Abstract
The human cytomegalovirus (HCMV) maturational proteinase is synthesized as an enzymatically active 74-kDa precursor that cleaves itself at four sites. Two of these, called the maturational (M) and release (R) sites, are conserved in the homologs of all herpesviruses. The other two, called the internal (I) and cryptic (C) sites, have recognized consensus sequences only among cytomegalovirus (CMV) homologs and are located in the 28-kDa proteolytic portion of the precursor, called assemblin. I-site cleavage cuts assemblin in half without detected effect on its enzymatic behavior in vitro. To investigate the requirement for this cleavage during virus infection, we used the CMV-bacterial artificial chromosome system (E. M. Borst, G. Hahn, U. H. Koszinowski, and M. Messerle, J. Virol. 73:8320-8329, 1999) to construct a virus encoding a mutant I site (Ala143 to Val) intended to be blocked for cleavage. Characterizations of the resulting mutant (i) confirmed the presence of the mutation in the viral genome and the inability of the mutant virus to effect I-site cleavage in infected cells; (ii) determined that the mutation has no gross effect on the rate of virus production or on the amounts of extracellular virions, noninfectious enveloped particles (NIEPs), and dense bodies; (iii) established that assemblin and its cleavage products are present in NIEPs but are absent from CMV virions, an apparent difference from what is found for virions of herpes simplex virus; and (iv) showed that the 23-kDa protein product of C-site cleavage is more abundant in mutant virus-than in wild-type virus-infected cells and NIEPs. We conclude that the production of infectious CMV requires neither I-site cleavage of assemblin nor the presence of assemblin in the mature virion.
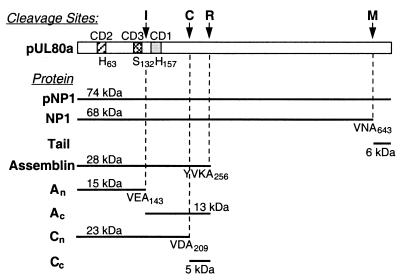
Herpesviruses encode a proteinase that functions during assembly and maturation of the capsid and that is essential for the production of infectious virus (13, 17, 24, 31, 41, 42, 53, 55). The human cytomegalovirus (HCMV) homolog is synthesized as an enzymatically active 74-kDa precursor that cleaves itself at two consensus sequences called the maturational (M) and release (R) sites (Fig. 1), which have counterparts among all herpesviruses (1, 12, 20, 55). M-site cleavage of the HCMV precursor eliminates its 6-kDa carboxyl “tail,” which mediates interaction with the major capsid protein (MCP; pUL86) at the earliest stages of subunit organization and capsid assembly (2, 37, 56). R-site cleavage releases the 28-kDa amino proteolytic domain from the carboxyl portion of the precursor, which targets the proteinase to its site of action within nascent intranuclear capsids (30, 39, 55, 56). The crystal structure of the proteolytic domain, called assemblin, established it as the prototype of a new clan of serine proteinases, distinguished by their Ser-His-His (rather than Ser-His-Asp/Glu) catalytic triad and different protein folds (7, 43, 46, 51). Assemblin is activated by dimerization (8, 9, 32) to constitute an enzyme with two separate catalytic sites and poor catalytic efficiency compared to other serine proteinases (38, 44, 48, 50, 53).
FIG. 1.
Autoproteolytic cleavages of HCMV maturational proteinase. Shown is a schematic representation of the HCMV maturational proteinase, indicating the following features: (i) The I, C, R, and M cleavage sites; (ii) three highly conserved domains (CD1 to CD3) (55), which contain the catalytic triad residues, H63, S132, and H157; (iii) abbreviations (left) used for some of the proteins derived by autoproteolyis of pNP1, the primary translation product of HCMV ORF UL80a; (iv) the computer-predicted size for each cleavage product; and (v) amino acids on the amino side of each cleavage site and the pUL80a residue number of the terminal Ala at each site.
In addition to the M and R sites common to the herpesvirus group, the HCMV proteinase has internal (I) and cryptic (C) cleavage sites within assemblin (Fig. 1) (1, 5, 28, 54). The I site has a recognized consensus sequence among other cytomegalovirus (CMV) homologs and has received comparatively more attention than the C site, which may be restricted to HCMV. The I site is located near the middle of assemblin, between Ser132 and His157 of the catalytic triad, yet its cleavage has no detected effect on the proteolytic behavior of the enzyme (22, 25, 28, 36), in contrast to the inactivating D-site cleavage of the Kaposi's sarcoma herpesvirus, human herpesvirus 8 (40). Furthermore, enzymatic activity can be constituted by coexpressing the separately cloned coding sequences for the two I-site cleavage fragments, An and Ac (22, 23), or by combining the independently expressed fragments in vitro (36). Moreover, an enzymatically active form of the herpes simplex virus (HSV) assemblin homolog, which has no I site, can be constituted by coexpressing the HSV sequences that are counterparts to CMV An and Ac (21). Thus, the significance of I-site cleavage has not been revealed by studies of the cloned enzyme expressed and tested in the absence of other viral proteins.
The work described here was done to test the requirement for I-site cleavage in the context of virus-infected cells by constructing and studying a mutant HCMV encoding a defective I site. Phenotypic characterizations were done to compare the I-site mutant and wild-type virus with respect to cytopathic effect (CPE), yield of infectious virus and noninfectious extracellular particles, I-site cleavage, and incorporation of the proteinase into virus particles. These comparisons establish that I-site cleavage, per se, is not essential for the production of infectious HCMV.
MATERIALS AND METHODS
Cells, viruses, and microscopy.
Viruses were propagated in human foreskin fibroblast (HFF) cells maintained in Dulbecco's modified Eagle medium containing high glucose, 10% fetal calf serum, and 100 U of penicillin and streptomycin/ml (culture medium) (16). HCMV strain AD169 was from the American Type Culture Collection (no. VR-538; Manassas, Va.). The AD169-derived HCMV-bacterial artificial chromosome (HCMV-BAC) system (3) was from Ulrich Koszinowski and Martin Messerle (Max von Pettenkofer Institute, Munich, Germany). Human retinal pigment endothelial (RPE) cells transformed with the reverse transcriptase subunit of human telomerase (hTERT-RPE1; no. C4000-1; BD Biosciences Clontech, Palo Alto, Calif) were used instead of HFFs for transfection, because of their comparatively better transfection efficiency (4; unpublished observations). However, because RPE cells yield much less virus than HFFs, virus stocks were prepared in HFF cells by cocultivation with HCMV-BAC-transfected RPE cells or infection with culture medium from the transfected RPE cells.
