG∗X cleavage motif (16). For the present study, P25K was used as the reporter substrate. P25K is the product of the L4R gene and was chosen because it is the smallest of the major core protein precursors and is relatively soluble. P25K contains two putative cleavage sites, a cryptic AG∗S site at amino acids 17 to 19 and the AG∗A site at amino acids 31 to 33 that is the authentic cleavage site (Fig. 1). The P25K precursor was tagged at the C terminus with an octapeptide epitope, FLAG (13), in order to monitor proteolytic cleavage of the substrate and distinguish it from the L4R gene product encoded within the viral genome. To further characterize the cleavage site, site-directed mutagenesis was used to create two mutations in the P25K ORF, altering the amino acids at the two cleavage sites. The first mutation involved changing amino acids 17 to 19 from AGS to IDI, and the second mutation involved changing amino acids 31 to 33 from AGA to RDP (Fig. 1). This assay system and these mutations were previously developed to allow a mutagenic analysis of the cis signals required for P25K processing (13).
FIG. 1.
Structures of the P25K:FLAG, I7L, and G1L expression vector plasmids.
Both the I7L and G1L gene products were expressed from the plasmid pRB21 that was constructed with a six-His tag fused to the C terminus and that has a synthetic early-late VV promoter. The six-His tag was included to facilitate subsequent purification of the enzymes in the event that they demonstrated activity in the assay. In Fig. 1, the proposed catalytic dyad of I7L is shown along with the conserved amino acids (His241, Asp248, Cys328), which are common to this family of cysteine proteinases. Also shown is the single amino acid that was found to be mutated in ts16 (Pro to Leu at position 344) and that conferred a temperature-sensitive phenotype on the enzyme. Similarly indicated are the active sites of G1L, an H-L-L-E-H motif at amino acids 40 to 44 which is inversely related to the conserved H-X-X-E-X active sites of metalloproteinases, similar to insulin-degrading enzymes (2), and the E-N-E metal-binding site at positions 112 to 114.
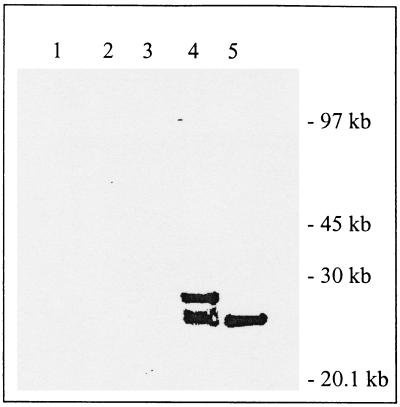
In order to test whether either of these enzymes is the vCPP required for cleavage of the core protein precursor P25K, a transient-expression assay was utilized in which cells were infected with VV and then transfected with plasmids encoding the P25K:FLAG reporter in the presence or absence of pRB21:I7L or pRB21:G1L. Under these conditions, the virus supplies both RNA polymerase and trans-acting factors necessary to drive expression of the substrate and enzyme in the cytoplasm of the infected cells. The initial experiments were carried out using ts16 as the source of superinfecting virus so that viral assembly would be blocked, perhaps providing essential cofactors to the processing reaction. Total cell extracts were prepared from the infected cells 24 h after transfection and subjected to immunoblot analysis using rabbit anti-I7L polyclonal antiserum (data not shown), anti-G1L antiserum (data not shown), or mouse anti-FLAG M2 monoclonal antibody (MAb) (Fig. 2). As expected in the uninfected cells, ts16-infected cells, or ts16-infected cells transfected with pRB21:I7L or pRB21:G1L, there was no specific substrate signal (Fig. 2, lanes 1 to 4). In the absence of a source of exogenous proteinase, P25K:FLAG appears as an unprocessed precursor protein with an apparent molecular mass of 28 kDa (Fig. 2, lane 5). When pRB21:I7L was cotransfected with p25K:FLAG, the P25K:FLAG substrate was completely cleaved to a 25-kDa species consistent with processing at the AG∗A site (Fig. 2, lane 6). In contrast, cotransfection of pRB21:G1L with p25K:FLAG resulted in no demonstrable cleavage of the P25K precursor. This result strongly suggests that at least in regard to P25K, I7L appears to be the vCPP. Furthermore, cleavage of P4b was also rescued by I7L in ts16-infected cells (data not shown).
