Abstract
Ljungan virus (LV) is a suspected human pathogen recently isolated from bank voles (Clethrionomys glareolus). In the present study, it is revealed through comparative sequence analysis that three newly determined Swedish LV genomes are closely related and possess a deviant picornavirus-like organization: 5′ untranslated region-VP0-VP3-VP1-2A1-2A2-2B-2C-3A-3B-3C-3D-3′ untranslated region. The LV genomes and the polyproteins encoded by them exhibit several exceptional features, such as the absence of a predicted maturation cleavage of VP0, a conserved sequence determinant in VP0 that is typically found in VP1 of other picornaviruses, and a cluster of two unrelated 2A proteins. The 2A1 protein is related to the 2A protein of cardio-, erbo-, tescho-, and aphthoviruses, and the 2A2 protein is related to the 2A protein of parechoviruses, kobuviruses, and avian encephalomyelitis virus. The unprecedented association of two structurally different 2A proteins is a feature never previously observed among picornaviruses and implies that their functions are not mutually exclusive. Secondary polyprotein processing of the LV polyprotein is mediated by proteinase 3C (3Cpro) possessing canonical affinity to Glu and Gln at the P1 position and small amino acid residues at the P1′ position. In addition, LV 3Cpro appears to have unique substrate specificity to Asn, Gln, and Asp and to bulky hydrophobic residues at the P2 and P4 positions, respectively. Phylogenetic analysis suggests that LVs form a separate division, which, together with the Parechovirus genus, has branched off the picornavirus tree most closely to its root. The presence of two 2A proteins indicates that some contemporary picornaviruses with a single 2A may have evolved from the ancestral multi-2A picornavirus.
Picornaviruses are small positive-stranded RNA viruses with a genome comprising approximately 7,500 to 8,200 nucleotides (nt) which is packed into a nonenveloped icosahedral capsid (51). The polyadenylated genome of most picornaviruses contains a single expressed open reading frame (ORF) flanked by 5′ and 3′ untranslated regions (UTRs). The 5′ UTR, comprising approximately 600 to 1,300 nt, contains replicative and translational regulator signals and is folded in a complex tertiary structure. Located in the 3′ half of the 5′ UTR is an internal ribosome entry site (IRES) crucial for the cap-independent initiation of translation. Three types of IRESs are recognized on the basis of structure and are conserved in different branches of picornaviruses (62). The ORF encodes a large protein precursor (polyprotein) whose domain backbone contains the following organization: NH2-L-VP0-VP3-VP1-2A-2B-2C-3A-3B-3C-3D-COOH, where VP0, VP3, and VP1 are paralogous proteins forming the capsid, with all other nonstructural proteins being primarily involved in the picornaviral replicative process. Among the nonstructural proteins, the leader (L) protein has been identified in some but not all picornaviruses.
The picornavirus polyprotein is autocatalytically processed at the conserved interdomain junctions by a proteolytic activity associated with the 3C moiety, which, depending on the individual picornavirus, may also be assisted by the (proteolytic) activities of L and/or 2A proteins with different specificities (14, 31, 43, 54). Additional cleavages of the polyprotein at a few alternative sites may take place, resulting in new products and some intermediate precursors, some of which are stable and/or functionally active. In most picornaviruses, VP0 is autocatalytically cleaved further into VP4 and VP2 proteins during the final stage of virion maturation.
Both the L and 2A proteins have been described as having four apparently unrelated structural forms (10, 24, 54, 64, 65), and this diversity sets them apart from all other proteins conserved across the entire picornavirus family. The conserved proteins include the multifunctional 2C ATPase (2CATPase), the main cysteine 3C protease (3Cpro), 3D RNA-dependent RNA polymerase (3Dpol), membrane-associated 2B and 3A proteins, and a small 3B protein (3BVPg) (51). 3BVPg serves as a primer for the RNA synthesis mediated by 3Dpol with the involvement of other nonstructural proteins and remains covalently linked to the 5′ end of plus- and minus-strand RNAs (46).
Picornaviruses infect mammals, including humans, and birds (28). Picornavirus-like viruses that infect invertebrates have also been identified (7). Depending on the nature of the individual picornavirus, the infection may cause severe ailments of the gastrointestinal tract and the respiratory, neural, hepatocellular, and cardiomuscular systems (23, 42). Likewise, the host range, progeny yield, and reproductive cycle mechanisms differ dramatically among picornaviruses. This phenotypic diversity of picornaviruses is ultimately linked to the plasticity of the picornavirus genome.
The family Picornaviridae was originally classified into four genera based on the antigenic and biophysical properties of the virions (35). Subsequent molecular analysis of the viral genomes supported this classification for the majority of picornaviruses. Such characterizations also led to the creation of two additional genera, Hepatovirus and Parechovirus, comprising two former enteroviruses which have diverged substantially from other picornaviruses (28). In order to accommodate the molecular diversity of newly recognized picornaviruses, three other genera have recently been introduced (49). The constant increase in the number of picornavirus genera, the current bias towards picornaviruses that infect humans and a few other species, and the huge disparity in the numbers of viruses assigned to different genera (28) indicate that the diversity of Picornaviridae is far from being fully described.
During a search for an infectious agent linked to myocarditis in humans, a new virus, Ljungan virus (LV), was recently isolated from bank voles (Clethrionomys glareolus) trapped near the Ljungan River in Medelpad County, Sweden (38, 39). The partial sequences of the LV prototype strain, 87-012, and two other serologically related LV isolates, 174F and 145SL, suggested that LV is most closely related to human parechoviruses (HPEVs) (39). In the present study, bioinformatic techniques were employed to analyze the newly determined, nearly complete viral genomes of three LV isolates. Evidence was obtained showing that LV is a prototype of a new picornavirus lineage clustering with, but distinct from, parechoviruses. The organization of the LV genome, a tentative polyprotein map of processing signals recognized by 3Cpro with a unique specificity, the unusual sequence characteristics of capsid proteins, and the unprecedented association of two different 2A proteins are also described. Due to their possession of these unique features, it is proposed that LVs be classified into a separate genus of the Picornaviridae.
MATERIALS AND METHODS
Viruses and cells.
LV was isolated from bank voles in BHK-21 cells as previously described (39). The 145SL and 174F isolates were passaged four times in A549 cells and then once in Vero cells, whereas strain 87-012 was passaged six times in Vero cells, causing a mild and delayed cytopathic effect (CPE).
Purification of LV RNA and cDNA synthesis.
Viral RNA was extracted from Vero cell culture-propagated LV stocks (clarified by slow-speed centrifugation) with Ultraspec II (Biotecx Laboratories, Inc., Houston, Tex.). The RNA was reverse transcribed into cDNA by using the RNase H-deficient reverse transcriptase Superscript II (Life Technologies, Täby, Sweden) and the primer Notdt25 (5′-ATAAGAATGCGGCCGCT25-3′) at 45°C for 2 h as previously described (33). After cDNA synthesis was completed, the RNA template was hydrolyzed by the addition of NaOH to a final concentration of 0.2 M and subsequent incubation at 37°C for 15 min. The samples were then neutralized with sodium acetate at a final concentration of 0.4 M, and single-stranded cDNA was precipitated with ethanol, dissolved in Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]), and stored at −20°C before amplification.