CPEs of the virus were monitored and recorded with a Nikon Eclipse TE200 inverted microscope equipped with a Sony DKC5000 imaging system. Contrast of the photomicrographs was adjusted in Adobe Photoshop, version 4.0.
Construction of I- site mutant virus.
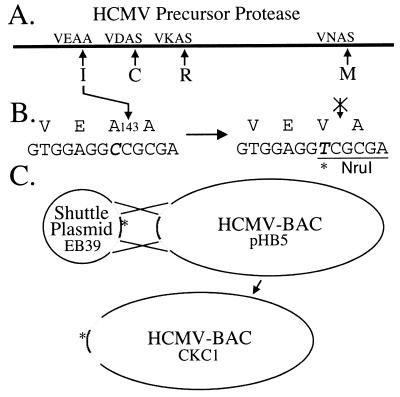
An outline of the procedure used to construct the I-site mutant HCMV is shown in Fig. 2. The HCMV proteinase-coding region (open reading frame [ORF] UL80a), flanked by ≈2 kb on each side, was recovered from AD169 virion DNA as the 5,986-bp NotI/NotI fragment (nucleotides 113129 to 119114) (6) and subcloned into pBluescript II SK(+) (no. 212205; Stratagene, La Jolla, Calif.) to make EB11. The A143V I-site mutation encoded by ORF UL80a, shown to block I-site cleavage of the protein expressed in bacterial (28, 36, 38, 43) and eukaryotic (28) cells, was made in EB11 by using the Quick Change system (no. 200518; Stratagene) with mutagenic primers 5′-GCG ACG ACG TGG AGG TCG CGA CGT CGC TTT CGG-3′ (sense; Val codon is underlined) and 5′CCG AAA GCG ACG TCG CGA CCT CCA CGT CGT CGT CGC-3′ (antisense). The entire mutant NotI fragment was sequenced and transferred from EB11 into shuttle vector pSTKS11 (modified to have a unique NotI cloning site, RC3) to give EB39. Aside from the intended mutation, only one other difference from the published sequence for AD169 was detected: thymidine at nucleotide 113423 instead of cytosine, which changes codon 167 of UL78 from a Ser codon to a Phe codon. This difference was also present in the parental wild-type HCMV-BAC and AD169 from the American Type Culture Collection but not in a clinical isolate (strain 751 [15]) tested for comparison (data not shown) and was not further investigated.
FIG. 2.
Construction of I-site mutant HCMV. (A) HCMV proteinase precursor, its four cleavage sites, and their P3 to P1′ cleavage sequences (1, 55). (B) Amino acid and corresponding nucleotide sequences of the native (left) and mutant (right) I sites, with the new NruI restriction enzyme site underlined. The Ala codon, GCC, was mutated to the Val codon, GTC, by a C-to-T change at nucleotide 115625 (italicized) in the AD169 sequence (6). (C) Homologous recombination between shuttle plasmid EB39 and HCMV-BAC, pHB5, that transfers the A143V-coding mutation into the viral genome to form mutant HCMV-BAC, CKC1. The asterisk denotes a mutation.
EB39 was then electroporated into CBTS bacteria transformed with the HCMV-BAC DNA (pHB5), and recombinant mutant HCMV-BACs were identified by their cleavage at an NruI site created by the A143V mutation (Fig. 2B). The gross integrity of the mutant HCMV-BAC selected for further study was confirmed by its fragment pattern following cleavage with HindIII and with EcoRI, and the presence of the mutation was verified by sequencing an inclusive PCR fragment.
HCMV-BAC DNAs were prepared by using Maxi-Prep columns (no. 12462; Qiagen, Valencia, Calif.) and transfected into RPE cells by using Cellphect (no. 27-9268-01; Amersham Pharmacia Biotech, Piscataway, N.J.). Approximately 10 days after transfection the RPE cells were removed from the culture dish by treatment with trypsin (0.05% in calcium- and magnesium-free phosphate-buffered saline [PBS] with 0.53 mM EDTA) and seeded in a ratio of ≈1:3 with trypsin-treated HFF cells. When wild-type HCMV-BAC DNA was used, the resulting cocultures generally showed extensive viral CPE within 2 weeks.
Assay of virus infectivity.
Virus titer was determined by an end point dilution assay for CPE with 96-well microtiter plates. Medium from cultures of infected cells was cleared of particulates by centrifugation at 16,000 × g for 2 min. Serial 10-fold dilutions of the clarified samples were made in culture medium, and 100 μl of each was combined in respective wells with 100 μl of HFF cells in culture medium (≈500 cells/ml). Cultures were incubated at 37°C for up to 8 weeks and monitored for the appearance of viral CPE. Culture medium was typically replaced every 2 weeks. Virus titer was calculated as the 10-fold sample dilution in the last well showing viral CPE.
Recovery of extracellular virus particles, polyacrylamide gel electrophoresis (PAGE), and Western immunoassay.
Extracellular virus particles were recovered from the culture medium by rate-velocity sedimentation in 20 to 50% (wt/wt) sucrose gradients (in 40 mM phosphate buffer containing 150 mM NaCl, pH 7.4), essentially as described before (16, 27). Following centrifugation (39,000 rpm, 4°C, 20 min, Beckman SW41 rotor), particles were visualized by light scattering and collected by needle aspiration through the wall of the centrifuge tube; diluted 1:1 with gradient buffer and concentrated by pelleting (35,000 rpm, 4°C, 2 h, Beckman SW55Ti rotor); suspended in 30 to 50 μl of protein sample buffer, a mixture containing 3 parts of 4× NuPAGE buffer (40% [wt/wt] glycerol, 8% [wt/wt] lithium dodecyl sulfate, 0.8% [wt/wt] EDTA, 0.075% [wt/wt] Serva blue G250, 0.025% [wt/wt] phenol red, 563 mM Tris base, 423 mM Tris-HCl) (no. NP0007; Invitrogen, Carlsbad, Calif.) and 2 parts of 1 M dithiothreitol; and stored at −80°C until analyzed.
