Abstract
The human cytotoxic T-lymphocyte (CTL) response to human immunodeficiency virus type 1 (HIV-1) has been intensely studied, and hundreds of CTL epitopes have been experimentally defined, published, and compiled in the HIV Molecular Immunology Database. Maps of CTL epitopes on HIV-1 protein sequences reveal that defined epitopes tend to cluster. Here we integrate the global sequence and immunology databases to systematically explore the relationship between HIV-1 amino acid sequences and CTL epitope distributions. CTL responses to five HIV-1 proteins, Gag p17, Gag p24, reverse transcriptase (RT), Env, and Nef, have been particularly well characterized in the literature to date. Through comparing CTL epitope distributions in these five proteins to global protein sequence alignments, we identified distinct characteristics of HIV amino acid sequences that correlate with CTL epitope localization. First, experimentally defined HIV CTL epitopes are concentrated in relatively conserved regions. Second, the highly variable regions that lack epitopes bear cumulative evidence of past immune escape that may make them relatively refractive to CTLs: a paucity of predicted proteasome processing sites and an enrichment for amino acids that do not serve as C-terminal anchor residues. Finally, CTL epitopes are more highly concentrated in alpha-helical regions of proteins. Based on amino acid sequence characteristics, in a blinded fashion, we predicted regions in HIV regulatory and accessory proteins that would be likely to contain CTL epitopes; these predictions were then validated by comparison to new sets of experimentally defined epitopes in HIV-1 Rev, Tat, Vif, and Vpr.
Strong cytotoxic T-lymphocyte (CTL) responses are likely to be an important aspect of an effective human immunodeficiency virus (HIV) vaccine, as their benefit has been established in simian immunodeficiency virus (SIV) vaccination studies and in natural infections (7, 14, 25, 32, 48). HIV-specific CTLs have been detected in HIV type 1 (HIV-1)-exposed persistently seronegative individuals, suggesting a protective effect (11, 35, 54). CTL responses coincide with the early containment of the virus (40, 52), and CTL escape mutations arise in conjunction with progression to AIDS (29, 36). Furthermore, depletion of CD8+ lymphocytes during primary HIV infection of monkeys results in poor containment of viral replication and early death (56).
Particular patterns of escape mutation in vivo depend on specific host-virus interactions (7, 23, 30, 36) and on a complex balance of functional constraints on viral proteins and immune pressure (7, 63, 64). In the controlled setting of a clonal SIV infection of Mamu A∗01 macaques, escape can be reproducible; both the initial dominant CTL response to a Tat epitope and the rapid emergence of escape variants occurred consistently during acute infection of different animals (3). In contrast, CTL escape mutations that unfold in a single patient with a specific human leukocyte antigen (HLA) genotype, naturally infected with a unique viral quasispecies, are difficult to extend to the population of infected individuals. Most CTL studies focus on summarizing individual responses; here we took a different approach, integrating the global HIV sequence and immunology databases to define broad correlates between natural variation in HIV-1 proteins and CTL epitope localization. We explored how escape mutations that could influence processing, HLA interactions, and T-cell recognition may impact the variability of the population of circulating viruses and in turn how HIV-1 variation may impact CTL responses.
Each step of CTL epitope generation and recognition has potential constraints imposed by sequence specificity. For viral proteins to be recognized by CTLs they must first be cleaved into short peptides; generally this step occurs in the cytosol and is due to the immunoproteasome (15, 20, 51) and often requires the additional trimming of epitope precursors by peptidases in the endoplasmic reticulum (ER) (60). After cleavage, the transporter associated with antigen processing (TAP) protein translocates peptides into the ER (1) for loading onto HLA class I molecules (51, 53). The peptide binding groove of HLA class I molecules accommodates peptides of 8 to 12 amino acids, and peptide binding depends in particular on amino acids known as anchor residues (53). The complex of a peptide and class I molecule is then presented on the cell surface, allowing recognition by epitope-specific CTLs (51).
Mutations both within and proximal to epitopes can influence their immunogenic potential. Some amino acid substitutions within an epitope can be tolerated without inhibiting class I molecule binding or T-cell receptor (TCR) recognition (17, 19, 46), while others eliminate the CTL response or reduce its efficiency (65). There is evidence that TAP transports peptides selectively, as successful presentation of peptides by class I molecules was found to correlate with affinity for TAP (22). Mutations that influence the cleavage step of epitope processing can also be critical; for example, cases of immune escape due to mutations in an epitope's flanking regions have demonstrated that escape through inhibition of processing is of immunological significance (9, 21, 30). Cleavage sites generated by the immunoproteasome are sensitive to the surrounding amino acid sequence (15, 49), although no simple cleavage signal is apparent; in this study we show that HIV-1 known cleavage sites are associated with high cleavage prediction scores by using a neural network approach (37), which is a computational way to identify complex predictive patterns in data. Alternative cleavage or trimming pathways (28, 60) may confer an additional layer of sequence specificity. Thus, potential epitope boundaries may differ among diverse HIV-1 strains because of greater or lesser propensity to be cleaved. The majority of HIV CTL escape and cross-reactivity studies, however, focus on the influence of substitutions within an epitope that would influence only class I HLA and TCR interactions, by testing synthetic peptide variants for cross-reactivity in vitro (19). Thus the relative influence of processing escape mutations is not well understood.
At the time of this writing, the boundaries of 334 unique HIV CTL epitopes have been experimentally defined and described in the literature (38) and were available through the HIV Molecular Immunology Database, year 2000 edition (38). By unique epitopes we mean that either the optimal epitopes have distinct boundaries (although sometimes overlapping) or different HLA-presenting molecules if they share boundaries or both. (If an epitope was defined in multiple individuals and had the same precise boundaries and HLA-presenting molecule, it was considered only once.) There are regions in HIV proteins that are rich in experimentally defined unique CTL epitopes and other regions where no epitopes have yet been identified despite repeated and thorough searching by many groups using overlapping peptides that span the complete proteins to test for reactive epitopes (31, 38, 64). The biology underlying this highly nonuniform distribution is not yet understood, although some correlations have been noted. In particular an association between conserved regions in the variable proteins and high epitope density was noted for p17 and Nef (42). Studies of major histocompatibility complex binding motifs in HIV-1 proteins revealed that the binding motifs tend to cluster in relatively short regions and that regions with low motif density tend to be the most variable protein regions (24, 47, 67).
In this study we systematically explored the relationship between the distribution of experimentally defined CTL epitopes and HIV-1 protein sequences. We confirmed and extended observations indicating there are very few HIV experimentally defined epitopes in the most variable regions of the virus (42) by using the much larger current data sets. Viral variation can influence a CTL response via processing, presentation, or recognition of an epitope. We examined variation in processing using a neural network approach to immunoproteasome cleavage sites in proteins. Instead of simply basing our predictions on a single protein, we extended the method to analyze full protein alignments to determine the levels of conservation of predicted cleavage sites in the viral population. Our method showed a very strong correlation between conserved high cleavage prediction scores and boundaries of known epitopes. We then explored HLA binding potential through estimating the frequency of amino acids that do not serve as C-terminal anchor residues comparing protein subregions that either carry or do not carry epitopes. Finally, we successfully used the sequence characteristics associated with epitope density to predict where new epitopes would be localized on regulatory proteins and compared our predictions to new experimentally defined epitopes.
MATERIALS AND METHODS
Protein alignments.
Protein alignments containing one protein sequence per individual were initially taken from the year 2000 alignments of the HIV database (41) (www.hiv.lanl.gov). To make the alignments comparable, each protein alignment was pared down to contain 101 protein sequences from different individuals with similar subtype distributions and all subtypes represented. (Protein translations of the full-genome alignments would have been more directly comparable, but many of these sequences do not include complete nef gene sequences. To enable the inclusion of nef, the compromise of using data sets comparable in terms of subtype distribution was used. Generally, information concerning a given patient's therapy status is not provided with full-length protein sequences, and we are not certain to what extent drug resistance mutations might contribute to the variation found in reverse transcriptase (RT) or protease. We ascertained that none of the B subtype isolates with known years of sampling (19 out of 27) were obtained after 1996, so these sequences were not subject to evolutionary pressure due to the use of highly active antiretroviral therapy, although certainly some of the patients received sequential therapies. The other 74 protein sequences were derived from other clades and were generally sampled in regions of the world where therapy is rarely available. Sequences that did not span full-length proteins were not included. The alignments are available from the authors upon request.
