Abstract
To examine whether the exonuclease activity intrinsic to the polymerase (Pol) of herpes simplex virus type 1 can influence the mutational spectra, we applied the denaturing gradient gel electrophoresis (DGGE) system combined with sequencing to characterize thymidine kinase mutants isolated from both the wild-type virus and a mutant deficient in exonuclease activity, Y7. Wild-type viruses produced predominately frameshift mutations (67%), whereas Y7 replicated a significantly lower proportion of frameshifts (21%; P < 0.005). Furthermore, the majority of substitutions were transitional changes in both groups, although they distributed differently. The implications of these findings are discussed.
The fidelity of DNA replication is regulated by several different mechanisms (reviewed in reference 14 and references therein). In particular, the selection of correct nucleotides by DNA polymerase (Pol) serves a major role in preserving the integrity of the genome. Furthermore, the excision of misinserted bases by the exonucleolytic proofreading activity of the Pol also directly contributes to the high level of fidelity. While exonucleolytic proofreading is important for the integrity of the genetic materials, its association with most Pols adds the complexity of the mechanisms by which Pol regulates the accuracy of nucleotide incorporation. For example, the polymerization reaction acts to extend the nascent primer strand in the 5′-to-3′ direction, whereas the exonuclease activity reacts in an opposite direction to remove the newly incorporated nucleotide. Although it is known that each polymerization involves multistep reactions to achieve Pol selectivity against insertion of incorrect nucleotides, the contribution of each individual step to the fidelity requires further detailed characterizations. Exonucleolytic proofreading contributes on average about 100-fold to fidelity. However, the contribution of exonuclease activity to base substitution fidelity can vary over a wide range, and the degree of its contribution is enzyme and sequence specific. This is based on the studies demonstrating that the “frayed” single-stranded primer due to a terminal mispair will preferentially bind to the exonuclease site and that the double-stranded terminus will preferentially bind to the Pol active site. Therefore, the stability of the duplex region of the template-primer will depend on its DNA sequence, and proofreading efficiency will vary in different sequence contexts. Furthermore, the degree of fraying required for single-stranded DNA to bind to the exonuclease active site may vary, depending on the distance between the Pol and exonuclease active sites. Therefore, it is also enzyme specific (for details see review in reference 19).
Use of the herpes simplex virus (HSV) is advantageous, given that its genome can be altered for biological, biochemical, and genetic studies in vitro and in vivo. We previously engineered two exonucleolytic-proofreading-deficient HSV type 1 (HSV-1) recombinants, Y7 and YD12, which are nonetheless competent in replication in cell cultures. These recombinants, as expected, are highly mutagenic (9), making them useful for studying the contribution of exonuclease activity to the fidelity of DNA replication. To examine whether exonuclease proofreading has effects on the mutation spectra, the thymidine kinase (TK) mutagenesis assay was applied to isolate drug-resistant tk mutants from both the wild-type strain KOS and the Y7 recombinant (6, 7, 9), and mutants were subjected to detailed characterizations for the purpose of identifying mutated nucleotide(s) within each tk mutant.
We applied the denaturing gradient gel electrophoresis (DGGE) technique (17) to screen for mutation(s) in the PCR-amplified DNA fragments of the tk gene. Subsequent sequencing of DNA fragments containing mutation(s) enabled the identification of altered bases in mutated tk genes. Results demonstrated that the HSV-1 Pol could be error prone in both base substitutions and frameshift mutations. However, frameshifts dominated in KOS-induced mutants (67%), whereas such mutations were found in a lower percentage (21%) of Y7-induced mutations. Furthermore, Y7 also induced different types of base substitutions between the tk and supF genes (10). This supports the belief that exonuclease proofreading is sequence dependent (reviewed in reference 19). Moreover, most KOS-induced substitutions were not found in Y7-induced mutants, thereby refuting the assumption that most substitutions observed in KOS mutants would also be isolated in Y7-induced mutants, if Y7 Pol is defective only in proofreading activity. Nevertheless, this suggests that Y7 Pol may also exhibit altered Pol activity, consistent with our previous finding that Y7 exhibits altered binding affinities to certain nucleoside analogs (12). Finally, the HSV-1 Pol might evolve base substitution mainly by misinsertion during DNA chain elongation.