For reverse transcription-PCR, several different types of primers were initially used in this study. The oligo(dT)-rich Notdt25 primer was used for both oligo(dT)-primed reverse transcription and for amplification of the 3′ end of the genome by use of the 3′ RACE (rapid amplification of cDNA ends) technique (3). For amplification of the 5′ end of the genome, two HPEV-based primers covering the 17 or 25 5′-proximate nt (PaLV5nc17 [5′-GATCTTAATTAATTTGAAAGGGGTCTCCT-3′] and PaLV5nc25 [5′-GATCTTAATTAATTTGAAAGGGGTCTCCTGGTGGGGT-3′], respectively, with the underlined sequences being those derived from the parechovirus nucleotide sequence) were used. A number of primers were derived from previously published partial sequences of LV, which include a 2,118-nucleotide segment that covers part of the 5′ UTR, VP0, and VP3 of strain 145SL and a 264-nucleotide segment that covers part of the 5′ UTR of strains 87-012 and 174F (39). Primer design was also based on comparisons with HPEV sequences listed in the GenBank database. Several overlapping PCR fragments were generated by using a primer walking strategy. All PCR amplifications were performed by using a PCR protocol previously described by Lindberg et al. (33), with a mixture of 2.5 U of Thermoprime Plus DNA polymerase (Abgene, Epsom, United Kingdom) and 0.05 U of Deep Vent DNA polymerase (New England Biolabs, Beverly, Mass.).
DNA sequencing.
The nucleotide sequences of the LV genomes were determined with purified PCR products (QIAquick gel extraction kit; QIAGEN GmbH) in a cycle sequencing reaction by using an ABI Prism BigDye terminator cycle sequencing ready reaction kit according to the manufacturer's instructions (PE Biosystems, Stockholm, Sweden). Each nucleotide was determined at least twice in each direction, except for the 3′ ends of the genomes, which were sequenced multiple times in the same direction. To avoid the possible inclusion of mutations generated by the amplification process, the final genome sequences were derived from sequences determined for at least two batches of viral RNAs extracted and amplified independently. Sequence data were recorded with an ABI Prism 310 genetic analyzer (PE Biosystems) and assembled and edited by using Sequencher 3.0 software (Gene Codes, Ann Arbor, Mich.).
Computer-aided comparative sequence analysis.
Sequence alignments were generated by using the Clustal 1.81 program, a windows-based interface of the Clustal W program (59, 60), and the Dialign2 program (36). Following protein comparisons, the Blossum matrix of the scoring interresidues tables (22) was used. To assess and illustrate the conservation of selected gap-free alignments, the sequences were converted into logos presentations (55). Prior to the phylogenetic analysis, the alignments were manually edited and all positions containing a gap(s) were removed by using the Data Analysis in Molecular Biology and Evolution (DAMBE) package (version 4.0.30; X. Xia, Department of Ecology and Biodiversity, University of Hong Kong). To investigate whether a data set contained phylogenetic signals corresponding to tree-, star-, or net-like evolution, a likelihood mapping analysis of the aligned sequences was performed (57). Phylogenetic reconstruction was conducted by employing the maximum-likelihood method using quartet puzzling as implemented in Tree-Puzzle version 5.0 software (58) and the Proml program of an alpha release of the PHYLIP version 3.6a2 package (J. Felsenstein, Department of Genetics, University of Washington, Seattle). The significance of the inferred phylogeny was evaluated by bootstrap analysis with 100 pseudoreplicas as implemented in the PHYLIP SeqBoot program (16). The resulting trees were visualized by using the Treeview program (41). The RNA secondary structures were predicted by using the RNAGA consensus program (5) and the MFOLD 3.1 program (67).
Nucleotide and protein sequences.
The genome sequences, and the polyproteins derived from them, that were used for comparisons in this study were from the following picornaviruses (with their GenBank accession numbers): Aphthovirus genus, foot-and-mouth disease virus (FMDV) (MJ10975) and equine rhinitis A virus (ERAV) (L43052); Cardiovirus genus, encephalomyocarditis virus (EMCV) (M22457) and Theiler's murine encephalomyelitis virus (TMEV) (M20301); Enterovirus genus, poliovirus type 1 strain Sabin (PV1S) (V01150) and A-2 plaque virus (A2pV) (AAF85765); Erbovirus genus, equine rhinitis B virus (ERBV) (X96871); Hepatovirus genus, hepatitis A virus (HAV) (M14707 and M59810) and avian encephalomyelitis virus (AEV) (AJ225173); Kobuvirus genus, Aichi virus (AiV) (AB010145); Parechovirus genus, HPEV1 strain Harris (HPEV1H) (S45504), HPEV2 strain Williamson (HPEV2W) (AJ005695), and HPEV2 strain CT86-6760 (HPEV2C) (AF055846); Rhinovirus genus, human rhinovirus 2 (HRV2) (X02316); and Teschovirus genus, porcine teschovirus 1 (PTV1) (AJ011380). The protein sequences of two insect viruses, infectious flacherie virus (InFV) (AB000906) and sacbrood virus (SBV) (AF092924), that are distantly related to picornaviruses were also used as out-groups in the phylogenetic analysis.
Nucleotide sequence accession numbers.
The genome sequences of LV strains 87-012, 174F, and 145SL described in this study have been submitted to GenBank and have been assigned accession no. AF327920, AF327921, and AF327922, respectively.
RESULTS AND DISCUSSION
Sequencing genomes of three LV isolates.
To determine the genomic sequence of LV, three field isolates were propagated through several different cell cultures (see Materials and Methods). LV replication in Vero cells induced a delayed and less pronounced CPE than that normally facilitated by many enteroviruses (our unpublished data). Despite serial passages in several cell lines, no evidence of adaptation of LV was observed within 2 weeks. The genomic LV RNA was isolated from infected cells and used to determine the nucleotide sequence from overlapping PCR-generated amplicons. Amplifications of the extreme 5′ UTR by use of different protocols of the RACE technique (15) were not successful, possibly due to a stable secondary structure of the 5′ terminal region, as has previously been suggested for parechoviruses (17) (Fig. 1). The most 5′-terminal LV sequences, which have been deposited in GenBank (17 nt for the prototype strain 87-012 and 25 nt for strains 174F and 145SL), remain of parechovirus origin and should be verified in the near future. Based on the results of the primer extension and Southern blot analyses (data not shown), the length of the amplified LV genomes reported here must be close or identical to the length of wild-type LV genomes. The LV 145SL sequence reported here and a fragment of it published previously (39) differ in four positions (A157G, T733C, T1894C, and G2267C, numbered according to our nucleotide sequence, AF327921), the last three of which are nonsynonymous. The newly described genomic sequences were determined from PCR-generated amplicons to ensure that they correspond to the dominant genotypes of the semiadapted LV present during propagation in Vero cells. In contrast, previously reported LV sequences were determined from PCR-generated individual clones (39) and may deviate from the consensus genotype. Indeed, at position 733, a nucleotide determined in the previous study (39) destroyed the initiator codon predicted here, which in the present analysis resides in an optimal Kozak context (29).