Measure of variability.
A Shannon entropy score (39) was calculated for each position in the protein alignment. Entropy is a measure of the amino acid variability at a given position that takes into account both the number of possible amino acids allowed and their frequency. (Entropy in each amino acid position is calculated as −ΣPaalog Paa, where Paa is the proportion of each amino acid in the respective position.) Positions where the majority of the protein sequences had gaps were excluded from consideration. When only a minority of protein sequences had gaps in a position, however, the position was included and the gaps were treated as separate symbols.
Measure of correlation between epitope distributions and amino acid variability.
To study whether the CTL epitope-rich regions correlate with the amino acid variation in protein sequences, we used the following statistical test (42). First, the density of unique CTL epitopes that contain a particular position in the protein was estimated by tallying for each position the number of distinct epitopes that span that position. Only well-characterized epitopes no longer than 11 amino acids with known HLA-presenting molecules were included in this study. Optimal epitopes were counted as distinct if the optimally defined boundaries of the epitopes differ or if different HLA molecules present them; epitopes with identical boundaries and with the same HLA-presenting molecule identified in multiple individuals were only counted once. Second, an entropy score was calculated for each position in a protein alignment. Entropy scores sometimes vary dramatically between contiguous sites. To identify regions of high variability or low variability, the entropy scores were smoothed by averaging over a window of nine amino acids, a typical epitope size, and both raw and smoothed scores were tested in statistical analyses.
Proteasome cleavage predictions.
The immunoproteasome cleavage predictions were made by using NetChop (www.cbs.dtu.dk/Services/NetChop) (37), an artificial neural network program that is trained on experimentally verified, naturally processed C termini of 1,110 known human CTL epitopes and peptides eluted from 59 HLA class I molecules and that models in vivo cleavage. HIV-1 epitopes were not included in the training set, so HIV-1 protein sequences could be used as a test set without bias.
NetChop uses a flanking region of eight residues on each side to predict the cleavage probability of a position in protein sequences (37). The input to the program is the protein sequence or, in our case, a protein alignment. The program assigns a value between 0 and 1 to each position in a protein, with higher values indicating sites that are most likely to be cleaved and serve as C termini of epitopes.
Other versions of NetChop trained on different data sets are available. For example, another network was trained to predict N termini of epitopes, but this network performed much more poorly than the network trained to predict C-terminal cleavage sites (37). Similarly we found that predictions of N termini were not correlated with actual N termini of observed HIV epitopes (results not shown). One explanation for the poor performance of the neural network for predicting N termini is possible serial trimming of N termini made by endopeptidases in ER, which does not seem sequence specific but rather time dependent (60).
Cleavage predictions for HIV-1 proteins were also made by using a version of NetChop trained on yeast enolase (61) and bovine beta-casein (26) degradation data obtained by in vitro cleavage with the constitutive proteasomes (37), not the immunoproteasomes. This network, however, did not do as well in terms of predicting the C termini of HIV-1 epitopes (see the legend for Fig. 5). The poor performance using this training set may be due to immunoproteasomes, not constitutive proteasomes, that generate most of the major histocompatibility complex ligands (20, 59, 62), and the two kinds of proteasomes recognize different patterns of amino acids surrounding cleavage sites (37). As a consequence of the two considerations above, we limited our conclusions in Results to the predictions of C termini of epitopes by the version of NetChop trained on in vivo cleavage.
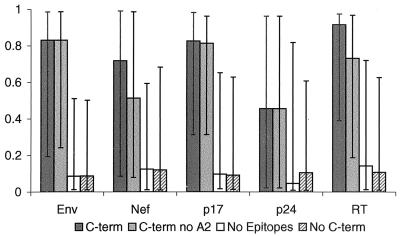
FIG. 5.
Proteasome cleavage predictions. For each protein and for each sequence in the alignment site-specific prediction scores were computed with NetChop (www.cbs.dtu.dk/Services/NetChop) (37) by using a neural network trained with HLA ligands (modeling in vivo degradation; see Materials and Methods). Then for each site of the alignment the site-specific predictions were calculated as the medians of the predictions from all protein sequences in the alignment. Site-specific predictions were then organized into four groups for each protein: group 1 (C-term), prediction scores at the sites corresponding to known C termini of experimentally defined epitopes; group 2 (C-term no A2), subset of group 1 excluding sites corresponding to C termini of HLA-A2 epitopes so we could establish that the NetChop program wasn't simply recognizing a common anchor motif; group 3, (no epitopes), predictions at all sites taken from epitope-lacking regions; group 4 (no C-term), predictions at all sites which do not serve as C termini of experimentally observed HIV epitopes. The bars in the figure show the medians of the distributions for each group for each protein. Error bars, 25th and 75th percentiles of the distributions. The nonparametric Mann-Whitney test was used to compare scores for known C-terminal positions; scores for group 1 were compared to those for groups 3 and 4, and those for group 2 were also compared with those for groups 3 and 4. For all five proteins the prediction scores for C termini of all experimentally observed HIV epitopes and for the subset excluding HLA-A2 binders were found to be statistically significantly higher than prediction scores for the epitope-lacking regions (for p24 and Nef, P = 0.002 for comparison of groups 1 and 3 and P = 0.0005 for comparison of groups 2 and 3; for p17, P = 0.002; for RT, P < 0.0001; for Env, P < 0.0001) and for positions that are not C termini of experimental epitopes (for p24 and Nef, P = 0.007 and 0.001, respectively, for comparison of groups 1 and 4 and 0.0003 for comparison of groups 2 and 4; for p17, P = 0.001; for RT, P < 0.0001; for Env, P < 0.0001). A different strategy for training NetChop to recognize cleavage sites, based on relative frequency of cleavage events in vitro observed in the yeast enolase and bovine beta-casein proteins (see Materials and Methods) rather than known epitopes, gave a statistically significant difference in the prediction scores between C-terminal positions and epitope-lacking regions for Env (P = 0.0012) and P24 (P = 0.0006) and a trend for RT (P = 0.08), but not for p17 (P = 0.67) and Nef (P = 0.67).
Statistics.
Statistical tests were performed with GraphPad Prism software, version 3.0, or with the R Package for Statistical Computing (http://www.r-project.org/).
RESULTS
Classification of HIV proteins by their variability.
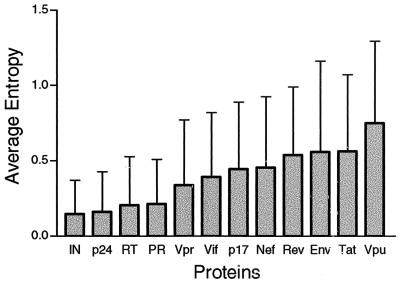
The relative variability of HIV proteins was assessed by estimating the average entropy (see Materials and Methods) of all positions within each protein by using matched sets of 101 aligned amino acid sequences to allow for direct comparison among the proteins. Here we are not attempting to assess the evolutionary distances between proteins; rather we are trying to obtain a cross-sectional measure of the amino acid variability in the population of infected individuals. Entropy is a simple measure that takes into account both the frequency and spectrum of amino acids at each position. HIV proteins have a range of variability (Fig. 1). Integrase has the lowest, and Vpu has the highest. Gag is usually considered a highly conserved polyprotein; however, the entropy-based classification clearly distinguishes conserved Gag p24 from the more-variable Gag p17. Env is considered to be the most variable HIV protein, yet it doesn't have the highest average entropy score. This is because Env is also subject to rapid and dramatic change by frequent insertions and deletions and by the gain and loss of potential N-linked glycosyation sites (the amino acid motif NX[S or T], where X can be any amino acid) in hypervariable domains. These changes are likely to have profound impact on the antigenic variability of Env, but they would not be captured in these simple entropy measures, which incorporate only amino acid substitutions. Because of these considerations, the entropy measure does not reflect the full potential antigenic diversity of Env.