Y7 is highly mutagenic in the tk gene.
We prepared 66 independent virus stocks each for wild-type KOS strain and its derivative Y7 recombinant, which conferred defective exonuclease-proofreading activity (9). Each viral stock was prepared by inoculating less than 50 PFU of the virus into 105 Vero cells. The low virus input is critical for excluding the potential of inoculating preexisting tk mutant(s), which would create biased mutagenesis data (6). The TK mutagenesis assay was then performed to isolate tk mutants by using either 10 μM ganciclovir (Roche) as previously described (6, 7, 9). Consistent with our previous reports, Y7 was highly mutagenic in the tk gene, with an average mutation frequency of 4 × 10−2, which is about 700-fold higher than the average mutation frequency of 6 × 10−5 exhibited by the KOS strain.
Characterization of tk mutants by DGGE and sequencing.
At the beginning we characterized 15 tk mutants by either directly cloning the mutated tk genes into plasmid vector pGEM3Zf(+) as described previously (8) or by PCR amplification of mutated tk genes for subsequent sequencing analysis as described previously (7). Although these traditional approaches are the preferred methods for identifying mutation(s) in the tk gene, they are too labor intensive and cumbersome. Therefore, we established the DGGE technique (17) to extend our ability to characterize a larger number of tk mutants. This method was then applied to analyze 57 KOS and 52 Y7 mutants, respectively. For statistical reasons, 9 KOS and 15 Y7 mutants characterized by traditional approaches were also included in this report. Thus, a total of 66 mutants each from KOS and Y7 were reported in this study.
The DGGE technique has proven to be very sensitive in detecting mutation(s) in PCR fragments with the size varying from 300 to 500 bp (1). For analyzing the tk gene, the established protocol was applied (17). Briefly, four overlapping DNA fragments, covering the entire tk open reading frame, were PCR amplified from crude viral DNA (8). Each PCR product was mixed with a corresponding DNA fragment of the wild-type product, denatured, and annealed to form DNA heteroduplexes. Samples were then loaded onto DGGE parallel gels containing 8% polyacrylamide and a defined gradient of denaturants. The gels were electrophoresed at 150 V at 60°C in 0.5× Tris-acetate-EDTA buffer for various times to obtain optimal resolution. DNA samples showing the heteroduplexes, which would indicate the presence of mutated nucleotide(s), were then sequenced as described earlier (8).
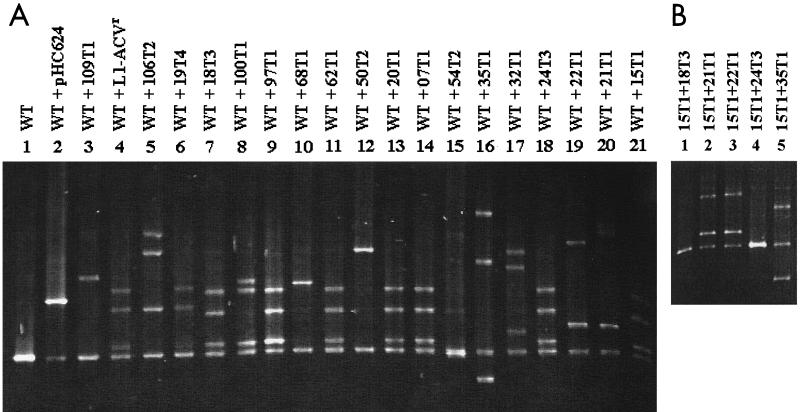
A DGGE gel showing 20 samples of PCR fragment II heteroduplexes is presented in Fig. 1A. This denaturing gradient gel contained 50 to 65% of denaturants and was electrophoresed for 7 h. Of the 20 tk mutants, two tk mutants, 32T1 (Fig. 1A, lane 17) and 35T1 (Fig. 1A, lane 16), and one plasmid clone, pHC624 (Fig. 1A, lane 2), had been previously characterized (Table 2 in reference 17). It was clear that each mutant formed heteroduplex DNA with altered mobilities, relative to the homoduplex sample of pSVTK1 (Fig. 1A, lane 1). It was also obvious that the different mobilities of the heteroduplexes were a direct consequence of different mutation(s). Among these 20 samples, seven (Fig. 1A, lanes 7, 9, 11, 13, 14, 18, and 21) exhibited identical DGGE profiles, and sequencing subsequently confirmed them to contain the same mutation (see below). Similarly, two other samples (Fig. 1A, lanes 4 and 6) that revealed identical DGGE profiles were shown to have the same mutation. All others shown in Fig. 1A revealed different DGGE profiles and contained different mutations.
