Abstract
The protein kinase C (PKC) pathway has been considered to be essential for activation of latent Epstein-Barr virus (EBV) into the lytic cycle. The phorbol ester tetradecanoyl phorbol acetate (TPA), a PKC agonist, is one of the best understood activators of EBV lytic replication. Zp, the promoter of the EBV immediate-early gene BZLF1, whose product, ZEBRA, drives the lytic cycle, contains several phorbol ester response elements. We investigated the role of the PKC pathway in lytic cycle activation in prototype cell lines that differed dramatically in their response to inducing agents. We determined whether PKC was involved in lytic cycle induction by histone deacetylase (HDAC) inhibitors. Consistent with prevailing views, B95-8 cells were activated into the lytic cycle by the phorbol ester TPA, via a PKC-dependent mechanism. B95-8 was not inducible by HDAC inhibitors such as n-butyrate and trichostatin A (TSA). Bisindolylmaleimide I, a selective PKC inhibitor, blocked lytic cycle activation in B95-8 cells in response to TPA. In marked contrast, in HH514-16 cells, the immediate-early promoters Zp and Rp were simultaneously activated by the HDAC inhibitors; TPA by itself failed to activate lytic gene expression. Inhibition of PKC activity by bisindolylmaleimide I did not block lytic cycle activation in HH514-16 cells by n-butyrate or TSA. In an extensive exploration of the mechanism underlying these different responses we found that the variable role of the PKC pathway in the two cell lines could not be accounted for by significant polymorphisms in the promoters of the immediate-early genes, by differences in the start sites of immediate-early gene transcription, or by differences in the nucleosomal organization of EBV DNA in the region of Zp or Rp. While B95-8 cells contained more total PKC activity than did HH514-16 cells in an in vitro assay, another EBV-transformed marmoset lymphoblastoid cell line, FF41, in which the lytic cycle was not inducible by TPA, contained comparably high levels of PKC activity. Moreover, two marmoset lymphoblastoid cells lines in which the lytic cycle could not be triggered by TPA maintained the same profile of EBV latency proteins as B95-8 cells. Thus, the profile of EBV latency proteins did not account for susceptibility to induction by PKC agonists. PKC activation is neither obligatory nor sufficient for the switch between latency and lytic cycle gene expression of EBV in many cell backgrounds. Lytic cycle induction by HDAC inhibitors proceeds by a PKC-independent mechanism.
The switch between latency and the productive lytic cycle of Epstein-Barr virus (EBV) has been studied extensively in cultured lymphoid cell lines, a system that is advantageous for study of the molecular mechanism of the switch. Viral genes that are expressed during latency rather than during the lytic cycle are well characterized (31). The major viral products that activate the lytic cycle, the transcriptional activators encoded by the genes BZLF1 and BRLF1, have been identified (10, 11, 26). A number of different stimuli have been shown to trigger the switch into the lytic cycle. These include halogenated pyrimidines (25), nutrient starvation (27), phorbol esters (61), calcium ionophores (16), transforming growth factor β1 (17), n-butyrate (36), specific histone deacetylase (HDAC) inhibitors such as trichostatin A (TSA) (9, 29), the demethylating agent azacytidine (5), cross-linking of surface immunoglobulin (53), superinfection with EBV stocks containing rearranged defective DNA (41, 43), and superinfection with human herpesvirus 6 (21). A general unanswered question is whether all inducing agents operate through a final common pathway.
The mechanisms mediating control of EBV lytic-cycle gene expression are understood in rough outline, but many important details are missing. During latency the genes BRLF1 and BZLF1, encoding the lytic-cycle activators, are repressed. No mRNA or proteins encoded by these genes are detectable during latency (55). Inducing stimuli that activate the lytic cycle lead to expression of BRLF1 and BZLF1. The kinetics of activation of the lytic cycle has been examined in detail only in the Akata Burkitt lymphoma (BL) cell line (54). When Akata cells are induced into the lytic cycle by treatment with anti-immunoglobulin, BRLF1 and BZLF1 are expressed simultaneously (13, 52). The protein products of BRLF1 (Rta) and BZLF1 (ZEBRA) autostimulate their expression and reciprocally activate each other (44). These processes are conditioned by the cell backgrounds in which the experiments are conducted. For example, Rta stimulates expression of ZEBRA and its own synthesis in epithelial cell-BL hybrids and in the HH514-16 clone of BL cells (44, 60). However, in other cell backgrounds, such as the Raji BL cell line, Rta does not mediate autostimulation or activation of ZEBRA (45). ZEBRA itself activates expression of Rta in Raji cells but fails to autostimulate in this cell background (32). Once ZEBRA and Rta are expressed to high levels, they activate downstream genes of the lytic cycle. Downstream lytic-cycle genes can be classified according to whether they respond primarily to Rta, to ZEBRA, or to a combination of the two activators (45).
Despite this impressive array of information, many important questions about the mechanism of lytic cycle activation remain unanswered. How are BRLF1 and BZLF1 repressed during latency? Repression may involve a pathway downstream of the EBV latency gene LMP2A, since EBV containing mutations or deletions in LMP2A enters the viral lytic cycle at a higher rate than the wild type (38). It is not known whether each activation stimulus has a distinct mode of action on the promoters of the immediate-early genes. It is also not yet known whether Rp, the promoter controlling the bicistronic BRLF1/BZLF1 transcripts, invariably responds to the same signals as Zp, the promoter controlling the monocistronic BZLF1 transcript. For example, in reporter-based assays, tetradecanoyl phorbol acetate (TPA) activates Zp but not Rp (52). It is not understood how cell background modulates the response to different inducing stimuli. Moreover, how cell background affects the autostimulatory or cross-stimulatory response to the Rta and ZEBRA proteins is unexplored. The physiologic stimuli which induce lytic-cycle viral gene expression in vivo remain mysterious.
Protein kinase C (PKC) has been assumed to play an essential role in the initiation of the lytic cascade of EBV (23, 24, 31). Phorbol esters, which can induce EBV lytic cycle expression in many cell backgrounds, activate PKC (8). Zp contains several DNA elements that mediate a response to PKC (7, 22). ZEBRA, an EBV lytic-cycle activator, shares structural features with members of the AP-1 family of bZIP proteins that mediate transcriptional activation in response to PKC (18, 32, 33, 56, 58). ZEBRA itself is a potential target for phosphorylation by PKC (4).
This report, which characterizes the pathway leading to lytic cycle gene expression in B-cell lines carrying EBV in a latent state, questions the assumption that PKC plays an obligatory role in lytic-cycle induction. We initially found that two prototype cell lines differed dramatically in their response to classical chemical inducing stimuli. While the PKC pathway was dominant in B95-8 cells, affecting primarily Zp, this pathway played no discernible role in lytic-cycle induction by HDAC inhibitors in HH514-16 cells. In extensive exploration of the potential mechanisms underlying this variant response to PKC agonists, we found that the differing response could not be explained by the origin of the cells, their profile of EBV latency proteins, their total PKC activity, or the nucleosomal configuration of Zp or Rp. Furthermore, in two other marmoset B-cell lines, FF41 and W91, TPA activated PKC but did not induce the EBV lytic cycle. These findings indicate that PKC activation is neither necessary nor sufficient for induction of the EBV lytic cycle.
MATERIALS AND METHODS
Cell lines.
B95-8 is a lymphoblastoid cell line established from peripheral blood lymphocytes of a cotton-top marmoset (Saguinus oedipus) following in vitro infection with EBV (40). A small fraction (1 to 3%) of current batches of B95-8 cells spontaneously enter the viral lytic cycle. FF41 and W91 are marmoset lymphoblastoid cell lines similarly established by exposure to EBV derived from patients with infectious mononucleosis or Burkitt's lymphoma, respectively (20, 39). HH514-16 is a subclone of the P3J-HR1K BL cell line. This subclone was selected for absence of spontaneous EBV lytic replication and a high level of chemically induced EBV lytic-cycle gene expression (28). All cell lines were grown in RPMI 1640 medium containing 8% fetal bovine serum and antibiotics. The expression of EBV latency proteins in these cell lines was detected by immunoblotting with a human antiserum that detects all of the latent EB nuclear antigens (50) and a 1:80 dilution of murine monoclonal antibody to LMP1 (M0897; Dako) followed by a rabbit anti-mouse immunoglobulin bridge (AXL232; Axcell).