FIG. 1.
M group protein variability. The variability was estimated by using Shannon entropy scores calculated for each position in a protein alignment (see Materials and Methods). The average entropies and standard deviations of all positions in the alignment of each of the 12 HIV proteins are shown. IN, integrase; PR, protease.
Correlation between occurrence of epitopes and HIV variability in proteins with highly variable domains, p17, Nef, and Env.
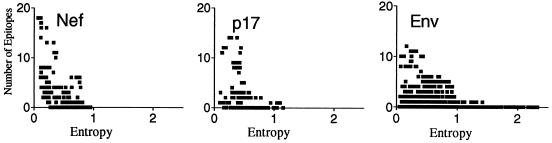
Both variability and epitope distributions are highly nonuniform within HIV proteins. Thus we compared the epitope distributions with regional entropy scores. The smoothed entropy (the entropy scores averaged over a nine-amino-acid window, the size of a typical CTL epitope) and the epitope density (the number of unique well-defined epitopes that overlap each amino acid position; see Materials and Methods) for the more-variable proteins, p17, Nef, and Env, are plotted for all amino acid positions in Fig. 2. The regions with no reported epitopes are associated with high entropy in p17, Nef, and Env, while the epitope-rich regions are relatively conserved. This observation was confirmed by using a nonparametric Spearman's rank correlation. Statistically significant negative correlations between entropy scores and the number of epitopes at each position were observed by using entropy measurements made both with and without smoothing (Fig. 2). The number of epitopes versus entropy was also plotted in the scatter diagrams of Fig. 3. Each point of the diagram corresponds to a position in a protein sequence. As shown in the diagrams, especially for Env, there are no experimentally defined epitopes found in highly variable positions and epitopes that have been experimentally defined are concentrated in conserved regions.
FIG. 2.
Correlation between entropy and epitope density for proteins that have been the focus of the most HIV-1 CTL studies. The plots show the numbers of defined CTL epitopes overlapping with each protein sequence position (red line; scale on the right axis) and Shannon entropy of each site in the protein alignments (black line; scale on the left axis). Protein sequence positions are given according to HXB2R sequence (accession no. K03455). The entropy data were smoothed by using a window of nine amino acids (the average size of a CTL epitope). The entropy scores for each site were calculated by using comparably diverse data sets for each protein (see Materials and Methods) and were plotted on the same scale for each protein, so that the figures can be directly compared. Spearman's correlation coefficient (r) and the P value for the correlation between number of epitopes and smoothed entropy are shown for each protein. r and P values for the correlation between the number of epitopes and nonsmoothed (raw) entropy scores are r = −0.36 and P < 0.0001 for Nef, r = −0.17 and P = 0.04 for p17, r = −0.12 and P = 0.0003 for Env, r = −0.1 and P = 0.3 for p24, and r = 0.1 and P < 0.001 for RT. Known secondary structural elements were assigned to positions based on crystal structures of each protein; gp160 was constructed by joining the structural models of gp120 and gp41, and Nef was constructed from N terminus and core Nef models. Alpha helices are blue, beta sheets are pink, and loops are left blank. Models used were as follows: Nef, 1AVV (5) and 1ZEC (6); p17, 1HIW (34); p24, 1E6J (12); gp120, 1G9M (43); gp41, 1DLB (58); RT, 1QE1 (55); and protease, 1HVK (66).
FIG. 3.
Scatter diagrams of the number of epitopes and entropy for each protein sequence position of the three most variable proteins, p17, Nef, and Env. Each point of the diagram corresponds to a protein sequence position to which two coordinates, entropy and number of epitopes, are assigned.
Because of the potential cross-reactivity of CTL epitopes, our method of determining the frequency of unique epitopes that overlap each amino acid position may be biased by the nonindependence of epitopes. Therefore, in addition to the above analysis, instead of epitope distributions, we defined two classes of protein regions: regions with experimentally defined epitopes and epitope-lacking regions. A protein sequence position belongs to an epitope-presenting region if it overlaps with at least one epitope. If no epitopes overlap with this position, it belongs to an epitope-lacking region. Epitopes may ultimately be discovered in some epitope-lacking regions, yet the current lack of epitopes in these regions suggests that epitopes are not readily detected in them. By this strategy, positions where epitopes overlap are considered equivalent to positions where only one epitope is found and bias due to potential cross-reactivity is minimized. A two-sided nonparametric Mann-Whitney test showed that Nef and Env entropy distributions in epitope-presenting and epitope-lacking regions were significantly different (Nef, P < 0.0001; Env, P = 0.0012), with the higher variability scores found in the epitope-lacking regions. For p17, the P value was 0.06, which is not statistically significant but which follows the same trend as Nef and Env. (The higher P value may be related to the relatively small size of p17, as there were only 132 amino acid positions to consider in p17, compared to 206 in Nef and 856 in Env.)
Weak correlations are found for the more-conserved proteins, p24 and RT.
p24 and RT do not have highly variable regions comparable to those found in Nef, p17, and Env (Fig. 2). Spearman's correlation between entropy and the number of epitopes is not significant for p24 (P = 0.3 on the basis of site-specific raw entropy, and P = 0.1 on the basis of smoothed entropy). The P value in the Mann-Whitney test comparing the distributions of entropy scores in epitope-presenting and epitope-lacking regions is 0.65. This lack of correlation may be related to the conserved nature of p24: p24 simply does not have regions variable enough to be free from defined epitopes, and epitopes are more evenly spread across the protein than across more variable proteins (Fig. 2).
Although RT is a conserved protein (Fig. 1), the defined epitopes are more sparsely distributed than in p24 (Fig. 2) (the median length of epitope-lacking regions in RT is 12 amino acid positions, and that in p24 is only 2 positions). Spearman's correlation between entropy values and epitope density is actually positive for RT (Fig. 2), the distributions of entropy scores in epitope-presenting and epitope-lacking regions are statistically distinct (P < 0.0001 in Mann-Whitney test). In contrast to what is found for other proteins, however, it is the most highly conserved positions that are associated with regions of no identified epitopes.
Interestingly, there are two short epitope-lacking regions in RT that are almost invariant (HXB2 positions 220 to 242 and 253 to 262). When we excluded these 33 amino acid positions from the RT comparison, the positive correlation between entropy scores and epitope density was lost and RT behaved like p24 in comparisons of epitope density versus entropy (P = 0.07 and 0.24 for the smoothed and raw entropy scores, respectively [Spearman's correlation test]) and in comparisons of entropy scores at epitope-presenting and epitope-lacking regions (P = 0.25; Mann-Whitney test). Similarly, in protease (Fig. 2), although only a few epitopes have been reported so far and thus there is inadequate data for statistical analysis, notably there are two essentially invariant regions (HXB2 positions 21 to 29 and 45 to 58) which to date are free from experimentally defined epitopes.
RT belongs to the polymerase superfamily, and HIV protease belongs to a family of aspartyl proteases, raising the possibility that the highly conserved regions in the proteins may be functional domains that have analogs in human proteins; these regions thus may be seen as “self” and be immunorefractive. To begin to explore this idea, we searched the Pfam database of protein domains (8) against the HXB2R protease and RT protein sequences and we examined the localization of conserved regions in the folded proteins to look for spatial proximity to the active sites. For protease, both of the conserved regions with no epitopes are proximal to the active site in the three-dimensional structure of the folded protein and one of these two regions (positions 21 to 29) contains the Asp cleavage residue (66) (Fig. 2). Both regions are in the top scoring domains from the Pfam alignment, which includes human proteases such as the renin precursor protein sequence angiotensinogenase from Homo sapiens (accession no. P00797). For RT, the conserved regions gave high scores in a Pfam alignment of the polymerase superfamily, but human proteins were not evident in the alignment.
Sequence variability within a single individual.