Proteins were separated by electrophoresis in polyacrylamide gels by using sodium dodecyl sulfate (SDS)-containing electrode buffers (SDS-PAGE). Specific conditions of SDS-PAGE are stated in the relevant figure legends. Proteins were either stained with Coomassie brilliant blue (CBB) (14) and imaged and quantified with a Kodak EDAS290 system or electrotransferred to Immobilon (no. IPVH0010; Millipore Corp., Bedford, Mass.) and subjected to Western immunoassay as described before (52, 54). The anti-minor capsid protein (mCP) antipeptide antiserum to HCMV mCP (pUL85) has been described previously (18). A rabbit antiserum to HCMV assemblin (i.e., residues 1 to 256 of HCMV pUL80a) that had been purified by Ni2+-chelate chromatography via an amino His/Trp handle (47) was prepared and subjected to SDS-PAGE. The assemblin-containing strip was cut from the gel, deionized, pulverized, and used as an immunogen (Hazelton Biologics, Lenexa, Kans.). Images of Western immunoassays were obtained and analyzed by using a Fuji BAS1000 phosphorimager and Fuji ImageGauge, version 3.3, software.
RESULTS
Cells transfected with I-site mutant HCMV-BAC developed typical CMV CPE.
Wild-type and I-site mutant HCMV-BAC DNAs were transfected into separate cultures of RPE cells, which were removed from the dish with trypsin after approximately 2 weeks and seeded together with HFF cells into new dishes. Within 2 weeks of coculture, CPE characteristic of CMV became apparent in both wild-type virus- and mutant virus-infected cells and progressed to involve the entire culture. When CPE was extensive, cells and medium were collected together and frozen at −80°C as primary stocks. Subsequent titrations of these stock preparations, cleared of cellular debris by centrifugation (16,000 × g, 2 min), gave virus titers of ≈107 PFU/ml for both after 4 weeks of incubation.
Wild-type and mutant viruses are produced at comparable rates.
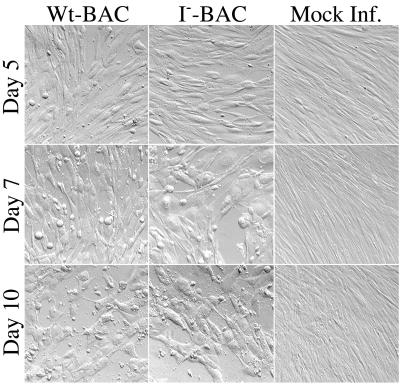
The time courses of virus production for wild-type and mutant viruses were compared by infecting 10-cm-diameter petri dish cultures of subconfluent HFF cells with 50 μl (multiplicity of infection ≈ 10) of the clarified stocks described above. One day after adding the inoculum, the culture medium was aspirated and replaced with 11 ml of fresh medium, and a 1-ml sample was removed and frozen at −80°C until assayed. On each of the following 7 days another 1-ml sample was removed and frozen, and 1 ml of fresh medium was added. Noninfected cells were treated in parallel for comparison. Photomicrographs of the cultures were taken on days 5, 7, and 10 (Fig. 3). CPEs were noted in both infected cultures by day 3 and were not distinguished in extent or appearance between wild-type virus- and mutant virus-infected cells. CPE became more extensive on days 7 to 10, but the overall appearance of the two cultures remained essentially indistinguishable.
FIG. 3.
CPE of wild-type and I-site mutant virus on HFF cells. HFF cells were infected with wild-type (Wt-BAC) or mutant (I−-BAC) HCMV-BAC virus, or not infected (Mock Inf.), and photographed 5, 7, and 10 days after infection. CPEs of CMV infection include cell rounding and enlargement and disruption of the “fingerprint” pattern of fibroblast orientation.
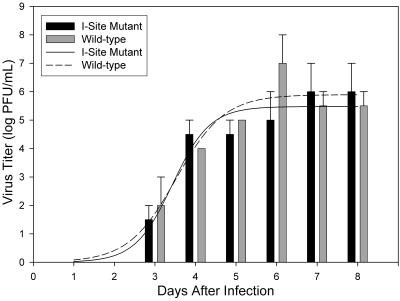
The infectious virus in the eight samples collected from these cultures was measured by an end point assay for CPE. This was done twice because of an irregular 100-fold “spike” in the wild-type virus titer on day 6 in the first assay. The averaged data illustrate that mutant and wild-type viruses yielded infectious progeny at similar rates and reached comparable final titers (Fig. 4). Virus was first detected 3 days after infection, increased through day 6 or 7, and plateaued at a titer of ≈106 to 107 PFU/ml, similar to the level observed before for both AD169 and its HCMV-BAC derivative, RVHB5 (3). The elevated titer in the wild-type virus, day 6 sample was seen in both assays and is attributed to a sampling irregularity (e.g., aspirating a clump of sloughed-off infected cells), rather than a difference between the viruses.
FIG. 4.
Growth kinetics of wild-type versus I-site mutant virus. Samples were collected and frozen on days 1 to 8 from HFF cultures infected with wild-type or I-site mutant HCMV-BAC virus and were titrated for infectivity as described in Materials and Methods. The titration was done twice and data from the two assays were averaged (error bars shown). Shown is a histogram of virus titer on each day of the time course. Curves are computer-derived best fits to data. Samples titered were from the cultures shown in Fig. 3.
Similar amounts of NIEPs, virions, and dense bodies are produced by wild-type virus- and mutant virus-infected cells.