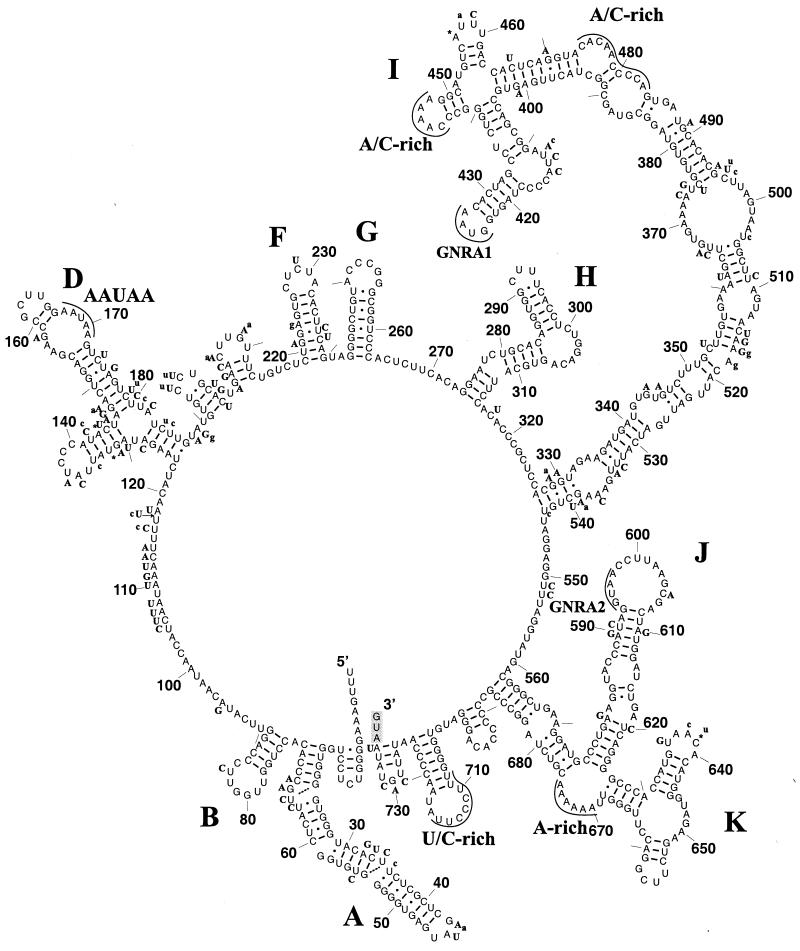
FIG. 1.
Predicted stem-loop structures of the LV 5′ UTR. Stem-loop structures in the 5′ UTR were predicted for three LV isolates as described in Materials and Methods. Illustrated is the predicted organization of the 5′ UTR of strain LV 87-012 drawn with the XRNA program (B. Wasei and H. F. Noller, unpublished data). The secondary-structure elements recognized in the 5′ UTRs harboring group II IRESs are labeled. Also highlighted are various conserved sequence elements discussed in the text. Stem-loop structures A through H were derived from the RNAGA consensus program (5) which compared LV strains and three parechoviruses. The folding of the I and J-K stem-loop regions was supported by the consensus analysis and was predicted using the MFOLD program (67). Replacements in strains 145SL and 174F are indicated in boldface uppercase and lowercase letters, respectively, and insertions and deletions are marked with asterisks and arrows, respectively. Base pairings predicted for LV 87-012 are indicated with continuous lines, and base pairings that are possible in either of the two other isolates are indicated with broken lines. The initiator codon is indicated with a shaded box.
The GC content of the LV genomes is 42%, which is similar to those of rhino- (39% for HRV2), hepato- (38% for HAV), and parechoviruses (39% for HPEV1). The major features of the polyproteins encoded by the genomes of the three LVs are summarized in Table 1. Excluding the poly(A) tract, the LV 87-012, 174F, and 145SL genomes comprise 7,606, 7,608, and 7,609 nt, respectively. The 5′ UTRs of the LV 87-012, 174F, and 145SL genomes consist of 733, 735, and 731 nt, respectively, and are followed by single ORFs encoding polyproteins containing 2,253, 2,253, and 2,256 amino acids (aa), respectively, while the 3′ UTRs possess 111, 111, and 107 nt, respectively, and poly(A) tails. The genomes and polyproteins of the LV isolates have 80 to 93% identical nt and 89 to 99% identical amino acid residues, respectively. Previously, based on their antigenic properties, LV 87-012 and 174F had been separated from 145SL (39); the genomic sequence analysis reported here confirms this grouping. A detailed domain comparison of LV with representatives of all genera of the Picornaviridae confirmed the clustering of LV with parechoviruses (Table 2).
TABLE 1.
Features of proteins of LV strains 145SL and 174F and the prototype strain 87-012a
| Protein | No. of amino acids in protein of strain:
|
Amino acid positions for the prototype protein | Predicted activity or function of viral protein | ||
|---|---|---|---|---|---|
| 145SL | 174F | 87-012 | |||
| VP0 | 259 | 259 | 259 | 1Met-Gln259 | Capsid protein |
| VP3 | 244 | 244 | 244 | 260Gly-Gln503 | Capsid protein |
| VP1 | 297 | 297 | 297 | 504Gly-Glu800 | Capsid protein |
| 2A1 | 20 | 20 | 20 | 801Met-Gly820 | Primary polyprotein processing |
| 2A2 | 135 | 135 | 135 | 821Pro-Gln955 | Inhibition of cell growth |
| 2B | 140 | 138 | 138 | 956Ser-Glu1093 | Membrane associated |
| 2C | 334 | 333 | 333 | 1094Gly-Asn1426 | ATPase |
| 3A | 130 | 130 | 130 | 1427Glu-Glu1556 | Membrane associated |
| 3B | 29 | 29 | 29 | 1557Arg-Glu1585 | Initiation of RNA synthesisb |
| 3C | 198 | 198 | 198 | 1586Ala-Gln1783 | Protease |
| 3D | 470 | 470 | 470 | 1784Gly-Asp2253 | RNA-dependent RNA polymerase |
The polyproteins of strains 145SL, 174F, and 87-012 contain 2,256, 2,253, and 2,253 amino acids in all, respectively.
Protein 3BVPg.
TABLE 2.