FIG. 1.
DGGE analysis. (A) Heteroduplex formation of DNA fragment II of mutant tk genes derived from Y7. The PCR products of DNA fragment II of the tk gene amplified from pSVTK1 (lane 1) were used as the wild-type (WT) control and were mixed with PCR samples amplified from cloned DNA of pHC624 (lane 2) and from DNA prepared from indicated virus-infected cells for analyses of heteroduplex formations (lanes 3 to 21). Lanes 7, 9, 11, 13, 14, 18, and 21 have an identical DGGE profile. Lanes 4 and 6 also have identical profiles. (B) Heteroduplex analyses of DNA fragment II of various tk genes. The PCR product of sample 15T1 was mixed with other PCR samples, as indicated, for heteroduplex analyses. Only one homoduplex band is found in lanes 1 and 4, whereas heteroduplex bands are revealed in lanes 2, 3, and 5.
TABLE 2.
Classification of substituted bases
| Type of substitution | No. of substitutions (%) derived from:
|
|
|---|---|---|
| KOS (n = 22) | Y7 (n = 64) | |
| Transition | 17 (77) | 40 (63) (P > 0.05) |
| G:C to A:T | 15 (68) | 28 (44) |
| A:T to G:C | 2 (9) | 12 (18.8) |
| Transversion | 5 (23) | 24 (37) (P > 0.05) |
| G:C to T:A | 3 (14) | 18 (28) |
| G:C to C:G | 0 | 4 (6) |
| A:T to T:A | 2 (9) | 0 |
| A:T to C:G | 0 | 2 (3) |
To demonstrate that samples showing identical DGGE profiles did indeed contain identical mutations, a sample of 15T1 was separately mixed with five different samples and was analyzed for heteroduplex formation. Figure 1B shows that sample 15T1 could only form a homoduplex band with two different samples (Fig. 1B, lanes 1 and 4), whereas it formed heteroduplexes with the three other samples (Fig. 1B, lanes 2, 3, and 5). Subsequent sequencing of all five samples confirmed mutation profiles, consistent with DGGE results.
It was notable from our previous report (17) that the plasmid pHC624 contained double mutations in fragment II and that one of the changes (G to A at position 370) was also found in all Y7 mutants but not in KOS mutants. However, this mutation did not have the drug-resistant phenotype, since the recombinant virus containing only this mutation exhibited ganciclovir sensitivity similar to that of the TK-positive KOS virus (Q. Lu and C. Hwang, unpublished data). Thus, this mutation presumably evolved prior to the establishment of Y7 viral stocks. Therefore, this mutation was excluded from statistical analysis in this study. Nevertheless, results demonstrate that DGGE is highly sensitive in detecting mutations in the DNA fragments of interest.
Among 52 Y7 tk mutants analyzed by DGGE, 14 different DGGE profiles of the DNA fragment II were observed. Sequencing analysis revealed that each of these mutants contained different mutations. In addition, mutants with identical DGGE profiles were randomly selected and sequenced to confirm that these mutants indeed contained the same type of mutation(s). Therefore, only 14 samples of fragment II were required for the identification of the mutations via the sequencing technique.