Chemical induction of the EBV lytic cycle.
Cells were subcultured at 3 × 105/ml. Chemical inducing agents were added when the cells were in logarithmic-phase growth, usually 24 to 72 h after subculture. The inducing chemicals included 20 ng of TPA (524400; Calbiochem) per ml, 3 mM sodium butyrate (B5887; Sigma), and 5 μM TSA (Wako BioProducts). Cells were assessed for the expression of mRNAs and proteins of the EBV immediate-early genes 17 to 24 h after chemical treatment or, when indicated (see Fig. 5), at different intervals after chemical treatment.
FIG. 5.
Kinetics of expression of BRLF1 and BZLF1 transcripts in chemically induced B95-8 (A) and HH514-16 (B) cells. Shown are results of RNase protection experiments. Cytoplasmic RNA was prepared at the indicated times from untreated cells or cells that were chemically induced into the lytic cycle. The RNA probe ΔZp was 315 nt long. It contained EBV sequences from −89 to +152 (Fig. 4A) and 73 nt of vector sequences. The bicistronic transcript is represented by a 242-nt protected RNA; the monocistronic transcript is represented by a 152-nt protected RNA. BUT, n-butyrate.
Detection of ZEBRA and Rta by immunoblotting.
Cells were resuspended in sodium dodecyl sulfate (SDS) sample buffer, and extracts were prepared by sonication and boiling. Each lane of a 10% polyacrylamide-SDS gel received protein extract equivalent to 3 × 106 cells. After electrophoresis the gel was transferred to nitrocellulose. The blot was blocked in 5% nonfat milk at 4°C overnight. Filters were incubated for 1 h at 25°C with a 1:200 dilution of rabbit antisera raised to TrpE/BZLF1 or to the N-terminal 320 amino acids of the BRLF1 protein, Rta (44, 55). Blots were also probed with antiserum to β-actin (A5316; Sigma). Blots were washed (10 mM Tris [pH 7.5], 200 mM NaCl, 0.05% Tween 20), incubated with 1 μCi of 125I-protein A for 1 h at 25°C, washed again, and exposed to XAR film for autoradiography. Autoradiographs were scanned with a Molecular Dynamics SI personal densitometer, and the data were quantitated with ImageQuant NT software.
Preparation of cellular RNA and Northern blotting.
Cytoplasmic RNA was prepared as described previously (32). Total RNA was prepared with RNeasy and Qia-shredder spin columns (Qiagen) or TRIzol reagent (GIBCO/BRL) according to the manufacturers' directions. Each lane of a 1% agarose-6% formaldehyde gel received cytoplasmic RNA from 2 × 106 cells or total RNA from 3 × 106 cells. Northern blots were probed with a 32P-labeled 623-bp BamHI-to-PstI subfragment of BZLF1 cDNA (37, 49). RNA loading was standardized by probing the blots with a 1.8-kbp portion of the β-actin cDNA or 370-bp NcoI/PstI fragment of the cDNA of the H1 RNA component of human RNase P (2). BRLF1 and BZLF1 mRNA levels were measured with a Molecular Dynamics PhosphorImager and ImageQuant software and standardized for the expression of mRNA for β-actin mRNA or the H1 component of RNase P.
Primer extension.
The start site of the BZLF1 mRNA was estimated by primer extension. The oligonucleotide primer 5′ TTGCGATGGCCCTCCCAGGTC 3′ was labeled with 32P by using T4 polynucleotide kinase and annealed to total cellular RNA (TRIzol) in hybridization buffer at 45°C for 16 to 18 h. Reverse transcriptase was added at 42°C for 2 h. The size of the extended cDNA product was estimated by electrophoresis through an 8% polyacrylamide-7 M urea gel. Markers were fragments of φX174 DNA digested with HaeIII.
RNase protection assay.
The expression of the bicistronic BRLF1/BZLF1 and monocistronic BZLF1 mRNAs was analyzed simultaneously by an RNase protection assay with a single RNA probe that overlapped the start of transcription of the BZLF1 gene. (See Fig. 4A for DNA sequences represented in the probe.) EBV DNA sequences (242 nucleotides [nt]) were cloned in the antisense direction into pBluescript SK−. The plasmid was linearized with EcoRI, adding 73 nucleotides to the template. RNA was transcribed in vitro with T3 RNA polymerase incorporating [32P]UTP. After electrophoresis, the radiolabeled RNA probe was isolated from an 8% polyacrylamide-7 M urea gel. The probe was hybridized overnight to cytoplasmic RNA in 5 M guanidium thiocyanate-0.5 M EDTA buffer at 45°C. Free probe was digested with RNase T1 and RNase A. The protected RNA fragments remaining were electrophoresed on an 8% polyacrylamide-7 M urea gel which was dried and exposed to XAR film. In this assay the free probe is 315 nt, the portion of the probe protected by the bicistronic mRNA is 242 nt, and the portion of the probe protected by the monocistronic mRNA is 152 nt.
FIG. 4.
Comparison of the transcriptional start site of the 1.0-kb BZLF1 mRNA in chemically induced B95-8 and HH514-16 cells. (A) Diagram of EBV DNA including Zp, the transcriptional start site, and a portion of the BZLF1 ORF. Two ZEBRA binding sites, ZIIIA and ZIIIB, are boxed. Also boxed is a region partially responsible for the TPA response of Zp, here designated TRE, also known as a ZII site (22). Five point mutations distinguish P3J-HR-1 Zp from B95-8 Zp (30). Those found in the HH514-16 genome are circled. The arrow indicates the start of transcription in B95-8 and HH514-16 cells, mapped by primer extension (see panel B). The location of the primer is indicated with a dashed underline. Δ, 5′ end of the B95-8 BZLF1 cDNA cloned previously (49); •, start of BZLF1 ORF. Brackets indicate the region encompassed in the probe ΔZp, used for RNase protection (Fig. 5). (B) Results of primer extension. RNA was prepared by the TRIzol method 24 h after induction of the EBV lytic cycle by TPA (B95-8 cells) or a combination of TPA and n-butyrate (T/B) (HH514-16 cells). The radioactive cDNAs synthesized from the primer shown in panel A were analyzed on a polyacrylamide-7 M urea gel. Markers were φX174 DNA digested with HaeIII.
Fluorescence in situ hybridization (FISH) for EBV DNA.
Untreated cells and cells that had been chemically induced 2 days previously were incubated with 0.1 μg of Colcemid per ml for 2 h at 37°C. Cells (3 × 106) were diluted in 15 ml of a hypotonic phosphate-buffered saline (PBS) solution (26 mM NaCl, 0.6 mM Na2HPO4, 0.6 mM NaH2PO4 [pH 7.4], 0.8% sodium citrate, 0.8 mM MgCl2, 0.8 mM CaCl2). After 10 min at 37°C the cells were resuspended in the same hypotonic solution at 3 × 106 cells/ml. Ten microliters of cells were spotted per well on eight-well microscope slides (60-5453-36; PGS Scientific), air dried for 20 min at room temperature, and fixed for 15 min at room temperature in 4% paraformaldehyde in PBS. Slides were rinsed briefly in PBS and stored in 70% ethanol at 4°C.
On the day of hybridization, the slides were pretreated with 0.5% Triton X-100 in PBS for 5 min at 4°C and washed for 5 min at room temperature in PBS and then for 5 min each in cold 70, 90, and 100% ethanol. Slides were immersed in 70% formamide (F4761; Sigma)-2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min at 93°C to denature the DNA and then for 5 min each in the 70, 90, and 100% ethanols.
Biotin-16-dUTP (109 3070; Boehringer/La Roche)-labeled probes, nick translated with a BioNick labeling system (18247-015; GIBCO), were prepared with 1 μg of pBR-BamHI W plasmid DNA. The efficiency of biotinylation of the DNA was verified by a colorimetric dot blot test using alkaline phosphatase, streptavidin-alkaline phosphatase, nitroblue tetrazolium, and 5-bromo-4-chloro-3-indolylphosphate. Ten micrograms of denatured salmon sperm DNA and yeast tRNA were added to 50 μg of biotinylated probe and dried. This was resuspended in 10 μl of formamide, heated to 70°C for 10 min, placed on ice for 5 min, and then combined with an equal volume of hybridization mix for a final concentration of 10% dextran sulfate-0.2% bovine serum albumin (nuclease free; 7114 54; Boehringer/La Roche) 2× SSC. Five microliters of this final mix was used per spot of cells. Coverslips were gently applied, and the hybridization proceeded overnight (∼16 to 20 h) at 37°C in a humid box.