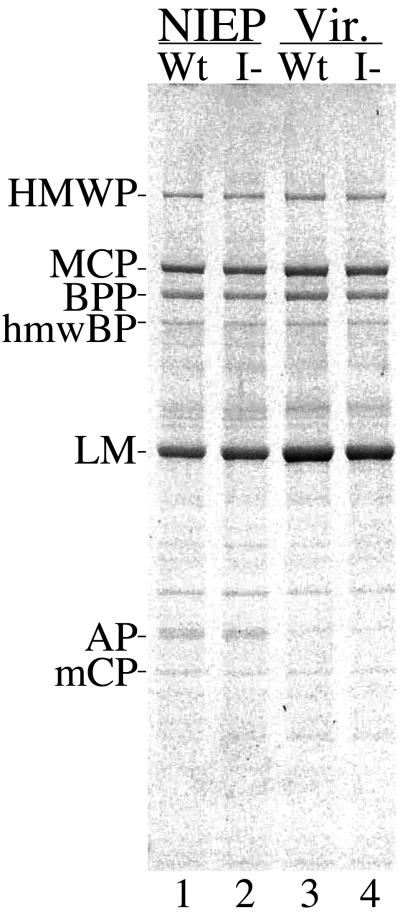
HCMV-infected HFF cells release three types of extracellular particles: virions and noninfectious enveloped particles (NIEPs), both of which contain a capsid, and dense bodies, which do not (26). To determine whether production of these particles was affected by mutating the I site, culture medium from wild-type virus- and mutant virus-infected cells was analyzed by rate-velocity sedimentation in sucrose gradients. The medium used was from the cultures photographed in Fig. 3 and titered in Fig. 4. All three particle types were produced by mutant-infected cells and in amounts comparable to those produced by wild-type virus-infected cells. The bands were collected from the gradients, concentrated, and subjected to SDS-PAGE followed by protein staining with CBB (Fig. 5). There was little difference between the wild type and mutant in either the gross (i.e., ±11%) or relative (i.e., ±5%) abundance of virions and NIEPs, as determined by the amount of MCP (constant number per virion or NIEP) present in these preparations (Table 1; Fig. 5; data not shown for dense bodies). At this level of sensitivity, the respective protein compositions of virions and NIEPs from wild-type and mutant viruses were indistinguishable (Fig. 5). Thus, both biological (Fig. 4) and physical (Fig. 5) measurements indicate that the mutant and wild-type viruses are produced in similar amounts.
FIG. 5.
Wild-type and I-site mutant give rise to similar extracellular particles. NIEPs and virions (Vir.) were collected from HFF cells infected with either wild-type (Wt) or mutant (I−) HCMV-BAC virus, concentrated, and subjected to SDS-PAGE in a NuPAGE 4 to 12% polyacrylamide gradient gel (no. NP0323; Invitrogen) run in 3-(N-morpholino)propanesulfonic acid buffer (no. NP0001; Invitrogen), and the proteins were stained with CBB. Shown is an image of the stained gel. Protein abbreviations are as follows: HMWP, high molecular weight protein (pUL48; 253,206 Da); MCP, major capsid protein (pUL86; 153,862 Da); BPP, basic phosphoprotein (pUL32; 112,680 Da); hmwBP, HMWP-binding protein (pUL47; 109,954 Da); LM, lower matrix protein (pUL83; 62,894 Da); AP, assembly protein (pUL80.5; 31,979 Da; migrates slower than expected); mCP, minor capsid protein (pUL85; 34,592 Da).
TABLE 1.
I-site mutation does not affect amounts of virions and NIEPs produced
| Particle | Optical absorbancea (arbitrary units) for:
|
I−/Wt | |
|---|---|---|---|
| Wtb | I−c | ||
| Virion | 9,236 | 8,250 | 0.9 |
| NIEP | 8,437 (1.1) | 7,816 (1.1) | 0.9 |
Values are from measurements of the CBB-stained MCP bands in the four preparations shown in Fig. 5. Values in parentheses are ratios of the values for virions to the values for NIEPs.
Virus particles from wild-type (Wt) HCMV-BAC, RVHB5.
Virus particles from I-site mutant (I−) HCMV-BAC, CKC1.
Nucleotide change confirmed in virion DNA of I-site mutant.
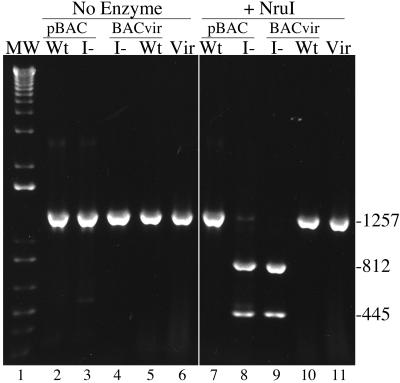
Sucrose gradient-banded virions of standard AD169 and of the wild-type and mutant HCMV-BAC viruses were used as in situ DNA sources to amplify by PCR a 1,257-bp fragment containing the I-site coding sequence. The resulting PCR products were tested for cleavage of the NruI site introduced by the mutation (Fig. 2B). As controls, I-site mutant HCMV-BAC and parental wild-type HCMV-BAC DNAs purified from Escherichia coli were similarly analyzed.
All DNAs yielded the expected 1,257-bp PCR product (Fig. 6, lanes 2 to 6). That from the I-site mutant virus was cleaved to the expected 812- and 445-bp fragments by NruI (Fig. 6, lane 9), indicating that the mutation is present in packaged genomic DNA from the mutant virus. This was confirmed by sequencing the mutant virion DNA PCR product. The positive control (i.e., mutant HCMV-BAC DNA; Fig. 6, lane 8) was also cleaved as expected, and the negative controls were not (i.e., from wild-type HCMV-BACmid DNA and AD169 and wild-type HCMV-BAC virions) (Fig. 6, lanes 7, 10, and 11). A fragment that included the entire UL80 ORF was also amplified by PCR from a preparation of mutant virions and determined to have no changes from the wild-type sequence, other than the intended C-to-T change (Fig. 2B).
FIG. 6.
DNA from I-site mutant virus contains mutagenic nucleotide change. A 1,257-bp sequence beginning at the 5′ end of UL80 was amplified by PCR from five DNA templates and analyzed following incubation without (No Enzyme) or with (+NruI) NruI. Shown is an image prepared from a 1% agarose gel containing the resulting DNA fragments stained with ethidium bromide, together with molecular weight markers (MW, lane 1). PCR templates were wild-type (Wt) and mutant (I−) HCMV-BACs (pBAC; lanes 2 and 7 and lanes 3 and 8, respectively), mutant and wild-type HCMV-BAC virions (BACvir; lanes 4 and 9 and lanes 5 and 10, respectively), and standard AD169 virions (Vir, lanes 6 and 11). Fragment sizes (base pairs) are indicated at the right.