To identify the mutations within fragments I, III, and IV, DGGE gels were also applied as described above, except that a 55 to 75% gradient of denaturants and an 8-h electrophoresis were used for analyzing fragments I and III. With this application, samples showing heteroduplex profiles in DNA fragments I, III, and IV were identified and classified into 9, 23, and 5 groups, respectively. This approach simplified the subsequent sequencing of only 37 samples of these fragments. Again several DNA fragments were randomly chosen and sequenced to demonstrate the absence of mutations within samples failing to show heteroduplex formations. Therefore, the application of DGGE provides a rapid screening for mutations within defined DNA fragments and avoids unnecessary sequencing analysis.
The DGGE system was also applied to examine 57 KOS mutants. Results demonstrated that five, five, six, and two types of mutations were present in DNA fragments I, II, III, and IV, respectively. Thus, only 18 different DNA fragments were sequenced to discern the altered sequences within 57 tk mutants. A total of 66 tk mutants derived from the KOS strain, including nine independent KOS mutants analyzed by the traditional approaches (7), are reported here.
It was also notable that most mutations located in the overlapping regions of different fragments were consistently identified in both fragments by DGGE, with the exception that mutations located immediately next to the GC-clamping primer failed to reveal heteroduplex formation, presumably due to the relatively high melting characteristics of sequences adjacent to the GC clamp. In summary, application of the DGGE technique can dramatically reduce the effort of sequencing the entire tk gene of each mutant; in this study, only 51 Y7 and 18 KOS DNA fragments required sequencing, which is an amount of labor equivalent to less than one-sixth of that dictated by traditional approaches.
Y7 induces altered spectra of tk mutations.
After DGGE analysis, DNA fragments displaying altered and unique DGGE profiles were sequenced to identify mutation(s). Table 1 shows the different types of mutations mediated by KOS and Y7 Pols. The KOS strain predominantly induced frameshift mutations (67%). In contrast, only 21% (P < 0.005) of tk mutants replicated by Y7 Pol were frameshifts (Table 1). Each frameshift mutation contained an insertion or deletion of a G:C base pair at repeating runs of G:C sequences, with the exception of two mutants induced by KOS virus: one with an insertion of 2 G's at the iterated 7-G sequences and the other with a deletion of one A within a repeat of 4 A's at nucleotides 184 to 187. In addition to the differences in the types of mutations, 11 Y7 mutants (17%) contained 2 or more substituted bases, whereas only one KOS mutant contained a substituted base and a deletion of the C:G base pair in repeat sequences (Table 1).
TABLE 1.
Classification of mutants
| Type of mutation | No. (%) of virus samples that induced mutation
|
|
|---|---|---|
| KOS (n = 66)a | Y7 (n = 66) | |
| Base substitution | 22 (33.3) | 52 (78.8) (P < 0.005) |
| Single change | 22 | 41 |
| Double changes | 0 | 10 |
| Triple changes | 0 | 1 |
| Frameshift | 45 (66.6) | 14 (21.2) (P < 0.005) |
| Insertion of G or C | 30b | 9 |
| Deletion of G or C | 14 | 5 |
| Deletion of A or T | 1 | 0 |
One mutant derived from KOS strain contains a base substitution and frameshift mutation in fragments II and III, respectively.\
One mutant contains an insertion of two G's at sequences of seven G's.