The coverslips were removed and the slides were washed at 37°C for 30 min each in 50% formamide-2× SSC and 2× SSC and then washed at room temperature for 30 min in 1× SSC and for 5 min in 4× SSC. They were blocked at 37°C for 20 min in 1% BSA-4× SSC. The slides were immediately incubated with fluorescein-conjugated avidin (100205; Boehringer/La Roche) at 2 μg/ml in 1% bovine serum albumin-4× SSC at 37°C for 30 min Slides were washed three times in 4× SSC at 37°C, 10 min each, and a final wash was done in 1× PBS for 5 min at room temperature. DABCO (0.2 M; diazabicyclo-[2.2.2] octane; D2522; Sigma ) in 0.02 M Tris (pH 8)-90% glycerol was used as an antifade mounting medium. Slides were examined with a Zeiss fluorescent microscope.
Effect of inhibition of PKC on lytic cycle induction.
Cells that had been chemically induced into the lytic cycle were treated simultaneously with bisindolylmaleimide I (B6292; Sigma), an inhibitor of the α, β, γ, δ, and ɛ isoforms of PKC, in doses ranging from 0.1 to 20 μM (57). The effects on expression of BRLF1 and BZLF1 mRNAs and on Rta and ZEBRA proteins were measured in the absence and presence of the PKC inhibitor.
In vitro PKC assays.
In initial experiments (see Fig. 9A and B), PKC activity was measured in cell extracts by using the PKC assay system (GIBCO/BRL). Each assay used 400 ng of cell protein, which was incubated for 5 min at 30°C with the substrate. The assay kit is based on the measurement of phosphorylation of an 11-residue peptide of myelin basic protein. PKC specificity was confirmed using a pseudo-substrate peptide which inhibits the α, β, and γ isozymes of PKC. Total PKC activity was measured in the cell lines by adding a lipid preparation containing TPA, phosphatidyl serine, and Triton X-100-mixed micelles to the cell extract. Specific PKC activity was determined by subtracting the activity in the presence of the inhibitor from the activity in the absence of inhibitor. PKC activity was also determined in cell extracts without the addition of the TPA, phospholipid, or mixed micelles. PKC activity was measured in untreated cells and in cells that had been treated in vivo for 24 h with different inducing chemicals. Specific endogenous PKC activity was obtained by subtracting activity in the presence of the inhibitor from activity in the absence of the inhibitor. Results are expressed as counts per minute of 32P incorporated into the substrate.
FIG. 9.
Comparison of PKC activity in cell lines. (A) Comparing total PKC activity in B95-8 and HH514-16 cell lines in four replicate experiments. (B) Effect of chemical treatment on PKC activity. The data are means of four experiments for untreated and TPA-treated cells and of two experiments for n-butyrate- and TSA-treated cells. (C) Comparison of total and spontaneous PKC activity in four lymphoid cell lines. The data are from three replicate experiments. (D) Comparison of spontaneous and TPA-inducible PKC activity in four lymphoid cell lines in three replicate experiments. The data for all experiments are expressed relative to total (A and C) and TPA-inducible (B and D) PKC activity in B95-8 cells.
In subsequent assays (see Fig. 9C and D), PKC activity in four cell lines was compared using an 11-amino-acid substrate (QKRPSQRSKYL) in a PKC assay kit obtained from Upstate Biotechnology (17-B9). Cell extract protein (8 to 38 μg) was incubated in the assay for 10 min at 30°C. Equal amounts of cell extract protein were used for each cell line. PKC activity was determined by subtracting activity in the absence of substrate from activity in the presence of substrate. Specific PKC activity was measured by determining the extent of kinase activity inhibited by 0.5 μM bisindolylmaleimide I. “Total” PKC activity is specific PKC activity in the presence of PKC lipid activator (phosphatidyl serine and diacyl glycerol). “Spontaneous” PKC activity is bisindolylmaleimide-inhibitable kinase activity in the absence of PKC lipid activator. PCK activity after treatment of cultured cells with TPA is referred to as “induced PKC activity.” The data from three replicate experiments were expressed relative to total activity of the B95-8 cell line.
Micrococcal nuclease digestion of cellular and EBV DNA.
HH514-16 and B95-8 cells were subcultured at 3 × 105/ml and incubated for 48 h. Between 1 × 107 and 5 × 107 cells were resuspended in permeabilization buffer (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 0.5 mM CaCl2). Cells were pelleted and resuspended in the same buffer containing 0.025% lysolecithin (L4129; Sigma) for 3 min at room temperature. The cells were resuspended in buffer (150 mM sucrose, 50 mM Tris [pH 7.5], 5 mM NaCl, 2 mM CaCl2) divided into aliquots and treated with 0 to 20 U of micrococcal nuclease (LS004797; Worthington Biochemicals) per ml for 5 min at room temperature. The cells were resuspended in stop solution (20 mM Tris [pH 8], 20 mM NaCl, 20 mM EDTA, 1% SDS, 0.6 mg of pronase per ml) and swirled briefly; an equal volume of 150 mM NaCl-5 mM EDTA was added. After overnight incubation at 37°C the DNA was precipitated with ethanol and resuspended in 10 mM Tris (pH 7.5)-1 mM EDTA. The DNA was electrophoresed through a 1% agarose gel and transferred to nitrocellulose. The blot was probed with 32P-labeled EBV BamHI W, the BamHI/DraI fragment of BamHI Z containing the BZLF1 promoter, Zp/BD (nt 103741 to 103226 of the EBV genome), or nt −589 to +68 of Rp (nt 106771 to 106114).
RESULTS
Induction of BZLF1 mRNA expression by TPA, butyrate, and TSA differs in two prototype laboratory strains.
The capacity of TPA and n-butyrate to induce the EBV lytic cycle in B95-8 and HH514-16 cells was assessed by examining the expression of mRNAs for the genes encoding BRLF1 and BZLF1, the lytic-cycle activators (Fig. 1A). These transcripts include two alternatively spliced bicistronic mRNAs, 4.0 and 3.0 kb in size, directed from Rp, the promoter of BRLF1, and a 1.0-kb monocistronic mRNA controlled from Zp, the promoter of BZLF1 (37). In the two cell lines the patterns of response to the two inducing chemicals were dramatically different. In B95-8 cells, TPA induced expression of the transcripts from both Rp and Zp (Fig. 1A, lane 3), while n-butyrate by itself had no effect (lane 2). In HH514-16 cells, n-butyrate activated both immediate-early lytic-cycle mRNAs (Fig. 1A, lane 7), while TPA was inactive (lane 8). Moreover, the response to the combination of TPA and n-butyrate also differed dramatically between the two cell lines. In B95-8 cells, n-butyrate reduced lytic cycle induction by TPA (Fig. 1A, lane 4), while in HH514-16 cells, TPA enhanced the response to n-butyrate (lane 9). These responses were consistent in the two cell lines over a period of 5 years.
FIG. 1.
Selective induction of BRLF1 and BZLF1 expression by inducing chemicals in B95-8 and HH514-16 cells. (A) Induction of mRNAs containing BRLF1 and BZLF1. At 20 h after treatment of the cells with TPA or n-butyrate, singly or in combination, RNA was prepared by the TRIzol method. RNA from 3 × 106 cells per lane was analyzed by Northern blotting with a probe prepared from a portion of the BZLF1 cDNA. This probe detects the 4.0- and 3.0-kb bicistronic (BRLF1 plus BZLF1) mRNA from Rp and the 1.0-kb monocistronic (BZLF1) mRNA from Zp (37). The blot was reprobed with β-actin to control for loading of RNA. (B) Comparison of the effects of n-butyrate (B) and TSA. Cytoplasmic RNA prepared 17 h after chemical treatment was analyzed by Northern blotting with the BZLF1 probe. The RNA blots were also probed for β-actin and the H1 RNA component of human RNase P to control for RNA loading.
n-Butyrate, an inhibitor of HDAC, is thought to activate gene expression by relief of chromatin repression (12, 46). However, its effects are pleiotropic. Therefore, we examined the response of B95-8 and HH514-16 cells to TSA, a more specific inhibitor of HDAC (59). The response of B95-8 and HH514-16 cells to TSA was similar to their response to n-butyrate (Fig. 1B). The lytic cycle was activated by both TSA and n-butyrate in HH514-16 cells (Fig. 1B, lanes 6 and 7) as judged by the induction of BZLF1 containing mRNAs. In HH514-16 cells there was 3- to 10-fold synergy between TPA and n-butyrate and between TPA and TSA, as assessed by measuring the levels of either the 1.0- or 3.0-kb BZLF1 transcript (data not shown). Neither TSA nor n-butyrate had any effect on the level of BZLF1 mRNA in B95-8 cells (Fig. 1B, lanes 2 and 3).