FIG. 2.
trans complementation of P25K:FLAG processing in ts16-infected cells. BSC-40 cells were infected with a temperature-sensitive VV mutant (ts16) and transfected with pRB21:17L, pRB21:G1L, p25K:FLAG, or a combination of these, and cleavage of the P25K:FLAG substrate was determined by Western blotting using anti-FLAG MAb. Lane 1, uninfected cells as a negative control; lane 2, ts16 virus alone; lane 3, ts16 with I7L; lane 4, ts16 with G1L; lane 5, ts16 with the P25K:FLAG substrate; lane 6, ts16 with I7L and P25K:FLAG; lane 7, ts16 with G1L and P25K:FLAG.
We next sought to determine whether the apparent I7L processing of the P25K:FLAG precursor could be observed in cells infected with wild-type VV instead of ts16 (Fig. 3). This experiment was performed to determine whether the reaction would proceed while viral maturation was occurring and to ensure that the result with ts16 was not the result of an unmapped second site mutation. As can be seen in Fig. 3, lane 4, partial conversion of P25K:FLAG to 25K:FLAG is observed in the absence of plasmid-derived I7L. Coexpression of pRB21:I7L with P25K:FLAG drove processing to completion and produced a product with the same apparent molecular weight (Fig. 3, lane 5). Analysis of this blot with anti-I7L antisera demonstrated the presence of the wild-type VV I7L as a 47-kDa band in lane 4, with an increased signal in lane 5 (data not shown), which is consistent with the extent of processing being dictated by the amount of I7L protein that was present.
FIG. 3.
trans complementation of P25K:FLAG processing in wild-type VV-infected cells. Cells were infected with wild-type VV and transfected with pRB21:17L, p25K:FLAG, or a combination of these, and cleavage of the P25K:FLAG substrate was determined by Western blotting using anti-FLAG MAb. Lane 1, cells alone; lane 2, wild-type VV; lane 3, VV with I7L; lane 4, VV with P25K:FLAG; lane 5, VV with I7L and P25K:FLAG.
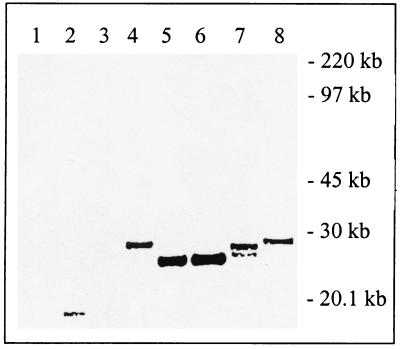
In order to demonstrate that the putative proteinase activity of I7L was directly involved in the processing of the P25K:FLAG precursor and that processing was occurring at the authentic A-G-A site, we utilized a site-specific mutagenesis approach in combination with the trans-complementation assay in ts16-infected cells incubated at the nonpermissive temperature (Fig. 4). As can be seen in Fig. 4, lanes 1 to 3, no anti-FLAG reactive proteins were detected in control cells, ts16-infected cells, or ts16-infected cells transfected with pRB21:I7L alone. Figure 4, lane 4, shows that a 28-kDa immunoreactive band is present when P25K:FLAG is transfected into infected cells and that this precursor is quantitatively converted to a 25-kDa species when pRB21:I7L is cotransfected (Fig. 4, lane 5). Processing of the P25K:FLAG precursor was not inhibited when the cryptic A-G-S was mutated to I-D-I (Fig. 4, lane 6), a mutation previously shown to inhibit cleavage by the G1L gene product (17). In contrast, when the authentic A-G-A site was mutated to R-D-P, there was only minimal processing and the size of the product was consistent with cleavage at the upstream A-G-S site (Fig. 4, lane 7). This cleavage activity could be mediated either by the I7L gene product or, more likely, by the G1L gene product as previously described. Also, as previously described, it should be noted that insertion of the proline residue results in slightly faster migration of the P25K:FLAG precursor, but this does not affect access to the A-G-S site (13). Finally, the effect of mutating the putative active site of the I7L protein was investigated. As shown in Fig. 4, lane 8, mutation of the histidine residue at position 240 in I7L to an alanine completely blocked I7L-mediated cleavage of the P25K:FLAG precursor. Taken together, these data support the conclusion that the I7L gene product is cleaving the P25K:FLAG precursor at the authentic A-G-A site and that this reaction requires the I7L gene product to be catalytically active. This conclusion was supported by the results of an experiment in which the replication of ts16 was rescued 15-fold by a plasmid copy of I7L whereas the mutant I7L (mutated in the active site) was incapable of rescuing replication (data not shown).