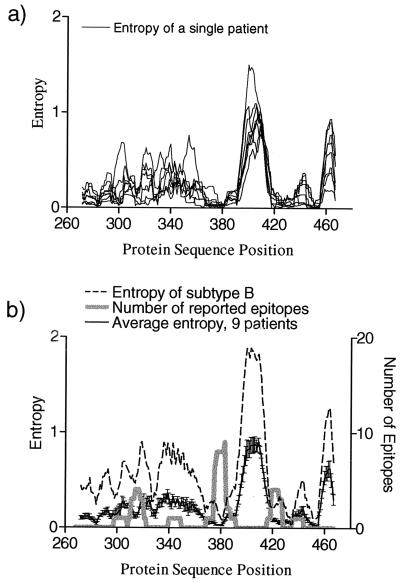
Results shown in the preceding sections were obtained by using HIV-1 protein sequences collected at the population level. To test whether these results can be extrapolated to the level of a single individual, we used partial Env gp160 sequences from nine patients collected over time from an extensive longitudinal study of HIV sequence variation (57). Figure 4a shows the smoothed entropy versus site for each patient separately. Figure 4b shows the averaged entropy of these nine patients superimposed with the entropy of subtype B protein sequences taken from the HIV database and the numbers of reported CTL epitopes observed in the literature at the respective protein sequence positions.
FIG. 4.
Variation of HIV protein sequences at the level of individual patients compared to the population. (a) Entropy scores for all positions in partial Env alignments were calculated for nine patients from the study of R. Shankarappa et al. (57). For entropy calculations each patient's protein sequences were combined into a single alignment and then aligned with the HIV database B subtype protein sequences. Protein sequence positions are given according to the HXB2R sequence (accession no. K03455). Smoothed entropies are shown. (b) Average of the individual smoothed entropies of nine patients (solid black line with error bars indicating standard errors), entropies of subtype B protein sequences taken from the HIV Sequence Database (each sequence corresponds to a different patient), and numbers of experimentally defined epitopes from HIV database overlapping with each position.
Individual sequence variability consistently tracks with the population level sequence variability (Fig. 4). This result concerns a small portion of Env and may not hold for other regions of HIV. Assuming, however, that it can be extrapolated, this result suggests that the locations of variable regions in individual patients and in the population are generally similar. The CTL epitopes described in the literature (Fig. 4b) also correspond to the more-conserved regions in each of these nine individual patient sequences, analogous to our finding at the population level.
Immunoproteasome cleavage predictions.
To test whether epitope clustering might be influenced by the proteasome specificity and to predict which sites in HIV-1 protein sequences are most likely to be cleaved by the proteasome and serve as C termini of CTL epitopes, we used cleavage prediction program NetChop (37). This predictive method is based on the use of artificial neural networks, a nonlinear classification technique. The neural network we used was trained on naturally processed C termini of known human CTL epitopes. To train the neural network, many examples of true, known cleavage sites embedded in protein sequences are used as positive examples. Likewise a negative training set is provided by the amino acid sequences that do not contain cleavage sites. The training set deliberately excluded HIV epitopes, so known HIV-1 epitope C-terminal positions could be used as an unbiased test set to examine the ability of the neural network to identify C-terminal cleavage sites in HIV-1. Predictions are made for each position in a protein sequence by using a symmetrical flanking region of eight amino acids, i.e., a 17-residue sliding window is used for each prediction. Prediction scores are between 0 and 1, with higher values indicating sites that are most likely to be cleaved and serve as C termini of epitopes (see reference 37 and Materials and Methods for more details). Since we are interested in learning about the tendency of a site to be cleaved at the population level, we used the median value of predictions over all sequences obtained for each site in each HIV-1 protein alignment to represent the population cleavage prediction score for the site.
The median prediction scores for true C termini of experimentally observed HIV-1 epitopes were found to be greater than those for all other remaining sites and those for sites from only epitope-lacking regions, and the difference was highly significant for each of the five proteins studied (P < 0.01; Mann-Whitney test; Fig. 5). NetChop predictions are of course imperfect; a particular site might be misclassified. Tested on other non-HIV data sets NetChop gave 69% specificity and 73% sensitivity (37). The highly significant correlation between C termini of identified CTL epitopes and conservation of predicted cleavage sites in HIV proteins (Fig. 5) shows that, at the population level (as opposed to just a single strain), NetChop can distinguish classes of positions that are typically favorable for cleavage and positions that are embedded in a context that makes cleavage very unlikely. Regional localization of low NetChop scores in areas where there are no defined CTL epitopes suggests that these protein subregions may have reduced epitope-processing potential and that this feature persists throughout the HIV-1 M group.
Being trained on C termini of HLA ligands, NetChop may capture the combined specificity of TAP (22) and HLA molecules (53) as well as immunoproteasome cleavage. Since training of the neural network was conducted on a training set of known epitopes and eluted peptide ligands from 59 different HLA class I molecules (see Materials and Methods), however, the chance of the neural network learning a certain HLA binding motif instead of the specificity of the immunoproteasome was minimized (37). To further address this issue, we took into account the possibility that the training set for NetChop could be biased toward HLA-A2 binding motifs because a large portion of CTL class I epitopes described in the literature are presented by HLA-A2, the most common HLA type in Caucasians (45). We therefore considered separately the subset of HIV epitope C terminus prediction scores excluding HLA-A2 epitopes. The Mann-Whitney test showed that these prediction scores were still highly significant (Fig. 5). This result supports the ability of the method to minimize the influence of certain HLA binding motifs, even the most highly represented one.
C-terminal amino acids unfavorable for epitopes.
Whether a peptide binds to an HLA class I molecule depends critically on the anchor residues (53). In particular, C-terminal positions of peptides bind to the F pocket of the peptide binding groove in the class I molecule. Different types of HLA molecules have different binding motifs. To account for anchor residues as generally as possible, we considered a set of particular amino acids which are rare among C-terminal anchor motifs of class I molecules (18). These amino acids are G, S, T, P, N, Q, D, E, and H (using the one-letter amino acid code). We checked their frequencies in experimentally observed HIV epitopes that are compiled in the HIV database (41). Indeed D and G do not appear at all at the C termini of 334 well-characterized HIV epitopes, and the each of the remaining amino acids from the list appears in less than 1.5% of defined epitopes. These amino acids that are rarely found as C termini tend to be small, polar, or negatively charged.
To test if these unfavorable amino acids are enriched in highly variable regions that contain no defined epitopes, we compared the frequencies of unfavorable amino acids in the M group consensus amino acid sequences for all five proteins in epitope-presenting and epitope-lacking regions using Fisher's exact test. The M group consensus was generated based on the consensus of each subtype, so it was not biased by subtypes with large numbers of available sequences. The epitope-lacking regions turned out to be enriched for unfavorable amino acids in Nef (P = 0.05), Env (P = 0.01), and p17 (P = 0.01) (by using a Bonferroni correction for multiple tests, the value for Nef no longer reaches significance but the trend is consistent with what we observed for Env and p17). In p24 and RT, which have no highly variable subdomains that might better tolerate escape, there was no statistical difference in the distribution of unfavorable amino acids between regions with epitopes and epitope-lacking regions (p24, P = 0.17; RT, P = 0.25).
Comparison of epitope locations with known secondary structure of the proteins.
It has been suggested that the folding of peptides into a native alpha-helical structure may be an important aspect of CTL antigenicity (10, 13). To study possible relationships between protein secondary structure and epitope locations, we mapped secondary structural information derived from HIV-1 crystal structures concerning whether or not a site is embedded in an alpha-helix, beta-sheet, or loop onto protein sequences (Fig. 2). We then correlated these secondary structural elements with epitope density (the number of unique epitopes that overlap a given site) or with the simpler classification of presence or absence of epitopes, summarizing the five HIV-1 proteins that have been the focus of HIV-1 CTL studies: RT, Gag p24 and p17, Nef, and Env gp160 core and gp41 (Table 1). We constructed a two-by-three contingency table tallying the number of protein sequence positions that were either in an epitope-presenting region or an epitope-lacking region and whether they were embedded in an alpha helix, beta sheet, or loop (Table 1). Fisher's exact test revealed a highly significant distinct distribution of epitopes in the different structural elements (P < 10−6). CTL epitopes are more highly concentrated in alpha-helical regions of proteins than in other regions; the percentage of positions that overlap with known epitopes is higher in alpha-helices than in loops or beta-sheets, both in summary of all five proteins (Table 1) and for each of the proteins separately (not shown).