Response of ZEBRA and Rta proteins to inducing stimuli.
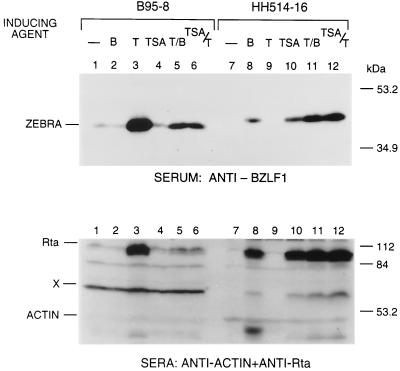
The two cell lines again differed markedly when the responses to TPA, n-butyrate, and TSA were assessed by measurement of the levels of ZEBRA and Rta proteins (Fig. 2). In B95-8 the two proteins were expressed spontaneously at a low level (Fig. 2, lane 1); expression of these proteins was not activated by n-butyrate or TSA (lanes 2 and 4). Only TPA stimulated ZEBRA and Rta protein expression in B95-8 cells (Fig. 2, lane 3); this effect was dampened when the HDAC inhibitors were included with TPA (lanes 5 and 6). In untreated HH514-16 cells (Fig. 2, lane 7) and in cells treated with TPA alone (lane 9), there was no detectable ZEBRA or Rta. Both HDAC inhibitors stimulated expression of the immediate-early proteins in these cells (Fig. 2, lanes 8 and 10). The addition of TPA to the HDAC inhibitors induced a level of ZEBRA and Rta protein that was only slightly greater than the level observed in the presence of the HDAC inhibitor alone (Fig. 2, lanes 11 and 12).
FIG. 2.
Induction of ZEBRA and Rta protein expression in B95-8 and HH514-16 cells. Extracts of 3 × 106 cells prepared 20 h after chemical induction were analyzed for expression of ZEBRA protein by probing an immunoblot with a 1:200 dilution of rabbit antibody raised to a TrpE-BZLF1 fusion protein expressed in Escherichia coli (55) or with a rabbit antibody to Rta (41). Antibody to β-actin and a cross-reactive band (X) served as controls for loading of cell protein.
Inducing chemicals increase the number of cells expressing early antigens and lytic viral DNA.
In the next series of experiments we investigated whether the dramatically different effects of inducing chemicals on B95-8 and HH514-16 cells extended to expression of early genes and lytic viral DNA replication, as assessed on a cell-by-cell basis (Table 1). About 3% of B95-8 cells spontaneously entered the lytic cycle, as judged by immunostaining with monoclonal antibody R3.1 to EA-D, the BMRF1 gene product (42). The fraction of EA-D-positive cells increased to about 25% of cells following TPA treatment but was unaffected by the HDAC inhibitors. No spontaneous early antigen expression was observed in HH514-16 cells. n-Butyrate and TSA both induced about 20% of the cells to express early antigen, but TPA by itself had no effect.
TABLE 1.
Correlation between number of cells induced into EBV early gene expression and number exhibiting lytic viral DNA synthesis
| Inducing stimulus | % of B95-8 cells expressing:
|
% of HH514-16 cells expressing:
|
||
|---|---|---|---|---|
| Early antigena | Lytic DNAb | Early antigena | Lytic DNAb | |
| None | 3 | 2 | <1 | 0 |
| n-Butyrate | 3 | 2 | 20 | 20 |
| TSA | 3 | 2 | 20 | 8 |
| Phorbol ester | 25 | 30 | <1 | 0 |
| Phorbol ester + n-butyrate | 25 | 30 | 30 | 25 |
Determined by indirect immunofluorescence using monoclonal antibody R3.1 to EA-D.\
Determined by FISH with a probe from BamHI W.
The number of cells expressing early antigens correlated with the number of cells induced into lytic viral DNA synthesis. When viral DNA was assayed by FISH, the EBV genomes in cells in latency appeared as small dots. Following lytic cycle induction there were two types of morphological changes, namely, the appearance of cells with brightly fluorescent coarse granules of viral DNA and the appearance of cells with larger coalescing globules of viral DNA (Fig. 3). The number of cells with coarse granules or globules of viral DNA, representing factories of lytic viral DNA, was similar to the number of cells that expressed EA-D (Table 1). There was a single exception; in repeated trials, TSA treatment of HH514-16 cells caused more cells to express EA-D than to undergo viral lytic DNA synthesis (Table 1 and data not shown).
FIG. 3.
FISH for EBV DNA in uninduced (A) and chemically induced (TPA-treated) (B) B95-8 cells. In cells in latency, the viral genomes appear as faint dots. In cells in the lytic cycle, viral genomes appear as brightly fluorescent coarse dots or globules.
Sequences of Rp and Zp of the two prototype strains and transcription start site of the 1.0-kb BZLF1 mRNA do not differ significantly.
The difference in the responses of B95-8 and HH514-16 cells to chemical inducers of the lytic cycle could be due to the presence or absence of binding sites for cellular proteins which affect the activity of Rp or Zp. These binding sites might be affected by polymorphisms in the DNA sequence of Rp and Zp. A previous study (45) compared the nucleotide sequences of Rp and Zp from B95-8 and HH514-16. The sequence of Rp in HH514-16 cells differs from that of B95-8 at only one location: at position −1 relative to the transcriptional start there is a C in B95-8 and an A in HH514-16. This difference might create a YY1 binding site in HH514-16 virus (51). Five point mutations distinguish Zp from P3HR-1, the parent of HH514-16, from Zp of B95-8 (30). Three of these mutations are present in HH514-16 (Fig. 4A), but none of the nucleotide differences within Zp of the two strains affected a known binding site for cellular or viral proteins.
A second explanation to account for the cell-specific response to inducing stimuli was that during entry into the lytic cycle a different transcriptional start site was utilized. In a primer extension experiment, the starts of transcription of BZLF1 mRNA from the two cell lines were analyzed side by side on the same gel (Fig. 4B). This experiment showed that the major BZLF1 transcription start site was identical in the two cell lines. This start site was approximately 26 nt downstream of the 5′ end of the putative TATA element (TTTAAA) in Zp and 51 nt upstream of the start of the BZLF1 open reading frame (ORF) (Fig. 4A).
Kinetics of expression of immediate-early mRNAs from Rp and Zp differ in the two cell lines.
Another scenario that might account for the different behavior of the two cell lines in response to inducing chemicals would be that in one cell line entry into the lytic cycle is preferentially controlled from Rp, while in the other cell line the lytic cycle is preferentially controlled from Zp. To determine whether the two cell lines differed in the utilization of these two promoters, the bicistronic and monocistronic transcripts were detected simultaneously with a single probe in an RNase protection experiment (Fig. 5). The results of this experiment demonstrated several clear differences between B95-8 and HH514-16 cells.
In B95-8 cells that were not treated with inducing chemicals there was constitutive transcription from Rp but little or no transcription from Zp (Fig. 5A). Within 2 h after addition of TPA, low levels of monocistronic transcripts from Zp were detected, as shown by the appearance of the 152-nt protected fragment. These Zp-driven transcripts were abundant 6 and 8 h after TPA treatment (Fig. 5A, lanes 8 and 10). The abundance of transcripts from Rp in B95-8 cells also increased at 6 and 8 h after TPA treatment, but to a lesser extent relative to their abundance in untreated cells.