Comparison of the LV prototype strain 87-012 with selected picornavirusesa
| Region |
Aphthovirus
|
Cardiovirus
|
Enterovirus
|
Erbovirus
|
Hepatovirus
|
Kobuvirus
|
Parechovirus
|
Rhinovirus
|
Teschovirus
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FMDV | ERAV | EMCV | TMEV | PV1S | ERBV | HAV | AEV | AiV | HPEV1 | HRV2 | PTV1 | |
| 5′ UTRb | 46 | 35 | 48 | 46 | 36 | 48 | 39 | 42 | 39 | 50 | 38 | ND |
| VP0 | 23 | 24 | 29 | 23 | 17 | 26 | 24 | 25 | 20 | 50 | 25 | 25 |
| VP3 | 22 | 25 | 25 | 27 | 19 | 22 | 26 | 25 | 27 | 56 | 21 | 25 |
| VP1 | 17 | 22 | 17 | 24 | 19 | 22 | 31 | 32 | 40 | 45 | 25 | 23 |
| 2A1 | 44 | 50 | 40 | 41 | ND | 50 | ND | ND | ND | ND | ND | 59 |
| 2A2 | ND | ND | ND | ND | ND | ND | 28 | 25 | 25 | 55 | ND | ND |
| 2B | 35 | 18 | 22 | 26 | 27 | 24 | 22 | 13 | 22 | 52 | 20 | 25 |
| 2C | 25 | 31 | 26 | 26 | 27 | 15 | 27 | 29 | 31 | 50 | 27 | 26 |
| 3A | 28 | 27 | 19 | 24 | 16 | 9 | 24 | 18 | 25 | 30 | 22 | 19 |
| 3B | 50c | 42 | 40 | 17 | 23 | 25 | 18 | 12 | 27 | 52 | 20 | 29 |
| 3C | 23 | 25 | 25 | 26 | 27 | 23 | 21 | 21 | 23 | 48 | 20 | 19 |
| 3D | 26 | 26 | 29 | 31 | 31 | 28 | 28 | 28 | 27 | 50 | 29 | 28 |
| 3′ UTR | 28 | 46 | 29 | 35 | 47 | 41 | 52 | 44 | 36 | 47 | 50 | 42 |
Values are percent amino acid identities and percent nucleotide identities (for the 5′ and 3′ UTRs) between 87-012 and the selected picornaviruses. Values were calculated by using the GCG GAP program. ND, not determined.
It should be noted that the true length of the extreme 5′ end has been estimated by Southern blots only.
FMDV protein 3B1 (VPg1) is used for comparison.
5′ UTR.
The 5′ UTR of LV precedes the putative initiation codons, which are located in an optimal Kozak context (ANNAUGG) (29) at positions 732, 734, and 736 in the 145SL, 87-012, and 174F isolates, respectively. The results of BLAST-mediated database searches and RNAGA-mediated secondary-structure predictions supported previous observations (39) that this region includes the type II IRES present in all picornaviruses except entero-, rhino-, and hepatoviruses (62). Specifically, counterparts to the three major 3′-located secondary-structure elements, I, J, and K, of the type II IRES were identified in the LV 5′ UTR (Fig. 1). The apical parts of the I domain, including the prominent GNRA tetranucleotide (GNRA1) and one of two A/C-rich loop regions, and J domain, as well as the A-rich loop at the junction of the J and K domains, were particularly well conserved at the sequence level (Fig. 1 and data not shown). Additionally, an 8-nucleotide oligopyrimidine tract (U/C-rich region) (26) was also recognized 19 nt upstream of the start codon in an atypically well-structured region. Interestingly, all LVs and parechoviruses contain a second copy of the GNRA tetranucleotide (GNRA2), which may be of possible functional significance in the apical 14-nucleotide loop of the J domain. The 5′ half of the 5′ UTR of LVs may also contain stem-loop structures found in other group II picornaviruses, although no equivalent to the element C was identified by our analysis. However, at the sequence level, conservation is limited, with a pentapeptide AAUAA present in a loop of the stem-loop D being the longest region conserved between LVs and parechoviruses. Stem-loop structures A, F, I, and J are supported by compensator mutations found in the three isolates of LV, while the element D may vary in LVs (Fig. 1).
3′ UTR.
The 3′ UTRs of picornaviruses vary in length and are organized in a higher-order RNA structure thought to be involved in RNA replication (19). The two most closely related LV strains, 87-012 and 174F, both have a 111-nucleotide-long 3′ UTR, while strain 145SL has a 4-nucleotide deletion in this region, corresponding to nt 7538 to 7541 in the prototype strain. This region seems to be unique for LVs, although it is predicted to fold into two hairpins (data not shown) structurally similar to those identified in the 3′ UTR of poliovirus and EMCV (1, 9, 48).
LV polyprotein processing.
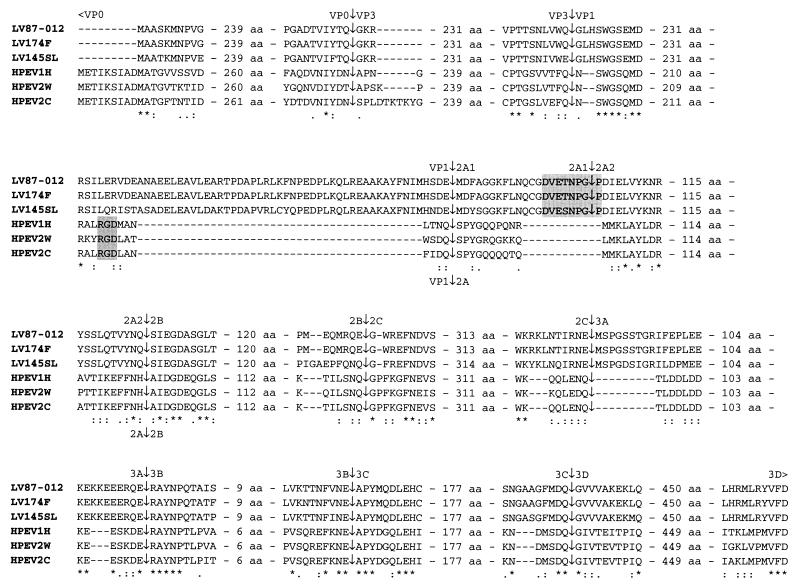
The polyprotein interdomain junctions processed by 3Cpro in all picornaviruses are confined to small regions that contain primary- and tertiary-structure elements recognized by 3Cpro (14, 31, 43, 54). An alignment of the polyproteins of LVs and their closest relatives, parechoviruses, was generated and analyzed for the presence of conserved 3Cpro sequence substrate determinants at interdomain junctions to predict the 3Cpro-mediated processing map of LV polyproteins (Fig. 2). It was noted that two previously determined sites, VP0|VP3 and VP3|VP1, and four predicted sites, 2B|2C, 2C|3A, 3A|3B, and 3C|3D, of parechoviruses (17, 25, 40) are also conserved in LVs (Fig. 2). In contrast, the previously predicted 3Cpro sites at the 2A|2B and 3B|3C junctions in parechoviruses were not conserved in LVs. However, two other dipeptides in these regions, which are conserved in LVs and parechoviruses and resemble 3C cleavage sites elsewhere, could actually be processed in these viruses (Fig. 2).
FIG. 2.
Comparison of the terminal regions and the predicted cleavage sites of the polyproteins of LV strains 87-012, 174F, and 145SL with those of parechoviruses (HPEV1H, HPEV2W, and HPEV2C). ∗, identical residues; : and ·, strong and weak conserved groups, respectively, of amino acid residues as defined by the Clustal X program (59, 60). The RGD motif in the C-terminal end of VP1 in HPEV is highlighted in the smaller gray box and is located in the last conserved region between HPEV and LV in VP1. The conserved DvExNPG|P motif putatively responsible for the LV 2A1 C-terminal polyprotein processing is highlighted in the larger gray box. Two of the previously predicted cleavage sites for parechoviruses, 2A|2B and 3B|3C, are revised here based on the now available LV amino acid sequences. The numbers of amino acids separating the amino acids shown in detail around the proposed scissile bonds are indicated in the alignments.