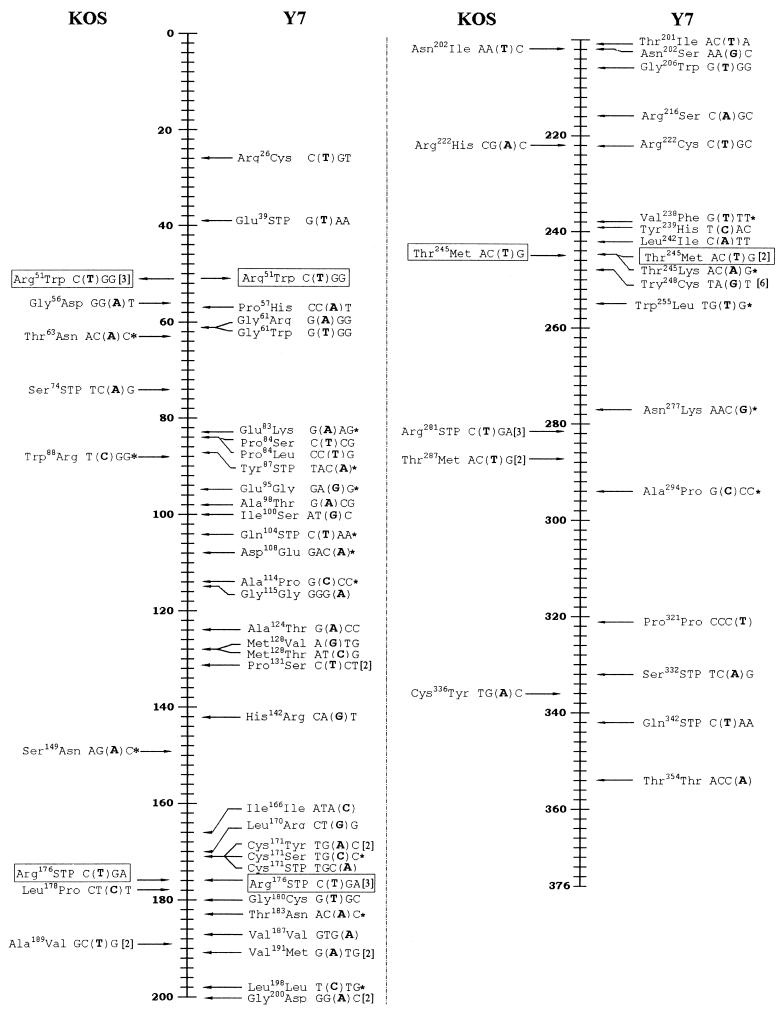
Assuming that Y7 Pol is defective only in exonuclease activity and retains the wild-type Pol activity, we expect most, if not all, misinserted bases found in KOS mutants to be present in Y7 mutants. Comparison of substituted bases induced by both the wild type and Y7 identified only three common substitutions (Fig. 2). Furthermore, KOS induced an AAC-to-ATC change (coding for Asn202) and a CGC-to-AGC change (coding for Arg222), whereas Y7 induced different substitutions at the corresponding residues (AAC to AGC and CGC to TGC). Although the lack of sufficient common mutations could be due to the limited number of samples analyzed, the observation that most Y7-induced substitutions are Y7 specific suggested that Y7 Pol might exhibit an altered Pol activity in addition to the loss of its exonuclease activity. Consistent with this proposition is the observation that the Y7 recombinant exhibited different binding affinities to certain nucleoside analogs (12). However, the extents of the altered activity contributing to the accuracy of Pol's activity and whether it is essential for retaining Y7 Pol's activity in vivo require further study. Nevertheless, wild-type and Y7 Pols can replicate tk genes with different mutation spectra, especially in the types and distributions of mutations.
FIG. 2.
Distribution of substituted bases replicated by wild-type (KOS) and Y7 Pol. Vertical lines depict the residues of the TK polypeptide of HSV-1. Relative positions of substituted bases and the resulting change of the coding residues are shown. The boldfaced nucleotide in parentheses refers to the substituted base from its 5′ adjacent nucleotide. The number shown in bracket refers to the number of mutants containing the same mutation. STP, stop codon; mutation marked in box, common change in both group; ∗, “dislocation” mechanism applicable.
Transitions are the major types of substitutions.