The kinetic response of Rp and Zp to induction of the lytic cycle in HH514-16 cells thus differed from that of B95-8 cells in three respects (Fig. 5B). (i) HH514-16 cells were tightly latent and, unlike B95-8 cells, exhibited no detectable spontaneous expression from Zp or Rp. (ii) Induction of the lytic cycle proceeded more slowly in HH514-16 cells than in B95-8 cells. Significant induction of expression from Zp and Rp was first observed 6 h after chemical treatment of B95-8 cells, but immediate-early mRNA reached high levels only at 16 h postinduction in HH514-16 cells. (iii) In HH514-16 cells Zp and Rp were activated simultaneously and to approximately the same extent at all times after the lytic cycle was induced. By contrast, at early times in B95-8 cells, induction occurred primarily at Zp.
Effect of addition of a specific inhibitor of PKC on induction of the EBV lytic cycle in B95-8 and HH514-16 cells.
We next determined whether PKC activation was required for lytic cycle induction in the two cell lines. To explore this point, we examined the effect of bisindolylmaleimide I, a specific inhibitor of PKC (57), on expression of BRLF1 and BZLF1 mRNAs (Fig. 6) and Rta and ZEBRA proteins (Fig. 7). In B95-8 cells, TPA-mediated induction of the immediate-early mRNAs was exquisitely sensitive to inhibition by bisindolylmaleimide I (Fig. 6A). Increasing doses of the inhibitor caused progressive blockade of expression of the TPA-induced immediate-early mRNAs: 0.1 μM caused 33% inhibition, 1 μM caused 67% inhibition, and 10 μM caused more than 85% inhibition of expression of both the bicistronic and monocistronic mRNAs (Fig. 6B). By contrast, the same amounts of bisindolylmaleimide I did not affect the stable levels of BRLF1/BZLF1 mRNAs that were induced by treating HH514-16 cells with the HDAC inhibitor TSA (Fig. 6C and D).
FIG. 6.
Effect of bisindolylmaleimide I, an inhibitor of PKC, on induction of expression of BRLF1 and BZLF1 mRNAs by TPA and TSA. Lytic-cycle induction was carried out in the presence of three concentrations of the PKC inhibitor (0.1, 1.0, and 10 μM) or no inhibitor. RNAs prepared 22 h after induction were analyzed by Northern blotting. (A and B) B95-8 cells were untreated or treated with 20 ng of TPA per ml. (C and D) HH514-16 cells were untreated or treated with 5 μM TSA. (A and C) Northern blots; (B and D) quantitation of results by phosphorimaging of mRNAs.
FIG. 7.
Effect of an inhibitor of PKC on expression of ZEBRA (A and C) and Rta (B and D) proteins following TPA and n-butyrate treatment. B95-8 (A and B) and HH514-16 (C and D) cells were induced into the lytic cycle in the absence or presence of increasing concentrations of bisindolylmaleimide I. After 24 h, cell extracts were analyzed for ZEBRA or Rta protein by immunoblotting with monospecific antisera. Results were quantitated by densitometry of the autoradiograph. β-Actin was used to control for protein loading. The immunoblots for each cell line were all run in the same experiment, but the lanes were spliced to remove unrelated data.
Bisindolylmaleimide I treatment also markedly reduced the levels of expression of ZEBRA and Rta proteins present in extracts of B95-8 cells treated with TPA (Fig. 7A and B). At the highest dose of inhibitor the levels of ZEBRA and Rta proteins were similar to that present in uninduced cells. The effect of the PKC inhibitor on ZEBRA protein was more dramatic than on Rta because TPA treatment of B95-8 cells leads to a more profound activation of expression of ZEBRA (50-fold) than of Rta (6-fold) (Fig. 5A). In HH514-16 cells, the PKC inhibitor did not block expression of ZEBRA or Rta that was inducible by n-butyrate (Fig. 7C and D) or TSA (data not shown).
These experiments showed that PKC acts positively in the pathway leading to induction of the lytic cycle following TPA treatment of B95-8 cells. However, PKC is not required for lytic-cycle induction of HH514-16 cells by HDAC inhibitors.
Exploration of the mechanism underlying the different response of B95-8 and HH514-16 cells to chemical induction.
The next group of experiments explored a series of hypotheses that might account for the mechanism underlying a preferential role of the TPA-inducible PKC pathway in lytic cycle induction of B95-8 cells. (i) Inducibility by TPA might be a general feature of marmoset lymphoblastoid cell lines. (ii) Inducibility by TPA might reflect the EBV latency program of the cells. (iii) Inducibility by TPA might correlate with the level of total PKC activity available in different cell backgrounds. (iv) Inducibility by TPA and a lack of effect of HDAC inhibitors in B95-8 cells might reflect different nucleosomal organization of Zp and Rp in B95-8 versus HH514-16 cells (15).
TPA does not induce the EBV lytic cycle in other lymphoblastoid cell lines derived from cotton-top marmosets.
To determine whether TPA inducibility was a general feature of EBV-transformed marmoset lymphoblastoid cell lines, we studied two additional marmoset cell lines: FF41, which, like B95-8, was transformed by EBV from a patient with infectious mononucleosis, and W91, which was transformed by EBV from a patient with Burkitt's lymphoma (20, 38). FF41 spontaneously expressed the lytic cycle (Fig. 8A). Expression of ZEBRA protein could be induced about threefold by the HDAC inhibitors n-butyrate and TSA, but ZEBRA expression was unaffected by treatment with TPA (Fig. 8A). W91 was tightly latent and not inducible by either TPA or HDAC inhibitors (data not shown). B95-8 and HH514-16 were induced by TPA and HDAC inhibitors, respectively, as shown in Fig. 1. Thus, the pattern of response to chemical inducing agents could not be correlated with the species of origin of the cell lines.
FIG. 8.
Pattern of latent gene expression and lytic cycle induction in the FF41 marmoset cell line. B95-8, FF41, and HH514-16 cells were untreated or treated with n-butyrate (B), TPA (T), or TSA in the same experiment. Cell extracts prepared 24 h after treatment were analyzed by immunoblotting for expression of LMP1, EBNA1, and EBNA2 (B) and ZEBRA (A). The immunoblots were probed for β-actin to control for protein loading.
Lack of correlation between the pattern of latent gene expression and chemical induction of the lytic cycle.
We attempted to learn whether the response to lytic-cycle-inducing agents correlated with the pattern of latent EBV gene expression (Fig. 8B). Both B95-8 and FF41 express the full panel of latent proteins; for example, LMP1, EBNA1, and EBNA2 can be detected (Fig. 8B) and both cell lines are therefore in latency III. In both cell lines the HDAC inhibitors increased the expression of LMP1, which is known to be activated during the lytic cycle (6). These results showed that the pattern of lytic-cycle inducibility of the two marmoset cell lines was independent of the pattern of latent gene expression. Furthermore, B95-8 cells revealed an additional interesting discordance. In the B95-8 cell line, TPA induced ZEBRA and Rta (Fig. 1, 2, and 8A), but the HDAC inhibitors, which did not induce ZEBRA or Rta expression in B95-8 cells, still induced LMP1 (Fig. 8B).
Latent gene expression in HH514-16 cells is affected by the deletion of the EBNA2 gene. However, these cells spontaneously express leader protein (48) and low levels of the LMP1 protein (Fig. 9 B, lanes 9 and 11). As in the marmoset cell lines, LMP1 expression was induced in HH514-16 cells by the HDAC inhibitors (Fig. 8B, lanes 10 and 12). The low level of spontaneous LMP1 expression did not account for the lack of response of HH514-16 cells to TPA. Transfection of HH514-16 cells with an LMP1 expression vector did not rescue the ability of TPA to induce ZEBRA expression (data not shown).
Comparing the content of PKC in different cell lines.