TABLE 1.
Protein sequence positions embedded in alpha helices, beta sheets, and loops with regard of whether they overlap epitope-presenting or epitope-lacking regionsa
| Secondary structure | No. (%) of protein sequence positions embedded in protein regions:
|
|
|---|---|---|
| Epitope lacking | Epitope presenting | |
| Alpha helix | 122 (30.1) | 283 (69.9) |
| Beta sheet | 123 (50.4) | 121 (49.6) |
| Loop | 292 (44.5) | 364 (55.5) |
P < 10−6 by Fisher's exact test, summary for RT, Gag p24, and p17, Nef, and Env gp120 gp41.
For the statistical analysis above, we used the simple classification of epitope-presenting regions and epitope-lacking regions to avoid bias of cross-reactive epitopes, but there is also an interesting trend when one considers the underlying structure in conjunction with the number of unique epitopes that overlap a given position in a protein (Table 2). There is a distinct tendency for sites with large numbers of overlapping epitopes to be found in either alpha helices or loops; these positions are embedded in highly immunogenic regions and are less commonly found in beta sheets.
TABLE 2.
Distribution of protein sequence positions with regard to the protein secondary structure and number of CTL epitopes overlapping with a given positiona
| Secondary structure | No. of epitopes overlapping with position:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| Alpha helix | 82 | 88 | 30 | 22 | 8 | 6 | 9 | 7 | 6 | 5 | 9 | 7 | 4 | |||||
| Beta sheet | 40 | 31 | 26 | 4 | 8 | 5 | 6 | 1 | ||||||||||
| Loop | 145 | 55 | 57 | 23 | 10 | 17 | 13 | 13 | 7 | 2 | 6 | 4 | 2 | 4 | 2 | 2 | 2 | |
Summary for RT, Gag p24 and p17, Nef, and Env gp120 and gp41.
Predictions of regions favorable for epitopes in regulatory and accessory proteins.
Based on our findings for p17, Nef, Env, p24, and RT, for which many epitopes have been described, we developed a general strategy to predict regions of epitope localization. Through incorporating quantitative information about entropy measurements, proteasome cleavage prediction scores, and frequency of unfavorable anchor residue amino acids into a summary graphic, we could predict regions in new protein alignments that would be likely to carry epitopes. We applied this strategy to regulatory proteins Tat and Rev and to accessory proteins Vpr, Vif, and Vpu. Few epitopes in these proteins have previously been defined, and we made our predictions blinded with regard to any known epitopes.
Regions designated likely to have epitopes were those that were conserved, contained sites with high proteasome cleavage scores (the median NetChop score for the alignment was high), and had a low proportion of unfavorable amino acids. The unlikely regions are either highly variable regions or regions in which there are no strong cleavage predictions or in which the proportion of unfavorable amino acids is high in every position or all of the above. To make predictions, the strategy was to move along the protein from right to left (from C terminus to N terminus). When a position was located in a conserved region, was predicted to be a C terminus of an epitope by NetChop, and had no or a low proportion of unfavorable amino acids, we included that position and 10 amino acids to the left in the epitope-favorable category. When a position within an already-marked promising region had analogous features, we again calculated 10 amino acids to the left of this position and elongated the promising region appropriately. With this approach the boundaries of the predicted epitope regions could be roughly defined. Tat, Rev, and Vif are variable enough to make predictions using this strategy, but Vpr is too conserved.
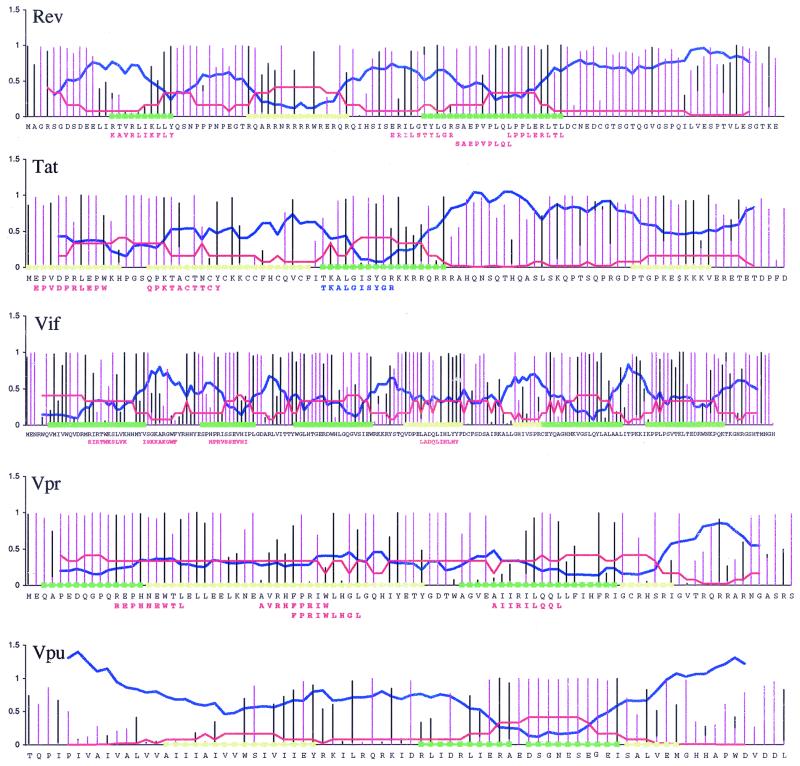
To assess whether epitope localization could be predicted accurately, our predictions were sent for comparison to the Partners AIDS Research Center at Massachusetts General Hospital, where a number of optimal CTL epitopes in Tat, Rev, Vif, and Vpr have recently been defined experimentally (2, 4; M. M. Addo and M. Altfeld, unpublished data). Nine out of 11 new epitopes in Tat, Rev, and Vif were identified in regions that we predicted would carry epitopes, and a 10th overlapped such a region. The correlation between our predicted epitope regions and the location of actual epitopes is highly significant (Fig. 6). At the time of this writing there is no experimental CTL epitope data available for Vpu; however, our predictions may be of interest for future experimental studies, so they are provided in Fig. 6.
FIG. 6.
Predictions of regions likely to hold epitopes in regulatory and accessory proteins Rev, Tat, Vif, Vpr, and Vpu and the localization of newly defined epitopes relative to these regions. Quantitative values used to predict regions likely to contain epitopes are plotted relative to the HXB2R reference strain. (Note that HXB2R strain is shown only for orientation, and predictions are done based on alignments described in Materials and Methods.) Green, regions deemed favorable for finding CTL epitopes; yellow, regions less likely but still promising. Black bars, site-specific proteasome cleavage prediction scores calculated as for Fig. 5 (high is favorable); pink bars, proportions of unfavorable amino acids at each site of the alignment (low is favorable); blue lines, smoothed entropy (low is favorable). To estimate the likelihood of finding an epitope in a region as a function of entropy, the entropy range was divided into 10 equal intervals and the ratio of the number of epitopes that fall into each interval out of all epitopes from Nef, p17, p24, Env, and RT was calculated (red lines; high is favorable) and can be considered as an estimate of the probability of finding an epitope given the entropy. The experimental epitopes defined by M. M. Addo and M. Altfeld (Partners AIDS Research Center, Massachusetts General Hospital) are shown by red letters below strain HXB2R. A Tat peptide that is the most highly recognized peptide in Tat (no optimal epitope was available) is indicated in blue below the HXB2R reference sequence. Since our analysis cannot discriminate well between epitope-presenting and epitope-lacking regions in conserved proteins (e.g., p24), our predictions for potential epitope locations within conserved Vpr are rather wide and cover about 74% of the protein. For more-variable Vif and highly variable Tat and Rev, the regions where we anticipated finding experimental epitopes span about 54% of the proteins considered. Experimental CTL epitopes in these proteins were found in regions spanning 25% of the positions, and 82% of experimental epitope positions were in regions that we predicted would carry epitopes. The predictions for Tat, Rev, and Vif were highly significant by Fisher's exact test (P < 0.001).