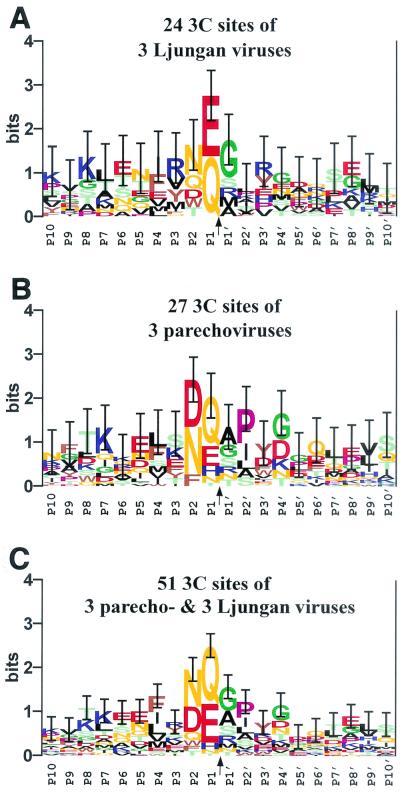
Like 3Cpros of other picornaviruses (4), the LV main protease is predicted to prefer Gln and Glu at the P1 position and a small amino acid residue at the P1′ position, although the latter requirement seems not to be stringent (Fig. 3A). The 3Cpros of parechoviruses also have a similar albeit relaxed specificity with respect to the P1 position (Fig. 3B). These proteases may also have affinity to structurally similar Asn, Gln, and Asp at the P2 position. This specificity is unique among 3Cpros of picornaviruses, although 3C-like proteases of nido- and potyviruses and 2A proteases of entero- and rhinoviruses possess other restricted specificities at the same position (14, 54, 66). When the putative 3Cpro cleavage sites of LVs and parechoviruses were analyzed together, the affinities of the 3Cpros to the bulky hydrophobic amino acid residues at the P4 position became evident (Fig. 3C). This P4 substrate position is also recognized by other 3Cpros, e.g., poliovirus 3Cpro is known for its preference for Ala and Thr residues at this position (4). Comparison of LV- and parechovirus-specific logos (Fig. 3A and B, respectively) suggested that the 3Cpro of parechoviruses, but not of LVs, may also recognize the P2′ substrate position.
FIG. 3.
Conservation of 3Cpro sites in LV and parechovirus polyproteins. Three separate multiple, gap-free 20-aa-long alignments around the P10-P10′ positions of the sites (presumably) cleaved by 3Cpro domains of LVs (A), parechoviruses (B), or both groups of viruses (C) were converted into sequence logos presentations (4, 55). Due to a greater uncertainty of the prediction for the LV VP1|2A1 sites (see the text), they were not included in the analysis. In the logos, the height of each letter (amino acid residue) is proportional to its frequency at the specific position, and the letters are sorted so that the most common residue is on the top of the stack. The height of each stack is proportional to the information content (measured in bits) of the sequences at this position. The upper limit of information at any position (4.32 bits) is determined by the natural diversity of the 20 aa, which is expressed as a logarithm of 20. The most conserved and important positions are relatively high and easily recognized. Vertical bars, whose sizes are reversibly proportional to the sampling size, indicate 1 standard deviation of the information content at each position (55). The letters indicating amino acid residues are colored as follows: light green for S, T, and C; orange for N and Q; red for D and E; blue for K, R, and H; brown for W, F, and Y; black for A, L, I, V, and M; pink for P; and green for G.
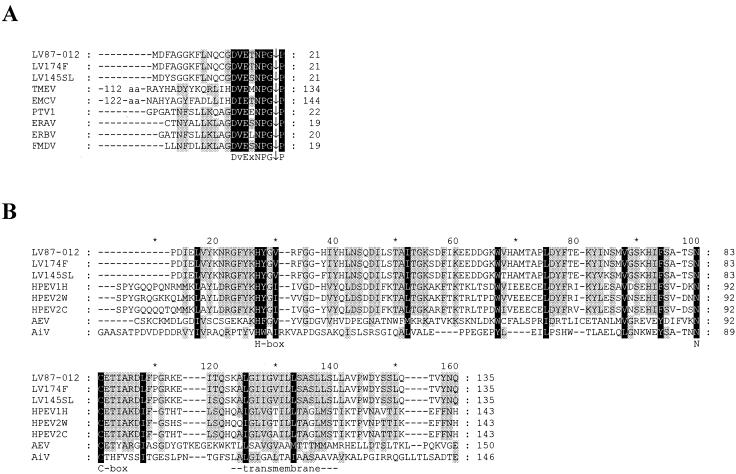
Molecular modeling of polyprotein processing at the LV VP1|2A1 border presented a special challenge. In HPEVs, this junction is likely to be processed by 3Cpro (Fig. 2). The 3C cleavage site separating VP1 and 2A in HPEVs resides in an ∼20-aa sequence corresponding to a larger region in LVs (∼70 aa) lacking reliable similarity (Fig. 2 and data not shown). A separate analysis of this region of LVs revealed that it, surprisingly, contains a homologue of a 17-aa peptide conserved at the 2A|2B junction in cardio-, erbo-, tescho-, and aphthoviruses. The C terminus of this peptide homologue conforms to the signature DvExNPG|P, with the uppercase letters denoting absolutely conserved residues (Fig. 4A) (2, 8, 10, 32, 47, 52, 63, 65). This peptide is sufficient to promote separation of the 2A and 2B moieties at the NPG|P junction in mengovirus and FMDV (11, 12, 21, 45) by a mechanism that is currently the subject of intensive debate (13, 20). In aphthoviruses and probably in erbo- and teschoviruses, this conserved peptide, excluding the most C-terminal Pro residue, is released as a mature 17- to 20-aa-long product through cleavage by 3C at the N terminus and autoprocessing at the C terminus (11). A similar 20-aa-long peptide could be produced in LVs assuming that 3Cpro cleaves at the N-terminal HSDE|M site (amino acid position 800) that conforms to the pattern conserved in other 3Cpro sites of LVs (Fig. 2 and 3A). This model implies that, unlike with all other picornaviruses, including parechoviruses, the LV polyprotein is processed at an extra site in a region between VP1 and 2B which generates two 2A proteins (2A1 and 2A2) rather than one. If LVs actually deviate further from picornaviruses, their 3Cpro might cleave another site (e.g., the FLNQ|C site, 11 aa further downstream [Fig. 2]) that would make the regulation of the expression of this region even more complex.
FIG. 4.
Multiple sequence alignments of 2A1 and 2A2 of LVs and selected picornaviruses. Black and gray backgrounds highlight alignment columns with 100 and 60% conserved residues, respectively, as defined in the GeneDoc default similarity groups (37). (A) Alignment of the LV 2A1 and the C-terminal regions of the picornavirus NPGP 2A protein family. (B) Alignment of the LV 2A2 and the picornavirus H-NC 2A protein family. The C-terminal border of parechovirus 2As is depicted as it was predicted (Fig. 2). The actual boundaries of 2A proteins from AiV and AEV were not defined. The conserved H-box and NC-box and a putative transmembrane domain made up of an approximately 20-aa-long hydrophobic region are labeled. ∗, each noneven count of 10.