Table 2 summarizes the types of substitutions induced by both wild-type and Y7 Pols. Seventeen out of 22 (77%) substitutions mediated by the wild-type Pol were transitions, including 15 G:C-to-A:T and 2 A:T-to-G:C changes (Table 2). Sixty-three percent of Y7-induced substitutions were transitions, including 44% of G:C-to-A:T and 19% of A:T-to-G:C mutations (Table 2). Therefore, the majority of point mutations found in the tk gene induced by HSV-1 Pols were transitions, although Y7 induced a substantial number of G:C-to-T:A transversions (28%) (Table 2). This result was different from that of our previous study, which demonstrated that both KOS and Y7 viruses induced primarily transversions in the supF gene in a transient DNA replication assay (Table 3 in reference 10). Therefore, the Y7 viruses replicate in infected cells with different mutation frequencies and spectra within the tk and supF genes (9, 10; this study), supporting the belief that exonuclease proofreading is sequence dependent (reviewed in reference 19). Therefore, like other Pols (reviewed in reference 5), the DNA replication fidelity of HSV-1 Pol can be dependent on the target gene, presumably due to the differences in sequence contexts. However, it is possible that the tk mutagenesis assay is not sensitive enough to detect all tk mutations, including silent mutations and certain missense mutations without altered TK activity.
We estimate that more than 56% of possible tk mutations can escape detection by the tk mutagenesis assay (11), based on the analysis of possible silent and missense mutations within 33 bases coding for amino acid residues 165 to 175, which form part of the nucleoside-binding site of the TK enzyme (3). Of 99 potential changes within these 33 bases, 34 are silent. Furthermore, these 11 residues can be replaced with amino acids having similar physical properties without losing TK activity (18). Assuming that each mutant contained only a single replaced base, 22 possible missense mutations within these 11 residues would be TK positive and would evade detection by TK mutagenesis screening. Thus, a total of 56 out of 99 possible mutations (56%) within this region are invisible to this assay. Moreover, the overall ratio of undetectable mutations within the entire tk gene is predicted to be higher, since other regions of TK may exhibit a much higher plasticity for substituted amino acids without altering TK activity. The supF mutagenesis, on the other hand, has been used to successfully isolate 96% of 245 (3 × 85 bases) possible mutations (4). Thus, the supF mutagenesis is much more sensitive than the TK mutagenesis, even though the difference between their replication modes (episomal verse genomic) may also affect the outcomes of DNA replication fidelity (10, 11). Nevertheless, the observed differences of mutational spectra induced by HSV-1 Pol between the supF and tk genes may be partially attributed to their different sequence contexts.
Mechanisms leading to mutations.
It has been suggested that the “template-primer slippage” mechanism (20, 21; reviewed in reference 14 and references therein) is responsible for the formation of frameshift changes, most commonly found in sequences containing homopolymeric nucleotides. The frequency of frameshift mutations due to template-primer slippage is also dependent on the length of repeating bases in that a sequence composed of longer repetitive nucleotides has a higher frequency of frameshift mutations. Consistent with this model, the majority of frameshift mutations (29 KOS mutants and 6 Y7 mutants) contained the insertion or deletion of G:C base pair(s) in a region containing 7 repetitive G nucleotides (coding for amino acids 144 to 146); 14 KOS mutants and 1 Y7 mutant contained an altered number of C:G base pairs within the 6-C region (amino acids 183 to 185); one KOS mutant contained a deletion of a C:G base pair within the 5-C sequence (amino acids 299 and 300); one Y7 mutant contained an inserted G:C base pair within the 5-G region (amino acids 206 and 207); one Y7 mutant contained an inserted G:C base pair within the 3-G sequence (amino acid 72); and one Y7 mutant contained a deletion of a C:G base pair within the 3-C region (amino acid 82). Additionally, one KOS mutant contained an A:T base pair deletion within the 4-A region (amino acids 62 and 63). Thus, similar to the behavior observed in other Pols, the longer the sequence of repetitive nucleotides, the higher the frequency of frameshift mutations (reviewed in reference 14). These results may also imply that HSV-1 Pol is error prone in the formation of misaligned intermediates in sequences containing three or more repeated nucleotides and subsequently allows the development of frameshift changes. While misaligned primer-templates may be corrected by the proofreading activity of Pol, the lack of exonuclease capability in Y7 Pol may be unable to excise such a misaligned primer-template containing only three repeating nucleotides. This is consistent with the finding that a Pol with defective exonuclease activity has more ability to form frameshifting within a short run of repetitive nucleotides than does the exonuclease-proficient Pol (16).