I site not cleaved in cells infected with the mutant.
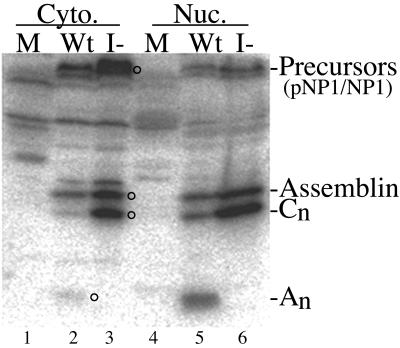
We used Western immunoassays to verify that I-site cleavage is blocked during replication of the mutant virus. HFF cells, infected with either wild-type or mutant virus or not infected, were collected 10 days after infection and were separated into nuclear and cytoplasmic fractions by treatment with NP-40 (16). The three cultures used for this experiment were those shown in Fig. 3, day 10. Samples of the cell fractions were combined with protein sample buffer and subjected to SDS-PAGE in duplicate. The proteins were electrotransferred to Immobilon, and the resulting two membranes were probed with (i) an antiserum to HCMV assemblin (anti-hNP1n) or (ii) a mixture containing antiserum to the mCP (anti-mCP) to permit normalization between samples. Precursor forms of the proteinase (e.g., 74-kDa pNP1 and 68-kDa NP1) were detected in the cytoplasmic fractions of both wild-type virus- and mutant virus-infected cells (Fig. 7, lanes 2 and 3) and in 2.3-fold-reduced amounts in the corresponding nuclear fractions (Fig. 7, lanes 5 and 6). The apparent size difference between mutant and wild-type precursors in the cytoplasmic preparations (Fig. 7, lanes 2 and 3) is seen in some gels but not others and is unexplained. Assemblin was also present in both wild-type virus- and mutant virus-infected cells and was about equally distributed between the respective cytoplasmic and nuclear fractions (Fig. 7). An, the 15.5-kDa amino half of assemblin resulting from I-site cleavage, was detected only in wild-type virus-infected cells and was 4.7-fold more abundant in the nuclear fraction than in the cytoplasmic fraction (Fig. 7). The absence of this diagnostic fragment in mutant virus-infected cells (Fig. 7A, lanes 3 and 6) is evidence that the A143V mutation blocked I-site cleavage, as intended (also see Fig. 8, lanes 5 and 6). A 23-kDa protein that became more abundant in mutant virus-infected cells (Cn, Fig. 7; see also Fig. 8, lanes 5 and 6) is the amino fragment of assemblin, resulting from its cleavage at Ala209 in the C-site sequence (Fig. 1). This assignment is based on (i) its comigration during SDS-PAGE with an assemblin truncation mutant (EB47) ending at Ala209 and (ii) its reaction with an antiserum to purified assemblin (anti-NP1n) but not with an antipeptide antiserum specific for its carboxyl end (i.e., anti-C2 [22]) (Western immunoassay data not shown). By normalizing to the amount of mCP (i.e., 1.04-fold more in mutant), we calculated that there is 2.1-fold more assemblin and 4.9-fold more of the Cn fragment in the nuclear fraction of cells infected with the I-site mutant than in cells infected with the wild type (Table 2; Fig. 7, lanes 5 and 6; Western immunoassay data with anti-mCP not shown).
FIG. 7.
I site is not cleaved in mutant virus-infected cells. Ten days after infection with wild-type (Wt) or mutant (I-) virus (M) or after mock infection, the cells shown in Fig. 3 were collected, separated into cytoplasmic (Cyto.) and nuclear (Nuc.) fractions with NP-40, and subjected to SDS-PAGE in a 12% polyacrylamide gel run in Tris-glycine buffer (29) followed by Western immunoassay using anti-hNP1n. Shown is a phosphorimage of the resulting membrane. Proteins of interest are indicated at the right; abbreviations are as in Fig. 1. Circles to the right of lanes 2 and 3 indicate protein bands corresponding to designations at the right.
FIG. 8.
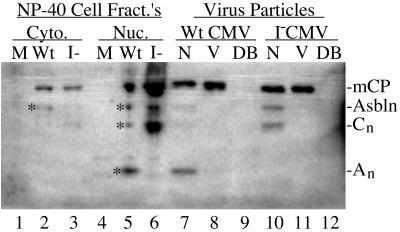
Wild-type and I-site mutant assemblin and specific cleavage products are present in wild-type and mutant NIEPs but not in virions or dense bodies. NIEPs (N), virions (V), and dense bodies (DB) were recovered from the maintenance medium of HFF cells infected with either wild-type (WtCMV) or mutant (I−CMV) HCMV-BAC virus, concentrated, and subjected to SDS-PAGE, as described in the legend for Fig. 5, and then Western immunoassay using a mixture of antisera to assemblin (i.e., anti-hNP1n) and to mCP (i.e., anti-mCP). Samples of cytoplasmic (Cyto.) and nuclear (Nuc.) fractions from NP-40-treated cells infected with wild-type (Wt) or mutant (I−) HCMV-BAC virus, or not infected (M) , were included for comparison (lanes 1 to 6; different samples from those in Fig. 7). Shown is a phosphorimage of the lower one-third of the resulting membrane (∼5- to 40-kDa range). Positions of proteins of interest are indicated at the right. Asbln, assemblin.
TABLE 2.
Amounts of mutant and wild-type proteinase and cleavage products in nuclei and NIEPsa
| Protein | NP-40 nuclei
|
NIEPs
|
||||
|---|---|---|---|---|---|---|
| PSL for:
|
I−/Wt | PSL for:
|
I−/Wt | |||
| Wtb | I− | Wtc | I− | |||
| Precursors (pNP1 + NP1) | 105 | 183 | 1.7 | |||
| Assemblin | 135 | 303 | 2.2 | 424 | 1,050 | 2.5 |
| Cn | 145 | 719 | 4.9 | 286 | 1,614 | 5.6 |
| An | 202 | 2,576 | ||||
| Totald | 482 | 1,022 | 2.1 | 3,286 | 2,664 | 0.8 |
Calculations were made from quantification of data shown in Fig. 7, lanes 4 to 6 (for NP-40 nuclei [pellet fraction of NP-40-treated cells]), and Fig. 8, lanes 1 and 4 (for NIEPs). Values are in units of photostimulated luminescence (PSL).