The pathway of activation of gene expression by phorbol esters such as TPA involves stimulation of PKC and thereafter activation of the AP-1 family of transcriptional activators (1). Since TPA by itself activated the EBV lytic cycle in B95-8 cells and not in HH514-16 cells, we initially explored the hypothesis that differences in PKC activity in the two cell lines might account for their variant response to TPA. We determined PKC activity in two ways. Total PKC activity was measured by adding TPA, phospholipid, and mixed micelles to a cell extract and then assaying the enzyme (Fig. 9A). This treatment activated any PKC that might be present in the cell extract. We also measured PKC activity in extracts of untreated cells and cells that had been treated in vivo with EBV lytic-cycle-inducing chemicals for 24 h (Fig. 9B). In the determination of induced PKC activity, no additional TPA, phospholipid, or mixed micelles were added to the cell extract before measurement of PKC activity. Thus, only PKC that was already activated was measured in this component of the assay. In both types of measurement, specific PKC activity was deduced by measurement of kinase activity that was removed by addition of a specific peptide inhibitor of PKC.
Figure 9A shows that untreated B95-8 cells reproducibly contained more total PKC activity than did HH514-16 cells. The mean ratio of total PKC activity in B95-8 cells to HH514-16 cells was 10 (range, 4.8 to 21). When no exogenous TPA, phospholipid, or mixed micelles were added to extracts of untreated cells prior to the enzyme assay, there was twofold more spontaneously activated PKC in B95-8 cells than in HH514-16 cells (Fig. 6B). In both cell lines, treatment with TPA for 24 h before the extracts were prepared increased the level of activated PKC. However, the level of PKC activity induced by TPA was 5.3-fold higher (range, 3.4 to 10.3) in B95-8 cells than in HH514-16 cells. Treatment of B95-8 cells with n-butyrate or TSA did not increase PKC activity above the level present in untreated cells. No PKC activity was present in extracts of HH514-16 cells that were treated with n-butyrate or TSA.
These results, showing that B95-8 cells contained considerably more PKC activity than HH514-16, suggested that the preferential susceptibility of B95-8 cells to lytic cycle induction by TPA and the refractoriness of HH514-16 cells to lytic cycle stimulation by the phorbol esters might be correlated with the amount of activatable kinase C. To explore this point further, we compared the levels of total and spontaneous PKC in two additional marmoset cell lines in which lytic-cycle induction was not triggered by TPA (Fig. 8A and 9C; also data not shown). The level of total PKC activity in FF41-1 cells, which were not inducible by TPA, was about 88% that of B95-8 cells. The level in W91 cells, which were refractory to lytic-cycle induction, was 43% that of B95-8 cells. All three marmoset lymphoblastoid cell lines contained four- to fivefold more spontaneously activated PKC than did HH514-16. When TPA was added to the cells in culture, B95-8 and FF41-1 contained approximately the same level of PKC (Fig. 9D). This level of PKC was approximately 2.2-fold higher than the level in W91 cells and 5-fold higher than the level in HH514-16 cells (Fig. 9D). n-Butyrate, which induced the EBV lytic cycle in FF41-1 cells, did not induce PKC activity (data not shown). These experiments showed that the levels of total, spontaneous, and TPA-inducible PKC activity did not correlate with TPA inducibility of the EBV lytic cycle.
Comparison of nucleosomal organization of EBV DNA in B95-8 and HH514-16 cells.
One hypothesis to account for the differing pattern of response of the two cell lines to lytic cycle inducing agents is that in B95-8 cells the EBV genome is in an “open chromatin” configuration, whereas in HH514-16 the EBV lytic genes are repressed by chromatin. This might explain why HDAC inhibitors are required in HH514-16 cells but not in B95-8 cells. To explore this hypothesis, we compared nucleosomal organization of EBV DNA in the two cell lines using the micrococcal nuclease technique (Fig. 10). Using a probe from the EBV large internal repeat we found a similar pattern of nucleosomal organization of EBV DNA in the two cell lines (Fig. 10A). Moreover, Zp, the promoter of the BZLF1 gene, was in a nucleosomal configuration in both B95-8 and HH514-16 cells (Fig. 10B) as was Rp, the promoter of the BRLF1 gene (Fig. 10C). The spacing and extent of nucleosomal organization of Zp and Rp were similar in the two cell lines. These experiments indicate that gross differences in the chromatinization of EBV DNA in the regions of Zp and Rp in the two cell lines did not account for their variant response to lytic-cycle-inducing agents.
FIG. 10.
Comparison of the nucleosomal organization of EBV DNA in B95-8 and HH514-16 cells. Cells were permeabilized with lysolecithin and incubated with increasing concentrations of micrococcal nuclease (concentrations are at the top of each gel, in units per milliliter). DNA was extracted and electrophoresed in agarose gels. The top panels show ethidium bromide-stained total cellular DNA. The bottom panels are Southern blots probed with BamHI W (A), with a BamHI/DraI subfragment containing the promoter of the BZLF1 gene Zp (BD) (B), or with Rp, nucleotides −589 to +68 relative to the start of transcription of BRLF1 (C).
DISCUSSION
These experiments describe profound differences in the EBV lytic-cycle induction pathway in well-characterized lymphoid cell lines and demonstrate that PKC activation is not obligatory for lytic-cycle induction. The lytic-cycle induction pathway was dependent on the virus and cell background. In each background there were distinguishable features of regulation of Rp and Zp, the two promoters controlling immediate-early gene expression. In B95-8 cells, these two promoters appeared to be under distinct and disconjugate regulation (Fig. 5A). However, in HH514-16 cells the two promoters were coordinately regulated (Fig. 5B). In B95-8 cells, expression from Zp was induced by TPA, whose effects were, as expected, mediated by PKC. However, in HH514-16 cells, inducibility of Zp and Rp by the HDAC inhibitors was independent of PKC (Fig. 6 and 7). Although PKC agonists could synergize with HDAC inhibitors in HH514-16 cells, PKC activation was not sufficient for activation of the lytic cycle in this cell background.
Mechanisms that may underlie the distinct response of cell lines to induction of the EBV lytic cycle.
What determines these profound biologic differences? We explored a number of possible mechanisms. (i) The start site of the BZLF1 mRNA was identical in the B95-8 and HH514-16 cell lines (Fig. 4B). (ii) Minor nucleotide alterations are found in the sequences of Zp and Rp from the two prototype strains (Fig. 4A) (42). These differences are unlikely to account for the marked variations in the response to inducing chemicals, but this point must be established by studying of the effects of specific Zp and Rp point mutations in recombinant viruses (19). (iii) Since marmoset lymphocytes containing the B95-8 strain allow spontaneous EBV replication and TPA inducibility while human lymphocytes harboring the same virus strain are often refractory to chemical induction (reference 36 and data not shown), it was feasible that some distinct feature of the interaction of the EB viral genome with the marmoset host cell facilitated lytic-cycle induction. However, TPA inducibility of the lytic cycle was not a general feature of marmoset lymphoblastoid cell lines; some of them were refractory to TPA but could be induced by HDAC inhibitors (Fig. 8A). (iv) Furthermore, marmoset lymphoblastoid cell lines which did respond to TPA and those which did not showed similar patterns of latent EBV protein expression characteristic of latency type III (Fig. 8B). (v) There was similar nucleosomal structure in the region of Zp and Rp from two cell lines which were sensitive or refractory to induction by TPA (Fig. 10 and data not shown). This result makes it unlikely that simple presence or absence of histone repression accounts for the variant response to chemical inducing agents in the B95-8 and HH514-16 cell backgrounds.
We explored in detail the importance of two notable differences between HH514-16 and B95-8 cells, namely, their level of expression of LMP1 and their level of activatable PKC. HH514-16 cells express a low level of LMP1 contingent on deletion of the EBNA2 gene (48) (Fig. 8B). However, overexpression of LMP1 in these cells did not allow them to be induced by TPA (data not shown). HH514-16 cells contained lower levels of spontaneously activated and total PKC activity than did the other cell lines examined (Fig. 9C). However, two marmoset cell lines with higher levels of spontaneous, total, and TPA-inducible PKC activity were also refractory to TPA induction (Fig. 8A, 9C, and 9D). Thus, there was no correlation between PKC activity and TPA inducibility of EBV lytic-cycle gene expression.
The variant response of different cell lines to lytic-cycle-inducing chemicals has potential implications for the goal of using activation of the EBV lytic cycle as oncolytic therapy (47). Since there are such profound differences in inducibility between cell lineages, each tumor would need to be examined individually for its response to lytic-cycle activators.
Relation between transcriptional response of immediate-early genes and downstream events.