Three mechanisms can lead to base substitutions: misinsertion of an incorrect nucleotide, dislocation mutagenesis (2, 15; reviewed in reference 14 and references therein), and spontaneous deamination (13). Among these three mechanisms, we favor the misinsertion mechanism, since the other two cannot satisfactorily explain the formation of the substitutions observed in this study. Perhaps the HSV-1 Pol has an intrinsic activity to insert incorrect bases and subsequently proofread certain errors, allowing for evolutional events. The lack of proofreading, therefore, can lead to an increased frequency of wrongly inserted bases. However, Y7 Pol may also have acquired altered Pol activity, leading to additional differences in the spectra and distributions of substitutions (Table 2 and Fig. 2).
“Dislocation” mutagenesis (2, 15) has been proposed as an important mechanism in the formation of base substitutions mediated by various Pols. In this model, dislocation between the primer and template strands will result in the formation of an unpaired base associated with the misalignment between the primer and template; the subsequent incorporation of correct nucleotides from the misaligned primer-template followed by a realignment will form a mispairing at the 3′ end of the primer; a continuous incorporation will result in the fixing of the mispaired nucleotide, resulting in base substitution. The nucleotide changed by this mechanism would be mutated to the base identical to its adjacent 5′ or 3′ nucleotide, depending on the dislocated base on the primer or template strand. Although this mechanism explains the formation of some (5 out of 22 KOS-induced substitutions and 16 out of 64 Y7-induced substitutions) substitutions (Fig. 2), it cannot account for all the other observed substitutions.
The deamination reaction can exclusively induce transitions (13). If the transitions observed in this study were the results of spontaneous deamination, we would expect to see similar distributions of transitions induced by both wild-type and Y7 Pols. This, however, is not the case. Indeed, only three common substitutions are found in both groups (Fig. 2). Furthermore, about one-quarter and one-third of substituions induced by the wild type and Y7, respectively, were transversions, which could not be formed by deaminations. Therefore, it is more likely that most of the base substitutions are the results of misincorporations.
The lack of sufficient common mutations between the two groups may also imply that the Y7 Pol has acquired a structural change at the microscopic level due to the mutation introduced within the Exo III motif (9). Such a change would further result in altered kinetics, which would then contribute to the induction of different spectra of mutations. However, we cannot rule out the possibility that the high mutagenicity of the Y7 recombinant may have caused the evolution of further mutations contributing to the differences in mutation spectra. Indeed, additional change(s) in pol and other genes have been identified in several progeny of Y7 recombinants (Q. Lu and C. Hwang, unpublished data). The effects of these mutations are presently under investigation. Furthermore, other proteins of the replication machinery and the postreplication repair system may also influence the replication fidelity and will require further study.
In summary, we applied the DGGE system to screen for the presence of mutations in tk fragments for the purpose of identifying mutated bases. Results from this study demonstrated that HSV-1 Pol's fidelity is target gene dependent. Furthermore, the exonuclease-deficient Y7 Pol replicates altered spectra of tk mutations, which can be attributed to a combination of the lack of proofreading and associated change(s) of the polymerization activity. To examine the specifics of HSV-1 Pol's exonuclease activity at the biochemical and molecular levels, it will be necessary to perform in vitro replication fidelity studies and kinetic examinations of the HSV-1 Pol and to acquire Pol mutants defective only in exonuclease activity.
Acknowledgments
We thank Roche for providing the ganciclovir. We are grateful to S. Kummur and J. Cuyl for critically reading the manuscript.
This study was supported by NIH grant DE 10051.