Values for the wild type (Wt) were increased 1.04 fold (1,547/1,484 PSL units) to normalize (relative to the amount of mCP) between the samples for NP-40 nuclei from Wt and mutant (I−) virus-infected cells (immunoassay data not shown).
Values for the Wt were increased 1.37 fold (7,665/5,610 PSL units) to normalize (relative to the amount of mCP) between the samples from Wt and I− NIEPs.
Total is sum of values for assemblin plus Cn plus An.
Proteinase cleavage products are present in NIEPs but not virions.
Assemblin is present in CMV B-capsids (e.g., 28-kDa protein; see Fig. 3 of reference 16 and Fig. 2 of reference 27), but neither it nor its I-site cleavage products have been reported in enveloped particles. We used the anti-hNP1n antiserum to HCMV assemblin to test for these proteins in extracellular virions, NIEPs, and dense bodies. All three particles from both wild-type virus- and mutant virus-infected cells were recovered by a single banding in sucrose gradients, concentrated, and subjected to SDS-PAGE. The proteins were then electrotransferred to Immobilon and subjected to Western immunoassay using anti-hNP1n, combined with anti-mCP for purposes of normalization. A phosphorimage prepared from the resulting membrane (Fig. 8) was used for quantification and showed the following. (i) None of the particles contained detectable amounts of the proteinase precursor (portion of membrane not shown). (ii) Both wild-type and mutant NIEPs contained assemblin (Fig. 8, lanes 7 and 10), but relative to mCP there was 2.5-fold more in mutant NIEPs, similar to the 2.1-fold difference in wild-type virus- and mutant virus-infected cells (Fig. 7; Table 2). (iii) Only wild-type NIEPs contained the I-site cleavage product, An (Fig. 8, compare lane 7 with 10), consistent with its absence from mutant virus-infected cells (Fig. 8, lanes 5 and 6, and Fig. 7, lanes 5 and 6). (iv) Mutant NIEPs contained 5.6-fold more of the 23-kDa fragment, Cn, than wild-type NIEPs, relative to mCP (Fig. 8, compare lane 7 with 10), similar to the 4.9-fold difference calculated above for Cn in virus-infected cells (Fig. 7; Table 2). (v) The total amounts of assemblin immunoreactivity (i.e., assemblin plus Cn plus An) in mutant and wild-type NIEPs were comparable (i.e., mutant/wild-type ratio = 0.8), even though the distributions of assemblin immunoreactivity for the three bands differed (Table 2). (vi) Neither assemblin nor its An and Cn cleavage products were detected in these or other CMV virion preparations (Fig. 8, lanes 8 and 11). (vii) Relative intensities of mCP in this assay indicate that there were 25 to 50% more virions than NIEPs in both preparations and ≈15% more mutant virus particles (virions plus NIEPs) than wild-type particles. (viii) Dense bodies, which have no capsid (26), contained none of these capsid-specific proteins (Fig. 8, lanes 9 and 12).
DISCUSSION
HCMV assemblin self-cleaves at an I site near its midpoint and just 11 residues from the nucleophilic serine of its catalytic triad, without known effect on enzymatic behavior (1, 5, 22, 25, 28, 36, 54). Although the assemblin homologs of other herpesviruses lack an I-site consensus sequence, its presence in most CMV homologs suggests a possible CMV-specific requirement. The study described here was done to test for such a requirement during virus infection that may have escaped detection in vitro. We used the HCMV-BAC system developed by Koszinowski and Messerle and colleagues (3, 33, 34) to make a mutant virus blocked for I-site cleavage and found that it produced infectious virus. Characterizations of the mutant showed that the blocked I site had little effect on infectivity or the ratio of virions to NIEPs but did result in an increased abundance of the assemblin C-site cleavage product, Cn. These changes in assemblin processing were reflected in the protein composition of mutant NIEPs, but neither assemblin nor its cleavage products were detected in CMV virions.
Outgrowth of virus from cells transfected with the mutant BAC DNA immediately suggested that I-site cleavage is not essential. Measurements of infectivity (Fig. 4) and particle yield (Fig. 5) showed that the mutation had little effect on either the rate of virus production or the amount of virus produced. A compensatory intragenic change does not account for this growth, as established by sequencing UL80 from mutant virion DNA. Nor does intergenic complementation seem a likely explanation, since (i) the mutant HCMV-BAC DNA was clonally derived, (ii) there was little, if any, difference in the time required for outgrowth of wild-type and mutant viruses following transfection, and (iii) the same phenotype (i.e., I site blocked; infectious virus produced) has been obtained from three separate DNA transfections.
We also tested and ruled out the possibility that the mutated I site was unexpectedly susceptible to cleavage in virus-infected cells by demonstrating that there is no detectable I-site cleavage product, An, in mutant-infected cells (Fig. 7 and 8). However, the increased abundance of a 23-kDa assemblin fragment in mutant-infected cells (Cn; Fig. 7 and 8) suggested that an alternate cleavage may circumvent the blocked I site. A corresponding fragment was identified in cells transfected with a plasmid encoding the same UL80 I-site mutation, and it was concluded that the fragment resulted from cleavage after Ala209 in the C-site sequence (Fig. 1) (28). Cleavage at this site also occurs during infection with wild-type HCMV (Fig. 7 and 8) (28), and it has been proposed that this cleavage represents an alternate processing pathway for HCMV assemblin (28). Although C-site cleavage may bypass the blocked I site, the accumulation of both assemblin (approximately twofold increased) and Cn (approximately fivefold increased) in cells infected with the I-site mutant (Fig. 7; Table 2) indicates that assemblin processing is slowed. If closely linked to virus production, the reduced cleavage efficiency of the mutant would be expected to lower virus yield. This was not detected by infectivity assays (Fig. 4) or by more-sensitive and quantitative particle comparisons (Fig. 5 and 8), suggesting that, even slowed by the A143V mutation, assemblin processing is not rate-limiting to virus production.