FIG. 4.
Mutational analysis of the enzyme and substrate requirements in the trans complementation of P25K:FLAG processing. Cells were infected with ts16 virus and transfected with pRB21:17L, p25K:FLAG, p25K:FLAG:IDI, p25K:FLAG:RDP, the pRB21:I7L mutant, or a combination of these, and cleavage of the P25K:FLAG substrate was determined by Western blotting using anti-FLAG MAb. Lane 1, cells alone; lane 2, ts16 alone; lane 3, ts16 with I7L; lane 4, ts16 with P25K:FLAG; lane 5 ts16 with I7L and P25K:FLAG; lane 6, ts16 with I7L and the P25K:IDI mutant; lane 7, ts16 with I7L and the P25K:RDP mutant; lane 8, ts16 with the I7L mutant and P25K:FLAG.
Previous studies in our laboratory have identified the unique cis signals required to direct endoproteolytic cleavage of core protein precursors (13), established the contextual requirements of core protein maturation (11), and suggested strongly that the proteinase which carries out this essential reaction is virus encoded. The transfection experiments have demonstrated that the gene product encoded by the I7L ORF is likely to be the proteinase that recognizes and cleaves the canonical A-G-A motif found in several of the major core protein precursors (Fig. 2 to 4). This conclusion is supported by the studies of Condit et al. (3) and Kane and Shuman (9), working with the ts16 mutant, who demonstrated that I7L was an essential late gene product. At the nonpermissive temperature, the core protein precursors are synthesized but not processed and no infectious progeny are produced, which is consistent with a proteinase-minus phenotype (5).
If it is assumed that the I7L gene product is the major vCPP involved in VV core protein maturation, then what is the function of G1L-encoded metalloproteinase and what are its in vivo substrates? Previous experiments demonstrated that the G1L gene product has proteolytic activity in vivo on an A-G-S site, and the present work conclusively shows that the I7L gene product recognizes and cuts at an A-G-A site. These data give rise to the hypothesis that both enzymes are involved in VV core protein maturation, with I7L recognizing the A-G-A motifs found in P25K and P4b and G1L recognizing the A-G-S, A-G-T, and A-G-K motifs found in P4a and P21K. There are several lines of evidence supporting the idea that there are two functionally different classes of A-G-X motifs: (i) A-G-A motifs are found near the N termini of precursor proteins, whereas A-G-S/T/K motifs are found within the interior of the precursor proteins; (ii) A-G-A sites are cleaved more rapidly than are A-G-S and A-G-T sites (15); and (iii) mutagenesis studies have shown that A-G-A sites are flanked by conserved features (upstream by a V or I residue at the −4 position, relative to the scissile bond, and downstream by a number of basic residues [R or K]) which are essential for efficient cleavage (12). A-G-S and A-G-T sites lack these features but still cleaved efficiently. Taken together, this suggests that there may be two subclasses of A-G-X sites within VV core protein precursors that are recognized by two separate and distinct proteinases. The potential biological relevance of this hypothesis for virion assembly remains to be examined, but the two enzymes may well play a regulatory role ordering processing reactions such as that proposed for the fast and slow cleavage sites within the potyvirus polyprotein (4).
Having two poxvirus proteinases apparently involved in virion maturation provides two targets for the development of drugs against poxviruses. This is highly advantageous for several reasons. First, not all targets are equally “drugable” due to the inherent structural features of the protein. Second, there can be specificity issues for any given target. For instance, I7L shares sequences with the African swine fever virus proteinase that processes proteins at G-G-X sites (1) and with the adenovirus protease that cleaves at G-G-A sites (14), and it shares the critical residues that surround the catalytic triad of a proteinase found in the ubiquitin pathway of protein degradation, SUMO-1 (1), as well as the YopJ proteinase of Yersinia pestis (14). Thus, I7L inhibitors could be broad-spectrum anti-infectives or they could lack the specificity required to be effective drugs. Finally, due to inherently high mutation rates, viruses have the ability to rapidly acquire resistance when exposed to drug selection. Having inhibitors directed at two targets, a cysteine proteinase (I7L) and a metalloproteinase (G1L), will enable the use of both in combination to achieve synergistic inhibition or allow one family of inhibitors to be held in reserve as a drug of last resort. Given the current concerns regarding smallpox as an agent of bioterrorism, it is essential that effective poxvirus antiviral drugs are developed and are available in our pharmaceutical repertoire to complement the existing vaccine. Furthermore, such drugs should be effective in the event that other orthopoxvirus pathogens find their way into the human population. Fortunately, the orthopoxviruses are highly related at the DNA level (e.g., 90% identity between variola virus and VV), making it likely that any antiviral agent developed would inhibit the replication of this entire group of viruses (6).