REFERENCES
- 1.Abrams, E. S., and V. P. Stanton, Jr. 1992. Use of denaturing gradient gel electrophoresis to study conformational transition in nucleic acids. Methods Enzymol. 212:71-104. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. [PubMed] [Google Scholar]
- 3.Brown, D. G., R. Visse, G. Sandhu, A. Davies, P. J. Rizkallah, C. Melitz, W. C. Summers, and M. R. Sanderson. 1995. Crystal structures of the thymidine kinase from herpes simplex virus type-1 in complex with deoxythymidine and ganciclovir. Nat. Struct. Biol. 2:876-881. [DOI] [PubMed] [Google Scholar]
- 4.Canella, K. A., and M. M. Seidman. 2000. Mutation spectra in SupF: approaches to elucidating sequence context effects. Mutat. Res. 450:61-73. [DOI] [PubMed] [Google Scholar]
- 5.Goodman, M. F., S. Creighton, L. B. Bloom, and J. Petruska. 1993. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28:83-126. [DOI] [PubMed] [Google Scholar]
- 6.Hall, J. D., D. M. Coen, B. L. Fisher, M. Weisslitz, S. Randall, R. E. Almy, P. T. Gelep, and P. A. Schaffer. 1984. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology 132:26-37. [DOI] [PubMed] [Google Scholar]
- 7.Hwang, C. B. C., and H. J. Chen. 1995. An altered spectrum of herpes simplex virus mutations mediated by an antimutator DNA polymerase. Gene 152:191-193. [DOI] [PubMed] [Google Scholar]
- 8.Hwang, C. B. C., B. Horsburgh, E. Pelosi, S. Roberts, P. Digard, and D. M. Coen. 1994. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc. Natl. Acad. Sci. USA 91:5461-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang, Y. T., B. Y. Liu, D. M. Coen, and C. B. C. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang, Y. T., B.-Y. Liu, C.-Y. Hong, E. J. Shillitoe, and C. B. C. Hwang. 1999. Effects of exonuclease activity and nucleotide selectivity of the herpes simplex virus DNA polymerase on the fidelity of DNA replication in vivo. J. Virol. 73:5326-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang, Y. T., B.-Y. Liu, and C. B. C. Hwang. 2002. Replication fidelity of the supF gene integrated in the thymidine kinase locus of herpes simplex virus type 1. J. Virol. 76:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang, Y. T., J. F. Smith, L. Gao, and C. B. C. Hwang,. 1998. Mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene can confer altered drug sensitivities. Virology 246:298-305. [DOI] [PubMed] [Google Scholar]
- 13.Kornberg, A., and T. Barker. 1992. DNA replication, 2nd ed., p. 772. W. H. Freeman & Co., New York, N.Y.
- 14.Kunkel, T. A., and K. Bebenek. 2000. DNA replication fidelity. Annu. Rev. Biochem. 69:497-529. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel, T. A., and P. S. Alexander. 1986. The base substitution fidelity of eukaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J. Biol. Chem. 261:160-166. [PubMed] [Google Scholar]
- 16.Longley, M. J., D. Nguyen, T. A. Kunkel, and W. C. Copeland. 2001. The fidelity of human DNA polymerase gamma with and without exonuclease proofreading and the p55 accessory subunit. J. Biol. Chem. 276:38555-38562. [DOI] [PubMed] [Google Scholar]
- 17.Lu, Q., Y. T. Hwang, and, C. B. C. Hwang. 2002. Detection of mutations within the thymidine kinase gene of herpes simplex virus type 1 by denaturing gradient gel electrophoresis. J. Virol. Methods 99:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Munir, K. M., D. C. French, D. K. Dube, and L. A. Loeb. 1992. Permissible amino acid substitutions within the putative nucleoside-binding site of herpes simplex virus type 1 encoded thymidine kinase established by random sequence mutagenesis. J. Biol. Chem. 267:6584-6589. [PubMed] [Google Scholar]
- 19.Roberts, J. D., and T. A. Kunkel. 1996. DNA replication in eukaryotic cells. Monograph 31, p. 217-248. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Streisinger, G., and J. Owen. 1985. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics 109:633-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streisinger, G., Y. Okada, J. Emrich, J. Newton, A. Tsugita, E. Terzaghi, and M. Inouye. 1966. Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 31:77-84. [DOI] [PubMed] [Google Scholar]