The finding that NIEPs contain assemblin follows from the proposition that they are enveloped B-capsids (26, 27), and B-capsids of both CMV (16, 27) and HSV (10, 11, 19, 45) contain assemblin. However, the absence of assemblin from virions of both simian CMV (SCMV) (16) and HCMV (Fig. 8, lanes 8 and 11), as well as its reduced abundance in DNA-containing C-capsids of SCMV (“28K” protein; see Fig. 3 of reference 16), suggests that encapsidation of DNA has a destabilizing effect on the association of assemblin with the CMV capsid. This is an apparent difference from the HSV homolog, VP24, which is present in DNA-deficient A- and B-capsids (11, 19, 35, 45), as well as DNA-containing C-capsids (11, 19, 45) and virions (19, 49). It remains to be determined whether this actually represents an evolutionary difference between the two viruses, but, if it does, it may result from a need for more space within the CMV capsid to accommodate its 34% longer DNA.
We conclude from this work that blocking I-site cleavage of HCMV assemblin has little effect on virus production. However, because cleavage at the downstream C-site continues undiminished in this mutant, the possibility remains that there is a requirement for maturational processing of assemblin that can be satisfied by either I- or C-site cleavage. A virus mutated at both the I and C sites would be of particular interest to test this possibility and the questions of whether and how these cleavages relate to the absence of assemblin from CMV virions. Such a mutant would be expected to be severely compromised for growth if at least one of these cleavages is needed for virus production.
Acknowledgments
Special recognition and thanks go to Ulrich Koszinowski and Martin Messerle and their colleagues for the generous and enabling gift of the HCMV-BAC system and for helpful advice and materials. We also thank Rebecca Casaday for plasmid RC3 and Prahant Desai for valuable discussions and generous help evaluating transfection methods. Jenny Borchelt provided technical assistance with cell cultures and Western immunoassays, and Matt Hall made the HCMV assemblin recombinant baculovirus and purified the HCMV assemblin used to prepare antiserum anti-hNP1n.
This work was aided by the National Institute of Allergy and Infectious Diseases through research grants AI13718 and AI32957 to W.G.
REFERENCES
- 1.Baum, E. Z., G. A. Bebernitz, J. D. Hulmes, V. P. Muzithras, T. R. Jones, and Y. Gluzman. 1993. Expression and analysis of the human cytomegalovirus UL80-encoded protease: identification of autoproteolytic sites. J. Virol. 67:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaudet-Miller, M., R. Zhang, J. Durkin, W. Gibson, A. D. Kwong, and Z. Hong. 1996. Virus-specific interaction between the human cytomegalovirus major capsid protein and the C terminus of the assembly protein precursor. J. Virol. 70:8081-8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E. M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 75:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burck, P. J., D. H. Berg, T. P. Luk, L. M. Sassmannshausen, M. Wakulchik, D. P. Smith, H. M. Hsiung, G. W. Becker, W. Gibson, and E. C. Villarreal. 1994. Human cytomegalovirus maturational proteinase: expression in Escherichia coli, purification, and enzymatic characterization by using peptide substrate mimics of natural cleavage sites. J. Virol. 68:2937-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 7.Chen, P., H. Tsuge, R. J. Almassy, C. L. Gribskov, S. Katoh, D. L. Vanderpool, S. A. Margosiak, C. Pinko, D. A. Matthews, and C.-C. Kan. 1996. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell 86:835-843. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. L. 1996. Characterization of human cytomegalovirus protease dimerization by analytical centrifugation. Biochemistry 35:15601-15610. [DOI] [PubMed] [Google Scholar]
- 9.Darke, P. L., J. L. Cole, L. Waxman, D. L. Hall, M. K. Sardana, and L. C. Kuo. 1996. Active human cytomegalovirus protease is a dimer. J. Biol. Chem. 271:7445-7449. [DOI] [PubMed] [Google Scholar]
- 10.Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J. Gen. Virol. 73:2709-2713. [DOI] [PubMed] [Google Scholar]
- 11.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115-124. [DOI] [PubMed] [Google Scholar]
- 12.DiIanni, C. L., D. A. Drier, I. C. Deckman, P. J. McCann, F. Liu, B. Roizman, R. J. Colonno, and M. G. Cordingley. 1993. Identification of the herpes simplex virus type 1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J. Biol. Chem. 268:2048-2051. [PubMed] [Google Scholar]
- 13.Dunn, W., P. Trang, U. Khan, J. Zhu, and F. Liu. 2001. RNase P-mediated inhibition of cytomegalovirus protease expression and viral DNA encapsidation by oligonucleotide external guide sequences. Proc. Natl. Acad. Sci. USA 98:14831-14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fairbanks, G., T. L. Steck, and D. F. Wallach. 1971. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10:2606-2617. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, W. 1983. Protein counterparts of human and simian cytomegaloviruses. Virology 128:391-406. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, W. 1981. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology 111:516-537. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, W., M. K. Baxter, and K. S. Clopper. 1996. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J. Virol. 70:7454-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, W., A. R. Welch, and M. R. T. Hall. 1995. Assemblin, a herpes virus serine maturational proteinase and new molecular target for antivirals. Perspect. Drug Discov. Des. 2:413-426. [Google Scholar]
- 21.Hall, M. R. T., and W. Gibson. 1997. Assemblin homolog of herpes simplex virus type 1 retains proteolytic activity when expressed as a recombinant two-chain enzyme. Virology 227:160-167. [DOI] [PubMed] [Google Scholar]
- 22.Hall, M. R. T., and W. Gibson. 1996. Cytomegalovirus assemblin: the amino and carboxyl domains of the proteinase form active enzyme when separately cloned and coexpressed in eukaryotic cells. J. Virol. 70:5395-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, M. R. T., and W. Gibson. 1997. Independently cloned halves of cytomegalovirus assemblin, An and Ac, can restore proteolytic activity to assemblin mutants by intermolecular complementation. J. Virol. 71:956-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holwerda, B. C. 1997. Herpesvirus proteases: targets for novel antiviral drugs. Antivir. Res. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 25.Holwerda, B. C., A. J. Wittwer, K. L. Duffin, C. Smith, M. V. Toth, L. S. Carr, R. C. Wiegand, and M. L. Bryant. 1994. Activity of two-chain recombinant human cytomegalovirus protease. J. Biol. Chem. 269:25911-25915. [PubMed] [Google Scholar]