Acknowledgments
We thank R. C. Condit for providing us with the ts16 VV mutant.
This work was supported by NIH grant no. AI-2.
REFERENCES
- 1.Andres, G., A. Alejo, C. Simon-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 2.Becker, A. B., and R. A. Roth. 1992. An unusual active site identified in a family of zinc metalloendopeptidases. Proc. Natl. Acad. Sci. USA 89:3835-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condit, R. C., A. Motyczka, and G. Spizz. 1983. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology 128:429-443. [DOI] [PubMed] [Google Scholar]
- 4.Dougherty, W. G., and T. D. Parks. 1989. Molecular genetic and biochemical evidence for the involvement of the heptapeptide cleavage sequence in determining the reaction profile at two tobacco etch virus cleavage sites in cell-free assays. Virology 172:145-155. [DOI] [PubMed] [Google Scholar]
- 5.Ericsson, M., S. Cudmore, S. Shuman, R. C. Condit, G. Griffiths, and J. K. Locker. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esposito, J. J., and J. C. Knight. 1985. Orthopoxvirus DNA: a comparison of restriction profiles and maps. Virology 143:230-251. [DOI] [PubMed] [Google Scholar]
- 7.Hellen, C. U., and E. Wimmer. 1992. The role of proteolytic processing in the morphogenesis of virus particles. Experientia 48:201-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hruby, D. E., D. L. Lynn, and J. R. Kates. 1979. Vaccinia virus replication requires the active participation of the host cell transcriptional apparatus. Proc. Natl. Acad. Sci. USA 76:1878-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kane, E. M., and S. Shuman. 1993. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J. Virol. 67:2689-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, K. I., S. H. Baek, Y. J. Jeon, S. Nishimori, T. Suzuki, S. Uchida, N. Shimbara, H. Saitoh, K. Tanaka, and C. H. Chung. 2000. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275:14102-14106. [DOI] [PubMed] [Google Scholar]
- 11.Lee, P., and D. E. Hruby. 1995. Analysis of the role of the amino-terminal peptide of vaccinia virus structural protein precursors during proteolytic processing. Virology 207:229-233. [DOI] [PubMed] [Google Scholar]
- 12.Lee, P., and D. E. Hruby. 1994. Proteolytic cleavage of vaccinia virus virion proteins. Mutational analysis of the specificity determinants. J. Biol. Chem. 269:8616-8622. [PubMed] [Google Scholar]
- 13.Lee, P., and D. E. Hruby. 1993. trans processing of vaccinia virus core proteins. J. Virol. 67:4252-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orth, K., Z. Xu, M. B. Mudgett, Z. Q. Bao, L. E. Palmer, J. B. Bliska, W. F. Mangel, B. Staskawicz, and J. E. Dixon. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594-1597. [DOI] [PubMed] [Google Scholar]
- 15.VanSlyke, J. K., C. A. Franke, and D. E. Hruby. 1991. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J. Gen. Virol. 72:411-416. [DOI] [PubMed] [Google Scholar]
- 16.Whitehead, S. S., N. A. Bersani, and D. E. Hruby. 1995. Physical and molecular genetic analysis of the multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein. J. Gen. Virol. 76:717-721. [DOI] [PubMed] [Google Scholar]
- 17.Whitehead, S. S., and D. E. Hruby. 1994. A transcriptionally controlled trans-processing assay: putative identification of a vaccinia virus-encoded proteinase which cleaves precursor protein P25K. J. Virol. 68:7603-7608. [DOI] [PMC free article] [PubMed] [Google Scholar]