The comparisons of polyprotein sequences of LVs and other picornaviruses do not provide positive evidence for the presence of an L peptide at the N terminus or cleavage of VP0 into VP4 and VP2. Overall, the LV polyprotein appears to be processed into 11 mature products by two different proteolytic activities (Fig. 2).
Nonstructural proteins.
LV polyproteins possess a full complement of nonstructural proteins identified previously in parechoviruses (17, 25, 40). With the exception of 2A moieties, all nonstructural proteins are conserved across picornaviruses; however, they are larger in LVs than in parechoviruses, mostly because of extra sequences at one or both termini (Fig. 2). Conserved sequence motifs characteristic of picornavirus 2CATPase, 3BVPg, 3Cpro, and 3Dpol proteins (18) have been identified in the LV homologues without substitutions known to compromise their respective functions (data not shown). Hence, the core enzymatic mechanisms of replication and expression employed by picornaviruses appear to be conserved in LVs.
Likewise, the stringent sequence conservation that is suggestive of functional competence is evident for the 2A moieties. The N-terminal 2A1 is related to the NPGP family of 2As encoded by cardio-, erbo-, tescho-, and aphthoviruses (Fig. 4A) (2, 8, 10, 32, 47, 52, 63, 65), and the C-terminal 2A2 belongs to the H-NC family of 2As encoded by parechoviruses, kobuviruses, and AEVs (Fig. 4B) (25, 34, 64). It is predicted (see above) that 2A1 mediates the separation between 2A1 and 2A2. The function of the picornavirus H-NC 2As is yet to be established, but they, as well as the 2A2 of LVs, may be involved in an aspect of virus-host interaction related to cell growth control (24). An unprecedented association of two structurally different 2As may also result in a unique regulation of the life cycle of LVs. Future comparative studies of parechoviruses and LVs should be especially useful in helping us to understand the unique benefits to a picornavirus associated with a cluster of two 2A proteins.
Capsid proteins.
Alignment of the capsid proteins of LVs and other picornaviruses revealed the conservation of the major elements forming an eight-stranded antiparallel β-barrel structure (jelly roll) in VP0, VP3, and VP1 (53). The most pronounced divergence between LVs and parechoviruses has occurred outside the β-strand elements at the terminal regions that have accepted large insertions and deletions. These include the N termini of the capsid proteins and the C terminus of VP1 (Fig. 2). Compared to parechoviruses, LVs have a 30-aa-shorter N terminus of VP0 that does not contain the myristylation signal GXXX(S/T) (6), which is typical of the majority of picornaviruses. The latter feature puts them in one group with Parechovirus and Hepatovirus, whose virion morphogenesis may not rely upon the myristylation of VP0.
Unlike other picornaviruses, LVs and parechoviruses contain 18-aa-long and 24- to 30-aa-long extensions, respectively, that are enriched with basic residues at the N terminus of VP3 (Fig. 2). This region is immunogenic in parechoviruses (27) but is yet to be characterized in LVs. Furthermore, a functionally uncharacterized 10- to 11-aa insertion was found in the vicinity of the N terminus of VP1 in the LV isolates, while there was a major difference between parechoviruses and LVs at the C terminus of VP1 (Fig. 2). In this region, the parechoviruses contain an Arg-Gly-Asp (RGD) motif important for cell surface interactions (50, 56, 61), while LVs contain a unique 43-aa extension (Fig. 2).
Picornaviruses have evolved three paralogous copies of the jelly roll domain to take advantage of the hetero-oligomer multisubunit organization for virion-associated activities. This evolution was accompanied by a substantial divergence of VP0, VP3, and VP1 as each protein assumed specific functions (53). Surprisingly, LVs and parechoviruses possess atypically high sequence similarity between the paralogous C-terminal regions of VP0 and VP1 encompassing β-strands I (Fig. 5). This conserved region is flanked from the C terminus by a tetrapeptide CysProArgPro in VP0 of LVs. Remarkably, the same peptide is also conserved in the equivalent position of paralogous VP1 of a majority of picornaviruses (Fig. 5) and is considered the most conserved sequence of this protein (44; A. E. Gorbalenya, unpublished observations). These observations indicate that some conserved function(s) may be performed by different capsid proteins in LVs and other picornaviruses.
FIG. 5.
Conservation between the C-terminal regions of VP0 and VP1 of picornaviruses. Multiple alignments of VP0 and VP1 of a set of picornaviruses representing all genera and LVs were generated as described in Materials and Methods. Presented is an alignment of sequences in the vicinity of the β-strands I of VP0 and VP1 split into four virus-protein groups separated by horizontal spaces. The top group comprises VP0 of all picornavirus genera except those of LVs and parechoviruses, which form the second group; the bottom group comprises VP1 of all picornavirus genera except those of LVs and parechoviruses, which form the third group. The amino acid conservation is highlighted separately for the top, bottom, and two middle groups. Black, dark gray (with white letters) and light gray (with black letters) backgrounds highlight alignment columns with 100, 80, and 60% conserved residues, respectively, as defined in the GeneDoc default similarity groups (37). The CysProArgPro (CPRP) tetrapeptide is indicated by italic type. Arrows indicate the position of the scissile bond at the VP0-VP3 junction.
Evolution and taxonomy of LVs.
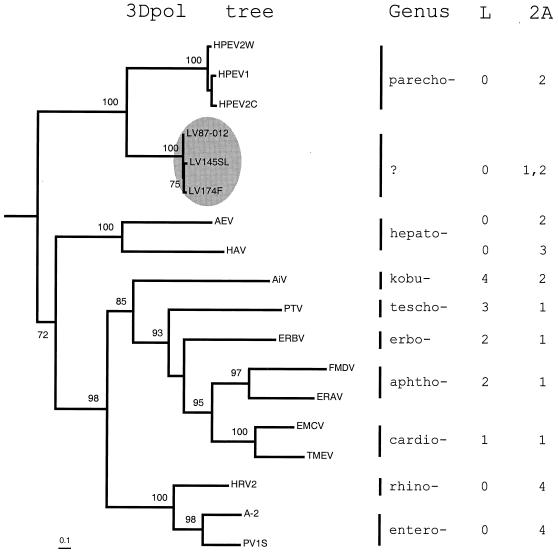
LVs and parechoviruses cluster together over the entire polyprotein (Table 2). To define the position of LVs among picornaviruses more precisely, a phylogenetic analysis of the 3Dpol proteins of LV and representatives of nine Picornaviridae genera was conducted. By using the maximum-likelihood criteria and including the 3D-like sequences of two picornavirus-related insect viruses as an out-group in the analysis, a largely resolved tree was inferred and successfully rooted (Fig. 6). The LVs comprise a compact division that, together with parechoviruses, is separated from other picornaviruses in a minor, poorly populated lineage close to the root of the Picornaviridae tree. The inferred topology of 3Dpol may accurately reflect the evolution of picornavirus genomes, since no evidence for intergenera recombination among picornaviruses has been presented to date.
FIG. 6.