DISCUSSION
We have shown, at the population level, using set of protein sequences from 101 different individuals with HIV-1 infection from viruses representing HIV-1 M group diversity and the database of hundreds of published CTL epitopes, that the presence of HIV-specific CTL epitopes is generally inversely correlated with protein sequence variability. One highly likely (and trivial) contributing factor to the inverse correlation between epitope density and protein sequence variability is an experimental selection effect due to the use of reference strain-based reagents for studying antiviral CTL activity that have limited cross-reactivity, i.e., CTLs that target variable regions may go undetected due to sequence substitutions in reagents used to define the CTL response. It is not possible at this time to measure the relative importance of this potential for lack of cross-reactivity, because experimental studies can only describe the epitopes they find, not those they miss; there are still very little data comparing early autologous isolates to reference strains. (Autologous sequences may help alleviate this problem for future studies; however, sequences from later time points in an infected individual may miss key early CTL responses, as these sequences may be preselected as escape mutants by the time they are sampled [50].) Despite possible reference strain experimental bias, our study strongly suggests that there are additional underlying immunological reasons for the clustering of epitopes, beyond reagent strain differences, that account for the paucity of epitopes in highly variable regions.
First, we have shown that variable, epitope-lacking regions of the virus have low median values of predicted proteasome cleavage sites; potential cleavage sites are infrequent and are not conserved. This observation goes beyond merely being an indication that the prediction method is working as anticipated. It indicates that the proteasomal cleavage of C termini contributes to the apparent clustering of HIV CTL epitopes: observed epitopes cluster in regions where most HIV protein sequences are predicted to carry C-terminal cleavage sites, and such cleavage sites tend to be selectively depleted from epitope-lacking regions. Our analysis also suggests that the C termini of epitopes that have been defined experimentally by using a few reference strains are well conserved through the spectrum of HIV-1 variants. Second, we have shown that epitope-lacking regions are enriched for amino acids that diminish HLA binding. Thus escape mutations that inhibit either processing or HLA binding occur more often in variable regions. Such mutations confer a deep and general form of CTL escape; if a cleavage site is lost or a peptide cannot bind to any class I molecule, then an accumulation of such mutations decreases the immunogenic potential of the regions where they are localized.
The decrease in the immunogenic potential of variable regions of the virus is a measurable trace of past CTL immune escape concentrated in regions of the virus that readily tolerate change. At a population level, this means that the virus may have evolved, within its structural and functional constraints, toward a state more refractive to the CTL response. This could be considered analogous to the increasing number of drug resistance mutations found among individuals with primary infections in populations where combination therapy is readily available (16, 44). It is not possible at this time to distinguish whether the accumulation of mutations that confer CTL resistance in variable domains can be associated with the infection of the human species or whether viral evolution to a state more refractive to the CTL response could have had an even earlier origin in a primate host; as more SIVcpz sequence data derived from chimpanzees accrue, it may become feasible to examine this question.
The variation of HIV-1 Env protein sequences within nine individual patients tracked with the B-subtype population variation (Fig. 3). Although the longitudinal history of the CTL response in these patients is not known, the CTL epitopes described in the literature correspond to the more conserved regions of these nine individual patient protein sequences. These observations are compatible with the notion that escape occurs within individual patients and influences detection of host-specific CTL responses, as well as the overall pattern of evolution of the virus.
Mutations that influence the level of processing of CTL epitopes may diminish a CTL response and be of importance in vivo. The magnitude of epitope-specific CTL responses is correlated with the expression level of the corresponding class I molecule-peptide complex on the surface of the infected cell (27). Moreover, it was recently found that antigenic stimulation of T cells can be induced by multiple short-lived contacts (on the scale of several minutes) and that repetitive short-term signaling might be necessary (33). Thus, the amount of a given peptide appearing on the surface of the infected cell and the frequency of HLA molecules loaded with this peptide are crucial in regulation of the immune response, and a mutant that can even only partially escape in vivo presentation (by reducing either processing or HLA binding) may have a large advantage compared to the wild type. Thus experiments that examine variant peptide CTL cross-reactivity in vitro may not reveal the actual immunogenic potential of the variant epitope in vivo. Protein domains that are highly variable at the population level are obviously regions that tolerate change. Since the variation of HIV-1 protein sequences within single patients tracks with the population variation (Fig. 3), there may be an additional explanation for the lack of observed epitopes in the variable domains. Mutations may in part arise stochastically simply because the domain tolerates variability. If this is the case, a CTL epitope may be present in only a fraction of the virus quasispecies, at a frequency that is inadequate for the stimulation of the CTL response.
Using entropy as a measure of regional variability, proteasomal cleavage prediction results, and the frequency of the C-terminal amino acids that inhibit HLA binding, we accurately predicted the epitope-promising regions in HIV regulatory and accessory proteins (Fig. 6). This has direct applications in vaccine development and further indicates that immunological features of the proteins that we have studied here are relevant to epitope clustering. A particular strength of the approach is the ability to predict regions likely to have CTL epitopes, regardless of the restricting HLA class I molecule. Our strategy is not generalizable to relatively conserved pathogens such as DNA viruses or human T-cell leukemia virus type 1. It could, however, potentially give good results if applied to other variable pathogens such as hepatitis C virus.
Our approach demonstrating immunological correlations with epitope clustering favors an alternative way to design polyepitope vaccines by inclusion of short regions where epitopes cluster and the proximal regions that might influence processing, rather than a string of single epitopes. Like a polyepitope string, these short regions may be safer than full-length proteins and would facilitate the inclusion of cocktails of variants. Additionally, these regions include overlapping epitopes with multiple HLA-presenting molecules, and natural processing signals of known epitopes would be included in a polyepitope construct.
Acknowledgments
We thank Bruce Walker for initially suggesting that we test our predictions on the set of epitopes in regulatory and accessory proteins newly defined in the Partners AIDS Research Center at Massachusetts General Hospital, Norman Letvin for suggesting the test of neural network predictions excluding HLA-A2 epitopes, and Philip Goulder for suggesting a list of amino acids which tend not to serve as C-terminal anchor residues. We also thank Vadim Zalunin for help in preparing the figures. Chang-Shung Tung advised us concerning protein structural considerations.
K.Y., B.T.K., V.D., and B.G. were supported by the Department of Energy under contract W-7405-ENG-3 by LDRD funding; K.Y. and B.T.K. were also supported by a Pediatric AIDS Foundation Elizabeth Glaser Award; M.M.A. was supported through the Doris Duke Charitable Foundation and the NIH (RO1 AI50429); M.M.A. was supported by Deutsche Forschungsgemeinschaft grant AD-171; and C.K. was supported by Dutch Science Foundation grant 809.37.009.