- 26.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 27.Irmiere, A., and W. Gibson. 1985. Isolation of human cytomegalovirus intranuclear capsids, characterization of their protein constituents, and demonstration that the B-capsid assembly protein is also abundant in noninfectious enveloped particles. J. Virol. 56:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, T. R., L. Sun, G. A. Bebernitz, V. P. Muzithras, H.-J. Kim, S. H. Johnston, and E. Z. Baum. 1994. Proteolytic activity of human cytomegalovirus UL80 proteinase cleavage site mutants. J. Virol. 68:3742-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London). 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Liu, F., and B. Roizman. 1993. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus 1 UL26 protein. J. Virol. 67:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, F., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 65:5149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margosiak, S. A., D. L. Vanderpool, W. Sisson, C. Pinko, and C. C. Kan. 1996. Dimerization of the human cytomegalovirus protease: kinetic and biochemical characterization of the catalytic homodimer. Biochemistry 35:5300-5307. [DOI] [PubMed] [Google Scholar]
- 33.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messerle, M., G. Hahn, W. Brune, and U. H. Koszinowski. 2000. Cytomegalovirus bacterial artificial chromosomes: a new herpesvirus vector approach. Adv. Virus Res. 55:463-478. [DOI] [PubMed] [Google Scholar]
- 35.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Boyle, D. R., K. Wager-Smith, J. T. Stevens, and S. P. Weinheimer. 1995. The effect of internal autocleavage on kinetic properties of the human cytomegalovirus protease catalytic domain. J. Biol. Chem. 270:4753-4758. [DOI] [PubMed] [Google Scholar]
- 37.Pelletier, A., F. Do, J. J. Brisebois, L. Lagace, and M. G. Cordingley. 1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J. Virol. 71:5197-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinko, C., S. A. Margosiak, D. Vanderpool, J. C. Gutowski, B. Condon, and C. C. Kan. 1995. Single-chain recombinant human cytomegalovirus protease. Activity against its natural protein substrate and fluorogenic peptide substrates. J. Biol. Chem. 270:23634-23640. [DOI] [PubMed] [Google Scholar]
- 39.Plafker, S. M., and W. Gibson. 1998. Cytomegalovirus assembly protein precursor and proteinase precursor contain two nuclear localization signals that mediate their own nuclear translocation and that of the major capsid protein. J. Virol. 72:7722-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pray, T. R., A. M. Nomura, M. W. Pennington, and C. S. Craik. 1999. Auto-inactivation by cleavage within the dimer interface of Kaposi's sarcoma-associated herpesvirus protease. J. Mol. Biol. 289:197-203. [DOI] [PubMed] [Google Scholar]
- 41.Preston, V. G., J. A. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston, V. G., F. J. Rixon, I. M. McDougall, M. McGregor, and M. F. Al Kobaisi. 1992. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy-terminal end requires the product of the whole of the UL26 reading frame. Virology 186:87-98. [DOI] [PubMed] [Google Scholar]
- 43.Qiu, X., J. S. Culp, A. G. DiLella, B. Hellmig, S. S. Hoog, D. A. Janson, W. W. Smith, and S. S. Abdel-Meguid. 1996. Unique fold and active site in cytomegalovirus protease. Nature 383:275-279. [DOI] [PubMed] [Google Scholar]
- 44.Sardana, V. V., J. A. Wolfgang, C. A. Veloski, W. J. Long, K. LeGrow, B. Wolanski, E. A. Emini, and R. L. LaFemina. 1994. Peptide substrate cleavage specificity of the human cytomegalovirus protease. J. Biol. Chem. 269:14337-14340. [PubMed] [Google Scholar]
- 45.Sheaffer, A. K., W. W. Newcomb, J. C. Brown, M. Gao, S. K. Weller, and D. J. Tenney. 2000. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 74:6838-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shieh, H.-S., R. G. Kurumbail, A. M. Stevens, R. A. Stegeman, E. J. Sturman, J. Y. Pak, A. J. Wittwer, M. O. Palmier, R. C. Wiegand, B. C. Holwerda, and W. C. Stallings. 1996. Three-dimensional structure of human cytomegalovirus protease. Nature 383:279-282. [DOI] [PubMed] [Google Scholar]
- 47.Smith, M. C., T. C. Furman, T. D. Ingolia, and C. Pidgeon. 1988. Chelating peptide immobilized metal ion affinity chromatography. A new concept in affinity chromatography for recombinant proteins. J. Biol. Chem. 263:7211-7215. [PubMed] [Google Scholar]
- 48.Smith, M. C., J. Giordano, J. A. Cook, M. Wakulchik, E. C. Villarreal, G. W. Becker, K. Bemis, J. Labus, and J. S. Manetta. 1994. Purification and kinetic characterization of human cytomegalovirus assemblin. Methods Enzymol. 244:412-423. [DOI] [PubMed] [Google Scholar]
- 49.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens, J. T., C. Mapelli, J. Tsao, M. Hail, D. O'Boyle, S. P. Weinheimer, and C. L. DiIanni. 1994. In vitro proteolytic activity and active-site identification of the human cytomegalovirus proteinase. Eur. J. Biochem. 226:361-367. [DOI] [PubMed] [Google Scholar]
- 51.Tong, L., C. Qian, M. J. Massariol, P. R. Bonneau, M. G. Cordingly, and L. Lagacé. 1996. A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease. Nature 383:272-275. [DOI] [PubMed] [Google Scholar]
- 52.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waxman, L., and P. L. Darke. 2000. The herpesvirus proteases as targets for antiviral chemotherapy. Antivir. Chem. Chemother. 11:1-22. [DOI] [PubMed] [Google Scholar]
- 54.Welch, A. R., L. M. McNally, M. R. T. Hall, and W. Gibson. 1993. Herpesvirus proteinase: site-directed mutagenesis used to study maturational, release, and inactivation cleavage sites of precursor and to identify a possible catalytic site serine and histidine. J. Virol. 67:7360-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch, A. R., A. S. Woods, L. M. McNally, R. J. Cotter, and W. Gibson. 1991. A herpesvirus maturational protease, assemblin: identification of its gene, putative active site domain, and cleavage site. Proc. Natl. Acad. Sci. USA 88:10792-10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]