Phylogenetic relationships among picornaviruses. A phylogenetic tree of picornavirus 3Dpol protein sequences (left) representing all genera and LVs was generated by using the Proml program in the PHYLIP 3.6a2 package as described in Materials and Methods. Prior to the tree inference, a likelihood mapping analysis of the 3Dpol alignment showed that the phylogenetic signal could be inferred by using bifurcating trees (data not shown). The tree was rooted by using two 3D-like sequences encoded by InFV and SBV as the out-group. All bifurcations supported in more than 70 of 100 bootstrap trials are labeled. Bar, 0.1 substitutions per site. The LV division is highlighted in the gray oval. The picornavirus genera are listed to the right of the tree. To illustrate the diversity of picornaviruses outside 3Dpol, structural variants of 2A and L proteins were arbitrarily assigned unique numbers, and these are listed along with their respective virus genera (right). The number 0 indicates the lack of L protein. Note that AEV and HAV of hepatoviruses encode different combinations of L and 2A proteins.
It is informative to compare the tree in Fig. 6 with a distribution of different molecular forms of 2A among picornaviruses. In this context, it becomes apparent that contemporary picornaviruses employing either the NPGP or H-NC types of 2A may have originated from the ancestral picornavirus having the LV-like organization of the 2A locus through selective and repeated loss of one of the 2As. Alternatively, the immediate ancestor of LV could have acquired the 2A1 gene from an outside source, although no specific sequence affinity was found between the 2A1 of LVs and any homologue of picornavirus or other origin (11) (data not shown). The generation of the entire molecular diversity of 2A(-like) proteins in picornaviruses and related viruses must have also involved other mechanisms in addition to that proposed above.
Regardless of whether LVs have inherited a cluster of two 2As from either a common picornavirus or an immediate LV ancestor, it is evident for the first time that, functionally, the two structural forms of 2A are not mutually exclusive. This compatibility of different 2As contributes to the evolutionary potential of picornaviruses (19), whose correct assessment may be vital for efforts to control picornavirus infections and could be used in the generation of new virus vectors.
Should a new LV-prototyped genus of the Picornaviridae be created? We are inclined to suggest so given the unique features of the genetic organization of LVs that correlate well with the substantial divergence of LVs from other picornaviruses. If the LV-based genus is established, it would join the majority of picornavirus genera (excluding hepatoviruses), which are characterized by a uniform combination of molecular forms of the L and 2A proteins (Fig. 6).
Overall, the analysis of the newly determined LV genomes presented here demonstrates that the basic picornavirus genetic plan has sufficient plasticity to evolve a cluster of two structurally different 2A proteins. The expression of the LV genomes is predicted to involve 3Cpro with a restricted specificity, which uniquely includes the P2 substrate position. Future studies should determine how these characteristics are translated into the biology of LV. The first step in this direction is to develop a cell culture system to support efficient multicycle production of LV progeny.
Acknowledgments
We are grateful to Anne Andersson, Thomas Elfström, and Viveka Svensson for technical assistance, J.-H. Chen for running the RNAGA program, and Darren Shafren for reviewing the manuscript.
This work was supported by grants from Nya Apodemus AB and the University of Kalmar. A.E.G. was supported with funds from the National Cancer Institute, National Institutes of Health, under contracts NO1-CO-56000 and NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
This paper is dedicated to the memory of Don Summers, a friend and colleague, who will be greatly missed.
REFERENCES
- 1.Auvinen, P., and T. Hyypiä. 1990. Echoviruses include genetically distinct serotypes. J. Gen. Virol. 71:2133-2139. [DOI] [PubMed] [Google Scholar]
- 2.Beard, C. W., and P. W. Mason. 2000. Genetic determinants of altered virulence of Taiwanese foot-and-mouth disease virus. J. Virol. 74:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertioli, D. 1997. Rapid amplification of cDNA ends, p. 233-238. In B. A. White (ed.), PCR cloning protocols: from molecular cloning to genetic engineering, vol. 67. Humana Press Inc., Totowa, N.J.
- 4.Blom, N., J. Hansen, D. Blaas, and S. Brunak. 1996. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 5:2203-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J. H., S. Y. Le, and J. V. Maizel. 2000. Prediction of common secondary structures of RNAs: a genetic algorithm approach. Nucleic Acids Res. 28:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 7.Christian, P., E. Carstens, L. Domier, K. Johnson, N. Nakashima, P. Scotti, and F. van der Wilk. 2000. Cricket paralysis-like viruses, p. 678-683. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 8.Cohen, S. H., R. K. Naviaux, K. M. Vanden Brink, and G. W. Jordan. 1988. Comparison of the nucleotide sequences of diabetogenic and nondiabetogenic encephalomyocarditis virus. Virology 166:603-607. [DOI] [PubMed] [Google Scholar]
- 9.Cui, T., and A. G. Porter. 1995. Localization of binding site for encephalomyocarditis virus RNA polymerase in the 3′-noncoding region of the viral RNA. Nucleic Acids Res. 23:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty, M., D. Todd, N. McFerran, and E. M. Hoey. 1999. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J. Gen. Virol. 80:1929-1941. [DOI] [PubMed] [Google Scholar]
- 11.Donnelly, M. L., L. E. Hughes, G. Luke, H. Mendoza, E. ten Dam, D. Gani, and M. D. Ryan. 2001. The “cleavage” activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring “2A-like” sequences. J. Gen. Virol. 82:1027-1041. [DOI] [PubMed] [Google Scholar]
- 12.Donnelly, M. L., D. Gani, M. Flint, S. Monaghan, and M. D. Ryan. 1997. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J. Gen. Virol. 78:13-21. [DOI] [PubMed] [Google Scholar]
- 13.Donnelly, M. L., G. Luke, A. Mehrotra, X. Li, L. E. Hughes, D. Gani, and M. D. Ryan. 2001. Analysis of the aphthoprotein 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translation effect: a putative ribosomal “skip”. J. Gen. Virol. 82:1013-1025. [DOI] [PubMed] [Google Scholar]
- 14.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumas Milne Edwards, J. B., O. Valdenaire, and J. Mallet. 1997. Anchoring a defined sequence to the 5′ ends of mRNAs, p. 261-278. In B. A. White (ed.), PCR cloning protocols: from molecular cloning to genetic engineering, vol. 67. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 16.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 17.Ghazi, F., P. J. Hughes, T. Hyypiä, and G. Stanway. 1998. Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J. Gen. Virol. 79:2641-2650. [DOI] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., and E. V. Koonin. 1993. Comparative analysis of the amino acid sequences of the key enzymes of the replication and expression of positive-strand RNA viruses. Sov. Sci. Rev. Sect. D 11:1-84. [Google Scholar]
- 19.Gromeier, M., E. Wimmer, and A. E. Gorbalenya. 1999. Genetics, pathogenesis and evolution of picornaviruses, p. 287-343. In E. Domingo, R. Webster, and J. Holland (ed.), Origin and evolution of viruses. Academic Press, San Diego, Calif.