REFERENCES
- 1.Abele, R., and R. Tampe. 1999. Function of the transport complex TAP in cellular immune recognition. Biochim. Biophys. Acta 1461:405-419. [DOI] [PubMed] [Google Scholar]
- 2.Addo, M. M., M. Altfeld, E. S. Rosenberg, R. L. Eldridge, M. N. Philips, K. Habeeb, A. Khatri, C. Brander, G. K. Robbins, G. P. Mazzara, P. J. Goulder, and B. D. Walker. 2001. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc. Natl. Acad. Sci. USA 98:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 4.Altfeld, M., M. M. Addo, R. L. Eldridge, X. G. Yu, S. Thomas, A. Khatri, D. Strick, M. N. Phillips, G. B. Cohen, S. A. Islam, S. A. Kalams, C. Brander, P. J. Goulder, E. S. Rosenberg, and B. D. Walker. 2001. Vpr is preferentially targeted by CTL during HIV-1 infection. J. Immunol. 167:2743-2752. [DOI] [PubMed] [Google Scholar]
- 5.Arold, S., P. Franken, M. P. Strub, F. Hoh, S. Benichou, R. Benarous, and C. Dumas. 1997. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure 5:1361-1372. [DOI] [PubMed] [Google Scholar]
- 6.Barnham, K. J., S. A. Monks, M. G. Hinds, A. A. Azad, and R. S. Norton. 1997. Solution structure of a polypeptide from the N terminus of the HIV protein Nef. Biochemistry 36:5970-5980. [DOI] [PubMed] [Google Scholar]
- 7.Barouch, D. H., and N. L. Letvin. 2000. DNA vaccination for HIV-1 and SIV. Intervirology 43:282-287. [DOI] [PubMed] [Google Scholar]
- 8.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beekman, N. J., P. A. van Veelen, T. van Hall, A. Neisig, A. Sijts, M. Camps, P. M. Kloetzel, J. J. Neefjes, C. J. Melief, and F. Ossendorp. 2000. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J. Immunol. 164:1898-1905. [DOI] [PubMed] [Google Scholar]
- 10.Berkower, I., G. K. Buckenmeyer, and J. A. Berzofsky. 1986. Molecular mapping of a histocompatibility-restricted immunodominant T cell epitope with synthetic and natural peptides: implications for T cell antigenic structure. J. Immunol. 136:2498-2503. [PubMed] [Google Scholar]
- 11.Bernard, N. F., C. M. Yannakis, J. S. Lee, and C. M. Tsoukas. 1999. Human immunodeficiency virus (HIV)-specific cytotoxic T lymphocyte activity in HIV-exposed seronegative persons. J. Infect. Dis. 179:538-547. [DOI] [PubMed] [Google Scholar]
- 12.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berzofsky, J. A., K. B. Cease, J. L. Cornette, J. L. Spouge, H. Margalit, I. J. Berkower, M. F. Good, L. H. Miller, and C. DeLisi. 1987. Protein antigenic structures recognized by T cells: potential applications to vaccine design. Immunol. Rev. 98:9-52. [DOI] [PubMed] [Google Scholar]
- 14.Betts, M. R., J. F. Krowka, T. B. Kepler, M. Davidian, C. Christopherson, S. Kwok, L. Louie, J. Eron, H. Sheppard, and J. A. Frelinger. 1999. Human immunodeficiency virus type 1-specific cytotoxic T lymphocyte activity is inversely correlated with HIV type 1 viral load in HIV type 1-infected long-term survivors. AIDS Res. Hum. Retroviruses 15:1219-1228. [DOI] [PubMed] [Google Scholar]
- 15.Bochtler, M., L. Ditzel, M. Groll, C. Hartmann, and R. Huber. 1999. The proteasome. Annu. Rev. Biophys. Biomol. Struct. 28:295-317. [DOI] [PubMed] [Google Scholar]
- 16.Brenner, B., M. A. Wainberg, H. Salomon, D. Rouleau, B. Spira, R. P. Sekaly, B. Conway, J. P. Routy, et al. 2000. Resistance to antiretroviral drugs in patients with primary HIV-1 infection. Int. J. Antimicrob. Agents 16:429-434. [DOI] [PubMed] [Google Scholar]
- 17.Buseyne, F., and Y. Riviere. 2001. The flexibility of the TCR allows recognition of a large set of naturally occurring epitope variants by HIV-specific cytotoxic T lymphocytes. Int. Immunol. 13:941-950. [DOI] [PubMed] [Google Scholar]
- 18.Calef, C., R. Thakalapally, D. Lang, C. Brander, P. Goulder, O. Yang, and B. Korber. 2000. PeptGen: designing peptides for immunological studies and application to HIV consensus sequences, p. I63-I67. In B. T. Korber, C. Brander, B. Haynes, R. Koup, J. P. Moore, C. Kuiken, B. D. Walker, and D. Watkins (ed.), HIV molecular immunology 2000. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 19.Cao, H., P. Kanki, J. L. Sankale, A. Dieng-Sarr, G. P. Mazzara, S. A. Kalams, B. Korber, S. Mboup, and B. D. Walker. 1997. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J. Virol. 71:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardozo, C., and R. A. Kohanski. 1998. Altered properties of the branched chain amino acid-preferring activity contribute to increased cleavages after branched chain residues by the “immunoproteasome.” J. Biol. Chem. 273:16764-16770. [DOI] [PubMed] [Google Scholar]
- 21.Chassin, D., M. Andrieu, W. Cohen, B. Culmann-Penciolelli, M. Ostankovitch, D. Hanau, and J. G. Guillet. 1999. Dendritic cells transfected with the nef genes of HIV-1 primary isolates specifically activate cytotoxic T lymphocytes from seropositive subjects. Eur. J. Immunol. 29:196-202. [DOI] [PubMed] [Google Scholar]
- 22.Daniel, S., V. Brusic, S. Caillat-Zucman, N. Petrovsky, L. Harrison, D. Riganelli, F. Sinigaglia, F. Gallazzi, J. Hammer, and P. M. van Endert. 1998. Relationship between peptide selectivities of human transporters associated with antigen processing and HLA class I molecules. J. Immunol. 161:617-624. [PubMed] [Google Scholar]
- 23.Day, C. L., A. K. Shea, M. A. Altfeld, D. P. Olson, S. P. Buchbinder, F. M. Hecht, E. S. Rosenberg, B. D. Walker, and S. A. Kalams. 2001. Relative dominance of epitope-specific cytotoxic T-lymphocyte responses in human immunodeficiency virus type 1-infected persons with shared HLA alleles. J. Virol. 75:6279-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Groot, A. S., A. Bosma, N. Chinai, J. Frost, B. M. Jesdale, M. A. Gonzalez, W. Martin, and C. Saint-Aubin. 2001. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine 19:4385-4395. [DOI] [PubMed] [Google Scholar]
- 25.Egan, M. A., W. A. Charini, M. J. Kuroda, J. E. Schmitz, P. Racz, K. Tenner-Racz, K. Manson, M. Wyand, M. A. Lifton, C. E. Nickerson, T. Fu, J. W. Shiver, and N. L. Letvin. 2000. Simian immunodeficiency virus (SIV) gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J. Virol. 74:7485-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emmerich, N. P., A. K. Nussbaum, S. Stevanovic, M. Priemer, R. E. Toes, H. G. Rammensee, and H. Schild. 2000. The human 26S and 20S proteasomes generate overlapping but different sets of peptide fragments from a model protein substrate. J. Biol. Chem. 275:21140-21148. [DOI] [PubMed] [Google Scholar]
- 27.Gallimore, A., H. Hengartner, and R. Zinkernagel. 1998. Hierarchies of antigen-specific cytotoxic T-cell responses. Immunol. Rev. 164:29-36. [DOI] [PubMed] [Google Scholar]
- 28.Geier, E., G. Pfeifer, M. Wilm, M. Lucchiari-Hartz, W. Baumeister, K. Eichmann, and G. Niedermann. 1999. A giant protease with potential to substitute for some functions of the proteasome. Science 283:978-981. [DOI] [PubMed] [Google Scholar]
- 29.Goh, W. C., J. Markee, R. E. Akridge, M. Meldorf, L. Musey, T. Karchmer, M. Krone, A. Collier, L. Corey, M. Emerman, and M. J. McElrath. 1999. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J. Infect. Dis. 179:548-557. [DOI] [PubMed] [Google Scholar]
- 30.Goulder, P., D. Price, M. Nowak, S. Rowland-Jones, R. Phillips, and A. McMichael. 1997. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol. Rev. 159:17-29. [DOI] [PubMed] [Google Scholar]
- 31.Goulder, P. J. 2000. Rapid characterization of HIV clade C-specific cytotoxic T lymphocyte responses in infected African children and adults. Ann. N. Y. Acad. Sci. 918:330-345. [DOI] [PubMed] [Google Scholar]
- 32.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 33.Gunzer, M., A. Schafer, S. Borgmann, S. Grabbe, K. S. Zanker, E. B. Brocker, E. Kampgen, and P. Friedl. 2000. Antigen presentation in extracellular matrix: interactions of T cells with dendritic cells are dynamic, short lived, and sequential. Immunity 13:323-332. [DOI] [PubMed] [Google Scholar]
- 34.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaul, R., T. Dong, F. A. Plummer, J. Kimani, T. Rostron, P. Kiama, E. Njagi, E. Irungu, B. Farah, J. Oyugi, R. Chakraborty, K. S. MacDonald, J. J. Bwayo, A. McMichael, and S. L. Rowland-Jones. 2001. CD8+ lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J. Clin. Investig. 107:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kesmir, C., A. K. Nussbaum, H. Schild, V. Detours, and S. Brunak. 2002. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 15:287-296. [DOI] [PubMed] [Google Scholar]
- 38.Korber, B. T., C. Brander, B. Haynes, R. Koup, J. P. Moore, C. Kuiken, B. D. Walker, and D. Watkins (ed.). 2000. HIV molecular immunology 2000. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 39.Korber, B. T., K. MacInnes, R. F. Smith, and G. Myers. 1994. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J. Virol. 68:6730-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchhan, J. Mellors, J. Mullins, S. Wolinsky, and B. T. Korber. 2001. HIV sequence compendium 2000. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 42.Kuiken, C. L., B. Foley, E. Guzman, and B. T. Korber. 1999. The determinants of HIV-1 protein evolution, p. 432-468. In K. A. Crandall (ed.), The evolution of HIV. The John Hopkins University Press, Baltimore, Md.