In general, there was good correlation between the level of induction of mRNAs controlled from Rp and Zp following exposure to a chemical induction signal and the expression of downstream events, such as Rta and ZEBRA protein expression, EA-D expression, and lytic viral DNA synthesis (Table 1). There were exceptions to this correlation, however. For example, the level of induction of BRLF1 and BZLF1 mRNAs observed after treatment with a combination of TPA with the HDAC inhibitors in HH514-16 cells was 3- to 10-fold greater than that observed with the HDAC inhibitors alone (Fig. 1A and data not shown). However, the levels of ZEBRA and Rta protein expressed following application of the combination of TPA with the HDAC inhibitors was only slightly greater in HH514-16 cells than with HDAC inhibitors alone (Fig. 2). Moreover, the combination of inducing agents was not more effective than HDAC inhibitors alone in activation of individual HH514-16 cells into the lytic cycle (Table 1). These findings suggest that, even with massive amplification of viral mRNA, viral protein synthesis may be limited. While TPA may synergize with HDAC inhibitors to increase the level of immediate-early transcripts it has relatively little effect on immediate-early protein. Furthermore, not all cells, even in these highly inducible cell backgrounds, may be competent to enter the lytic cycle.
The second discordant observation was that TSA, which was as potent as n-butyrate in stimulating expression of immediate-early mRNAs and proteins as well as early antigen, was less effective in activation of lytic viral DNA synthesis. This observation is consistent with the possibility that the two HDAC inhibitors may have different effects on the cells or virus.
Cell background affects the behavior of Rp and Zp.
In B95-8 cells, the two immediate-early viral promoters appear to be under different regulation. Rp is spontaneously active and, at early times after treatment with the inducing chemical, shows very little stimulation by TPA (Fig. 5A). Zp is more quiescent in B95-8 cells but responds strongly to TPA induction. (Fig. 5A). This difference in response of Rp and Zp to TPA in B95-8 cells is consistent with the finding that Zp contains TPA-responsive sequences whereas Rp does not (47). However, in HH514-16 cells the two promoters appear to be coordinately regulated, in a manner quite similar to what has been described for the Akata cell line (54). HH514-16 cells are tightly latent, and there is no detectable transcription from Zp or Rp in untreated cells. Once the HDAC inhibitors are added, both promoters are active. In a long series of experiments studying the kinetics of activation in HH514-16 cells, using a variety of assays for immediate-early mRNAs, including Northern blotting, S1 nuclease analysis, and RNase protection, there was never a time at which mRNAs were detectable from one of the two promoters and not detectable from the other promoter (D. Kwa, unpublished data).
One model to account for the different requirement of HDAC inhibitors to activate Zp and Rp in the two cell backgrounds is that the immediate-early promoters are repressed by histones in HH514-16 cells but are not so inhibited in B95-8 cells. One form of this model predicts that in B95-8 cells the immediate-early promoters might not be in a nucleosomal configuration and thus spared from repression by histones. However, our experiments to address this point (Fig. 10) did not reveal a difference in nucleosomal organization of Zp or Rp in the two cell lines. Further studies are needed to explore the extent of histone acetylation of Zp and Rp in different cell backgrounds.
Another model, which we currently favor, is that the differential action of TPA and the HDAC inhibitors in the two cell backgrounds is not due primarily to variations in the chromatin state of EBV but rather to indirect effects of the inducing agents on cell genes. For example, in B95-8 cells, a cell gene that makes a protein that acts positively on Zp may be constitutively expressed, but its product requires activation by TPA. In HH514-16 this cell gene may be repressed by chromatin, and its expression is activated by the HDAC inhibitors. TPA may provide further activation through phosphorylation of the cell protein once it is derepressed.
Role of PKC in EBV lytic cycle induction.
Ever since the discovery by zur Hausen et al. (61) that TPA could induce EBV lytic-cycle gene expression, considerable attention has focused on the potential role of PKC in the lytic induction pathway. Among various phorbol esters, the capacity to induce the EBV lytic cycle correlates with the ability to activate PKC (14). Phorbol esters that activate PKC stimulate expression of reporter genes containing Zp fused to chloramphenicol acetyltransferase or luciferase (22). The regions of Zp that mediate the response to phorbol esters have been extensively characterized (7, 22). The phorbol ester-responsive regions include the ZII domain, which contains binding sites for CREB/ATF as well as AP-1 like factors that may mediate transcription following activation of PKC (34). Zp also contains the ZI sites that appear to mediate both basal activity and phorbol ester responsiveness via Sp1 and Sp3 binding sites (35). ZEBRA, the main lytic cycle activator of EBV, is related to the AP-1 bZIP activators c-Fos and c-Jun. ZEBRA binds to some DNA sequences that are also bound by c-Fos and c-Jun, although ZEBRA and the AP-1 activators each bind to DNA elements that are not recognized by the other activator (56). Moreover, ZEBRA itself may be a target for phosphorylation by PKC (4). In two-hybrid assays, ZEBRA has been found to interact with RACK, a receptor of activated C kinase (3).
To this lengthy list of incriminating evidence, the present report adds several observations that confirm the importance of PKC in cell backgrounds such as B95-8. The inducibility of the EBV lytic cycle in these cells by TPA correlated with the high level of PKC measured in cell extracts (Fig. 9). Furthermore, induction of the lytic cycle by TPA in B95-8 cells was nearly completely inhibited by a highly specific inhibitor of PKC (Fig. 6 and 7).
PKC activation is not required for induction of the EBV lytic cascade.
Despite the impressive array of data implicating PKC in the switch between latency and reactivation of EBV, this report and related studies (A. El-Guindy et al., unpublished data) show that activation of PKC is not an obligatory event in this process. In HH514-16, FF41, and W91 cells, TPA, which was competent to activate PKC (Fig. 9), was not sufficient to disrupt latency (Fig. 8A). Chemical stimuli, such as n-butyrate and TSA, that activated the lytic cascade in HH514-16 and FF41-1 cells did not activate PKC (Fig. 9B and data not shown). Addition of the PKC inhibitor bisindolylmaleimide to HH514-16 cells did not block lytic-cycle induction by HDAC inhibitors (Fig. 6 and 7). Moreover, we have found that, although the ZEBRA protein itself can be phosphorylated by PKC in vitro, phosphorylation of ZEBRA by PKC in vivo is not required for disruption of latency (El-Guindy et al., unpublished). ZEBRA is a phosphoprotein in vivo, but its phosphorylation does not appear to be mediated by PKC. These findings indicate that PKC is just one of many mediators of EBV lytic-cycle induction.
Acknowledgments
This study was supported by grants CA12055 and CA16038 from the National Cancer Institute.
We thank Jill Countryman, Dan DiMaio, and Tobias Ragoczy for helpful discussions and critical readings of the manuscript. We are grateful to Nancy Raab-Traub for an LMP1 expression vector.