- 20.Hahn, H., and A. C. Palmenberg. 2001. Deletion mapping of the encephalomyocarditis virus primary cleavage site. J. Virol. 75:7215-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn, H., and A. C. Palmenberg. 1996. Mutational analysis of the encephalomyocarditis virus primary cleavage. J. Virol. 70:6870-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henikoff, S., and J. G. Henikoff. 1994. Position-based sequence weights. J. Mol. Biol. 243:574-578. [DOI] [PubMed] [Google Scholar]
- 23.Hollinger, F. B., and S. U. Emerson. 2001. Hepatitis A virus, p. 799-940. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Matrin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Hughes, P. J., and G. Stanway. 2000. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 81:201-207. [DOI] [PubMed] [Google Scholar]
- 25.Hyypiä, T., C. Horsnell, M. Maaronen, M. Kahn, N. Kalkkinen, P. Auvinen, L. Kinnunen, and G. Stanway. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. USA 89:8847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang, S. K., and E. Wimmer. 1990. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 4:1560-1572. [DOI] [PubMed] [Google Scholar]
- 27.Joki-Korpela, P., M. Roivainen, H. Lankinen, T. Pöyry, and T. Hyypiä. 2000. Antigenic properties of human parechovirus 1. J. Gen. Virol. 81:1709-1718. [DOI] [PubMed] [Google Scholar]
- 28.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-678. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 29.Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283-292. [DOI] [PubMed] [Google Scholar]
- 30.Kräusslich, H. G., M. J. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kräusslich, H. G., and E. Wimmer. 1988. Viral proteinases. Annu. Rev. Biochem. 57:701-754. [DOI] [PubMed] [Google Scholar]
- 32.Li, F., G. F. Browning, M. J. Studdert, and B. S. Crabb. 1996. Equine rhinovirus 1 is more closely related to foot-and-mouth disease virus than to other picornaviruses. Proc. Natl. Acad. Sci. USA 93:990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindberg, A. M., C. Polacek, and S. Johansson. 1997. Amplification and cloning of complete enterovirus genomes by long distance PCR. J. Virol. Methods 65:191-199. [DOI] [PubMed] [Google Scholar]
- 34.Marvil, P., N. J. Knowles, A. P. Mockett, P. Britton, T. D. Brown, and D. Cavanagh. 1999. Avian encephalomyelitis virus is a picornavirus and is most closely related to hepatitis A virus. J. Gen. Virol. 80:653-662. [DOI] [PubMed] [Google Scholar]
- 35.Melnick, J. L., V. I. Agol, H. L. Bachrach, F. Brown, P. D. Cooper, W. Fiers, S. Gard, J. H. Gear, Y. Ghendon, L. Kasza, M. LaPlaca, B. Mandel, S. McGregor, S. B. Mohanty, G. Plummer, R. R. Rueckert, F. L. Schaffer, I. Tagaya, D. A. Tyrrell, M. Voroshilova, and H. A. Wenner. 1974. Picornaviridae. Intervirology 4:303-316. [DOI] [PubMed] [Google Scholar]
- 36.Morgenstern, B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15:211-218. [DOI] [PubMed] [Google Scholar]
- 37.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNET News 4:1-4. [Google Scholar]
- 38.Niklasson, B., B. Hörnfeldt, and B. Lundman. 1998. Could myocarditis, insulin dependent diabetes mellitus, and Guillain-Barré syndrome be caused by one or more infectious agents carried by rodents? Emerg. Infect. Dis. 4:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niklasson, B., L. Kinnunen, B. Hörnfeldt, J. Hörling, C. Benemar, K. O. Hedlund, L. Matskova, T. Hyypiä, and G. Winberg. 1999. A new picornavirus isolated from bank voles (Clethrionomys glareolus). Virology 255:86-93. [DOI] [PubMed] [Google Scholar]
- 40.Oberste, M. S., K. Maher, and M. A. Pallansch. 1998. Complete sequence of echovirus 23 and its relationship to echovirus 22 and other human enteroviruses. Virus Res. 56:217-223. [DOI] [PubMed] [Google Scholar]
- 41.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 42.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses, p. 723-776. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Matrin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Palmenberg, A. C. 1990. Proteolytic processing of picornaviral protein. Annu. Rev. Microbiol. 44:603-623. [DOI] [PubMed] [Google Scholar]
- 44.Palmenberg, A. C. 1989. Sequence alignment of picornaviral capsid proteins, p. 211-241. In B. L. Semler and E. Ehrenfeld (ed.), Molecular aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 45.Palmenberg, A. C., G. D. Parks, D. J. Hall, R. H. Ingraham, T. W. Seng, and P. V. Pallai. 1992. Proteolytic processing of the cardioviral P2 region: primary 2A/2B cleavage in clone-derived precursors. Virology 190:754-762. [DOI] [PubMed] [Google Scholar]
- 46.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 47.Pevear, D. C., J. Borkowski, M. Calenoff, C. K. Oh, B. Ostrowski, and H. L. Lipton. 1988. Insights into Theiler's virus neurovirulence based on a genomic comparison of the neurovirulent GDVII and less virulent BeAn strains. Virology 165:1-12. [DOI] [PubMed] [Google Scholar]
- 48.Pöyry, T., L. Kinnunen, T. Hyypiä, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 49.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 50.Pulli, T., E. Koivunen, and T. Hyypiä. 1997. Cell-surface interactions of echovirus 22. J. Biol. Chem. 272:21176-21180. [DOI] [PubMed] [Google Scholar]
- 51.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Matrin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 52.Robertson, B. H., M. J. Grubman, G. N. Weddell, D. M. Moore, J. D. Welsh, T. Fischer, D. J. Dowbenko, D. G. Yansura, B. Small, and D. G. Kleid. 1985. Nucleotide and amino acid sequence coding for polypeptides of foot-and-mouth disease virus type A12. J. Virol. 54:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossmann, M. G., and J. E. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533-573. [DOI] [PubMed] [Google Scholar]
- 54.Ryan, M. D., and M. Flint. 1997. Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 78:699-723. [DOI] [PubMed] [Google Scholar]
- 55.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanway, G., N. Kalkkinen, M. Roivainen, F. Ghazi, M. Khan, M. Smyth, O. Meurman, and T. Hyypiä. 1994. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J. Virol. 68:8232-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 59.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Triantafilou, K., M. Triantafilou, Y. Takada, and N. Fernandez. 2000. Human parechovirus 1 utilizes integrins αvβ3 and αvβ1 as receptors. J. Virol. 74:5856-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wimmer, E., C. U. T. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 63.Wutz, G., H. Auer, N. Nowotny, B. Grosse, T. Skern, and E. Kuechler. 1996. Equine rhinovirus serotypes 1 and 2: relationship to each other and to aphthoviruses and cardioviruses. J. Gen. Virol. 77:1719-1730. [DOI] [PubMed] [Google Scholar]
- 64.Yamashita, T., K. Sakae, H. Tsuzuki, Y. Suzuki, N. Ishikawa, N. Takeda, T. Miyamura, and S. Yamazaki. 1998. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 72:8408-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zell, R., M. Dauber, A. Krumbholz, A. Henke, E. Birch-Hirschfeld, A. Stelzner, D. Parager, and R. Wurm. 2001. Porcine teschoviruses comprise at least eleven distinct serotypes: molecular and evolutionary aspects. J. Virol. 75:1620-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 67.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.