- 43.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leigh-Brown, A. J., H. M. Precious, J. M. Whitcomb, J. K. Wong, M. Quigg, W. Huang, E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, N. S. Hellmann, C. J. Petropoulos, D. D. Richman, and S. J. Little. 2000. Reduced susceptibility of human immunodeficiency virus type 1 (HIV-1) from patients with primary HIV infection to nonnucleoside reverse transcriptase inhibitors is associated with variation at novel amino acid sites. J. Virol. 74:10269-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsh, S. G. E., P. Parjam, and L. D. Barber. 2000. The HLA facts book. Academic Press, London, United Kingdom.
- 46.McAdam, S., P. Klenerman, L. Tussey, S. Rowland-Jones, D. Lalloo, R. Phillips, A. Edwards, P. Giangrande, A. L. Brown, and F. Gotch. 1995. Immunogenic HIV variant peptides that bind to HLA-B8 can fail to stimulate cytotoxic T lymphocyte responses. J. Immunol. 155:2729-2736. [PubMed] [Google Scholar]
- 47.Meister, G. E., C. G. Roberts, J. A. Berzofsky, and A. S. De Groot. 1995. Two novel T cell epitope prediction algorithms based on MHC-binding motifs; comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine 13:581-591. [DOI] [PubMed] [Google Scholar]
- 48.Musey, L., J. Hughes, T. Schacker, T. Shea, L. Corey, and M. J. McElrath. 1997. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 337:1267-1274. [DOI] [PubMed] [Google Scholar]
- 49.Niedermann, G., E. Geier, M. Lucchiari-Hartz, N. Hitziger, A. Ramsperger, and K. Eichmann. 1999. The specificity of proteasomes: impact on MHC class I processing and presentation of antigens. Immunol. Rev. 172:29-48. [DOI] [PubMed] [Google Scholar]
- 50.Novitsky, V., N. Rybak, M. F. McLane, P. Gilbert, P. Chigwedere, I. Klein, S. Gaolekwe, S. Y. Chang, T. Peter, I. Thior, T. Ndung'u, F. Vannberg, B. T. Foley, R. Marlink, T. H. Lee, and M. Essex. 2001. Identification of human immunodeficiency virus type 1 subtype c Gag-, Tat-, Rev-, and Nef-specific ELISPOT-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J. Virol. 75:9210-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pamer, E., and P. Cresswell. 1998. Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol. 16:323-358. [DOI] [PubMed] [Google Scholar]
- 52.Pantaleo, G., J. F. Demarest, H. Soudeyns, C. Graziosi, F. Denis, J. W. Adelsberger, P. Borrow, M. S. Saag, G. M. Shaw, and R. P. Sekaly. 1994. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature 370:463-467. [DOI] [PubMed] [Google Scholar]
- 53.Rammensee, H. G., J. Bachman, and S. Stevanovich. 1997. MHC ligands and peptide motifs. Landes Bioscience, Georgetown, Tex.
- 54.Rowland-Jones, S. L., S. Pinheiro, R. Kaul, P. Hansasuta, G. Gillespie, T. Dong, F. A. Plummer, J. B. Bwayo, S. Fidler, J. Weber, A. McMichael, and V. Appay. 2001. How important is the "quality' of the cytotoxic T lymphocyte (CTL) response in protection against HIV infection? Immunol. Lett. 79:15-20. [DOI] [PubMed] [Google Scholar]
- 55.Sarafianos, S. G., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 57.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shu, W., J. Liu, H. Ji, L. Radigen, S. Jiang, and M. Lu. 2000. Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides. Biochemistry 39:1634-1642. [DOI] [PubMed] [Google Scholar]
- 59.Sijts, A. J., S. Standera, R. E. Toes, T. Ruppert, N. J. Beekman, P. A. van Veelen, F. A. Ossendorp, C. J. Melief, and P. M. Kloetzel. 2000. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasomes in infected cells. J. Immunol. 164:4500-4506. [DOI] [PubMed] [Google Scholar]
- 60.Stoltze, L., M. Schirle, G. Schwarz, C. Schroter, M. W. Thompson, L. B. Hersh, H. Kalbacher, S. Stevanovic, H. G. Rammensee, and H. Schild. 2000. Two new proteases in the MHC class I processing pathway. Nat. Immunol. 1:413-418. [DOI] [PubMed] [Google Scholar]
- 61.Toes, R. E., A. K. Nussbaum, S. Degermann, M. Schirle, N. P. Emmerich, M. Kraft, C. Laplace, A. Zwinderman, T. P. Dick, J. Muller, B. Schonfisch, C. Schmid, H. J. Fehling, S. Stevanovic, H. G. Rammensee, and H. Schild. 2001. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 194:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van den Eynde, B. J., and S. Morel. 2001. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr. Opin. Immunol. 13:147-153. [DOI] [PubMed] [Google Scholar]
- 63.Wagner, R., B. Leschonsky, E. Harrer, C. Paulus, C. Weber, B. D. Walker, S. Buchbinder, H. Wolf, J. R. Kalden, and T. Harrer. 1999. Molecular and functional analysis of a conserved CTL epitope in HIV-1 p24 recognized from a long-term nonprogressor: constraints on immune escape associated with targeting a sequence essential for viral replication. J. Immunol. 162:3727-3734. [PubMed] [Google Scholar]
- 64.Walker, B. D., and B. T. Korber. 2001. Immune control of HIV: the obstacles of HLA and viral diversity. Nat. Immunol. 2:473-475. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, C. C., R. C. Brown, B. T. Korber, B. M. Wilkes, D. J. Ruhl, D. Sakamoto, K. Kunstman, K. Luzuriaga, I. C. Hanson, S. M. Widmayer, A. Wiznia, S. Clapp, A. J. Ammann, R. A. Koup, S. M. Wolinsky, and B. D. Walker. 1999. Frequent detection of escape from cytotoxic T-lymphocyte recognition in perinatal human immunodeficiency virus (HIV) type 1 transmission: the Ariel project for the prevention of transmission of HIV from mother to infant. J. Virol. 73:3975-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:543-585. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, C., J. L. Cornette, J. A. Berzofsky, and C. DeLisi. 1997. The organization of human leucocyte antigen class I epitopes in HIV genome products: implications for HIV evolution and vaccine design. Vaccine 15:1291-1302. [DOI] [PubMed] [Google Scholar]