REFERENCES
- 1.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 2.Bartkiewicz, M., H. Gold, and S. Altman. 1989. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 3:488-499. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, M., O. Gires, W. Kolch, H. Mischak, R. Zeidler, D. Pich, and W. Hammerschmidt. 2000. The PKC targeting protein RACK1 interacts with the Epstein-Barr virus activator protein BZLF1. Eur. J. Biochem. 267:3891-3901. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, M., H. Mischak, S. Dammeier, W. Kolch, O. Gires, D. Pich, R. Zeidler, H. J. Delecluse, and W. Hammerschmidt. 1998. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J. Virol. 72:8105-8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Sasson, S. A., and G. Klein. 1981. Activation of the Epstein-Barr virus genome by 5-aza-cytidine in latently infected human lymphoid lines. Int. J. Cancer 28:131-135. [DOI] [PubMed] [Google Scholar]
- 6.Boos, H., R. Berger, C. Kuklik-Roos, T. Iftner, and N. Mueller-Lantzsch. 1987. Enhancement of Epstein-Barr virus membrane protein (LMP) expression by serum, TPA, or n-butyrate in latently infected Raji cells. Virology 159:161-165. [DOI] [PubMed] [Google Scholar]
- 7.Borras, A. M., J. L. Strominger, and S. H. Speck. 1996. Characterization of the ZI domains in the Epstein-Barr virus BZLF1 gene promoter: role in phorbol ester induction. J. Virol. 70:3894-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castagna, M., Y. Takai, K. Kaibuchi, K. Sano, U. Kikkawa, and Y. Nishizuka. 1982. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J. Biol. Chem. 257:7847-7851. [PubMed] [Google Scholar]
- 9.Chang, L. K., and S. T. Liu. 2000. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 28:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Countryman, J., H. Jenson, R. Seibl, H. Wolf, and G. Miller. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cousens, L. S., D. Gallwitz, and B. M. Alberts. 1979. Different accessibilities in chromatin to histone acetylase. J. Biol. Chem. 254:1716-1723. [PubMed] [Google Scholar]
- 13.Daibata, M., S. H. Speck, C. Mulder, and T. Sairenji. 1994. Regulation of the BZLF1 promoter of Epstein-Barr virus by second messengers in anti-immunoglobulin-treated B cells. Virology 198:446-454. [DOI] [PubMed] [Google Scholar]
- 14.Davies, A. H., R. J. Grand, F. J. Evans, and A. B. Rickinson. 1991. Induction of Epstein-Barr virus lytic cycle by tumor-promoting and non-tumor-promoting phorbol esters requires active protein kinase C. J. Virol. 65:6838-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyson, P. J., and P. J. Farrell. 1985. Chromatin structure of Epstein-Barr virus. J. Gen. Virol. 66:1931-1940. [DOI] [PubMed] [Google Scholar]
- 16.Faggioni, A., C. Zompetta, S. Grimaldi, G. Barile, L. Frati, and J. Lazdins. 1986. Calcium modulation activates Epstein-Barr virus genome in latently infected cells. Science 232:1554-1556. [DOI] [PubMed] [Google Scholar]
- 17.Fahmi, H., C. Cochet, Z. Hmama, P. Opolon, and I. Joab. 2000. Transforming growth factor beta 1 stimulates expression of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA by an indirect mechanism which requires the MAPK kinase pathway. J. Virol. 74:5810-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer, D. K., G. Miller, L. Gradoville, L. Heston, M. W. Westrate, W. Maris, J. Wright, J. Brandsma, and W. C. Summers. 1981. Genome of a mononucleosis Epstein-Barr virus contains DNA fragments previously regarded to be unique to Burkitt's lymphoma isolates. Cell 24:543-553. [DOI] [PubMed] [Google Scholar]
- 21.Flamand, L., and J. Menezes. 1996. Cyclic AMP-responsive element-dependent activation of Epstein-Barr virus zebra promoter by human herpesvirus 6. J. Virol. 70:1784-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flemington, E., and S. H. Speck. 1990. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology: molecular biology, pathogenesis, and control, p. 236-284. ASM Press, Washington, D.C.
- 24.Gao, X., K. Ikuta, M. Tajima, and T. Sairenji. 2001. 12-O-tetradecanoylphorbol-13-acetate induces Epstein-Barr virus reactivation via NF-κB and AP-1 as regulated by protein kinase C and mitogen-activated protein kinase. Virology 286:91-99. [DOI] [PubMed] [Google Scholar]
- 25.Hampar, B., J. G. Derge, L. M. Martos, M. A. Tagamets, S. Y. Chang, and M. Chakrabarty. 1973. Identification of a critical period during the S phase for activation of the Epstein-Barr virus by 5-iododeoxyuridine. Nat. New Biol. 244:214-217. [DOI] [PubMed] [Google Scholar]
- 26.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henle, W., and G. Henle. 1968. Effect of arginine-deficient media on the herpes-type virus associated with cultured Burkitt tumor cells. J. Virol. 2:182-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heston, L., M. Rabson, N. Brown, and G. Miller. 1982. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature 295:160-163. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins, P. J., U. K. Binne, and P. J. Farrell. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenson, H. B., and G. Miller. 1988. Polymorphisms of the region of the Epstein-Barr virus genome which disrupts latency. Virology 165:549-564. [DOI] [PubMed] [Google Scholar]
- 31.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 32.Kolman, J. L., N. Taylor, L. Gradoville, J. Countryman, and G. Miller. 1996. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J. Virol. 70:1493-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouzarides, T., G. Packham, A. Cook, and P. J. Farrell. 1991. The BZLF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6:195-204. [PubMed] [Google Scholar]
- 34.Liu, P., S. Liu, and S. H. Speck. 1998. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J. Virol. 72:8230-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, S., A. M. Borras, P. Liu, G. Suske, and S. H. Speck. 1997. Binding of the ubiquitous cellular transcription factors Sp1 and Sp3 to the ZI domains in the Epstein-Barr virus lytic switch BZLF1 gene promoter. Virology 228:11-18. [DOI] [PubMed] [Google Scholar]
- 36.Luka, J., B. Kallin, and G. Klein. 1979. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 94:228-231. [DOI] [PubMed] [Google Scholar]
- 37.Manet, E., H. Gruffat, M. C. Trescol-Biemont, N. Moreno, P. Chambard, J. F. Giot, and A. Sergeant. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, C. L., A. L. Burkhardt, J. H. Lee, B. Stealey, R. Longnecker, J. B. Bolen, and E. Kieff. 1995. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 2:155-166. [DOI] [PubMed] [Google Scholar]
- 39.Miller, G., D. Coope, J. Niederman, and J. Pagano. 1976. Biological properties and viral surface antigens of Burkitt lymphoma- and mononucleosis-derived strains of Epstein-Barr virus released from transformed marmoset cells. J. Virol. 18:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, G., and M. Lipman. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA 70:190-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, G., M. Rabson, and L. Heston. 1984. Epstein-Barr virus with heterogeneous DNA disrupts latency. J. Virol. 50:174-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson, G. R., B. Vroman, B. Chase, T. Sculley, M. Hummel, and E. Kieff. 1983. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J. Virol. 47:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabson, M., L. Heston, and G. Miller. 1983. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc. Natl. Acad. Sci. USA 80:2762-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragoczy, T., and G. Miller. 1999. Role of the Epstein-Barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 73:9858-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riggs, M. G., R. G. Whittaker, J. R. Neumann, and V. M. Ingram. 1977. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 268:462-464. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, K. D., J. Barletta, D. Samid, and R. F. Ambinder. 1995. Pharmacologic activation of expression of immunodominant viral antigens: a new strategy for the treatment of Epstein-Barr-virus-associated malignancies. Curr. Top. Microbiol. Immunol. 194:145-154. [DOI] [PubMed] [Google Scholar]
- 48.Rooney, C., J. G. Howe, S. H. Speck, and G. Miller. 1989. Influence of Burkitt's lymphoma and primary B cells on latent gene expression by the nonimmortalizing P3J-HR-1 strain of Epstein-Barr virus. J. Virol. 63:1531-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowe, D. T., P. J. Farrell, and G. Miller. 1987. Novel nuclear antigens recognized by human sera in lymphocytes latently infected by Epstein-Barr virus. Virology 156:153-162. [DOI] [PubMed] [Google Scholar]
- 51.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 52.Sinclair, A. J., M. Brimmell, F. Shanahan, and P. J. Farrell. 1991. Pathways of activation of the Epstein-Barr virus productive cycle. J. Virol. 65:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takada, K. 1984. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt lymphoma lines. Int. J. Cancer 33:27-32. [DOI] [PubMed] [Google Scholar]
- 54.Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor, N., J. Countryman, C. Rooney, D. Katz, and G. Miller. 1989. Expression of the BZLF1 latency-disrupting gene differs in standard and defective Epstein-Barr viruses. J. Virol. 63:1721-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, N., E. Flemington, J. L. Kolman, R. P. Baumann, S. H. Speck, and G. Miller. 1991. ZEBRA and a Fos-GCN4 chimeric protein differ in their DNA-binding specificities for sites in the Epstein-Barr virus BZLF1 promoter. J. Virol. 65:4033-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Perret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771-15781. [PubMed] [Google Scholar]
- 58.Urier, G., M. Buisson, P. Chambard, and A. Sergeant. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida, M., and S. Horinouchi. 1999. Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann. N. Y. Acad. Sci. 886:23-36. [DOI] [PubMed] [Google Scholar]
- 60.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.zur Hausen, H., F. O'Neil, and U. Freese. 1978. Persisting oncogenic herpesviruses induced by the tumor promoter TPA. Nature 272:373-375. [DOI] [PubMed] [Google Scholar]