Abstract
By analogy with its human nectin1 counterpart, murine nectin1 serves as a cellular receptor for the entry of herpes simplex virus (HSV) into murine cells. HSV entry mediated by either receptor is dependent on the viral glycoprotein D (gD). Whereas human nectin1 binds gD at high affinity and in a saturable manner, murine nectin1 binds gD in a barely detectable fashion, depending on the sensitivity of the assay. The immunoglobulin type V domain of murine nectin differs from its human counterpart in 11 amino acids. To identify the key residues responsible for the high-affinity binding of gD to human nectin1, we replaced each of the 11 divergent amino acids with the human counterparts singly or in groups in an incremental manner. Replacement in murine nectin1 of six amino acids that lie within the gD binding region of human nectin1 (previously mapped to residues 64 to 94, likely the CC′C″ surface) increased the gD binding activity to a limited extent. In contrast, the single P138L substitution, which lies distal from the gD binding site, markedly increased gD binding. This substitution, when coupled with downstream substitutions, exerted the greatest effect. Three-dimensional modeling of the nectin1 V domain suggested that P138 in murine nectin1 might decrease the stability of the V domain by reducing the size of β-strand G. The results support the notion that the overall structure of V nectin1 plays a pivotal role in its ability to bind HSV gD.
Herpes simplex virus (HSV) enters cells by first attaching to cell surface heparan sulfate proteoglycans via the virion glycoproteins glycoprotein B (gB) and gC, after which gD interacts with one of the entry receptors (for reviews, see references 6 and 39). These receptors, which belong to three distinct molecular families, are the nectins1, which are members of the nectin family belonging to the immunoglobulin G (IgG) superfamily (10, 13, 20); HveA (herpesvirus entry mediator A), a member of the tumor necrosis factor receptor family (30); and modified heparan sulfate (38). Nectins1 are broadly expressed in a vast variety of human cells in culture and in human tissues, including tissues targeted by HSV in the course of human infection (10, 13). Nectins also mediate the cell-to-cell spread of HSV (9, 33). Three isoforms of human nectin1 are known (α, β, and γ), of which two contain transmembrane domains while the third is secreted (10, 13, 20, 21). All share the N terminus (the ectodomain in the transmembrane isoforms) constituted by three Ig domains, one of which is V-like and located N-terminally (V) and two of which are C2 type. V carries the region functional in HSV entry and in binding to gD (8, 18). Typically, V-like domains are composed of two β-sheets made of parallel and antiparallel β-strands (named A to G) and two additional C′C″ β-strands. Much of the current focus in elucidating the molecular events which lead to HSV penetration into the cell centers on the identification of key residues involved in the interaction between nectin1 and gD. Recently, we constructed a panel of chimeric receptors where progressively smaller portions of human nectin1 were transferred to the corresponding regions of poliovirus receptor (PVR), a member of the nectin family homologous to nectin1 that is nonfunctional in HSV entry (7). The site on human nectin1 where the HSV entry site and gD binding activities reside was located to the amino acid region spanning residues 64 to 94 (likely to encode the CC′C″ β-strands and intervening loops); within it lies the minimal entry site (residues 77 to 94). Competition binding between gD and a panel of antibodies to human nectin1 also pointed to this region as the gD binding site (17). The homologous region in human nectin2 carries part of the rid1/2 HSV entry site (23).
While humans are the natural hosts of HSV, the virus displays a broad animal host range, and mammalian cells may become infected through animal homologs of the nectin family (28). In particular, the mouse is the small animal model of choice in a variety of experiments, and there has been an interest in determining the molecular basis of its interaction with HSV. The V of the murine nectin1 shares with its human counterpart a 94% identity at the amino acid level (26, 37). Despite the high level of conservation, human and murine nectin1 differ with respect to their binding properties to virions and gD. Thus, the binding of the human isoform is readily detectable and saturable in a number of assays that included binding of soluble forms of receptor and soluble gD in enzyme-linked immunosorbent assays (ELISA), cell ELISA binding of receptor-expressing cells to soluble gD, binding of soluble nectin1 to virions, and gD pulldown by soluble nectin1. In the same assays, the binding of murine nectin was either weak and not saturable or below the limits of detection (25, 26). Studies from another laboratory reported the binding of murine nectin1 to gD (37). The allele examined in that instance (FVB/N) differed from the C3H allele cloned in our laboratory by three amino acids, of which one is P138 in the C3H allele and L138 in the FVB/N allele. Mutagenizing FVB/N to the C3H allele had no effect on gD binding activity (37). Inasmuch as differences between human and murine nectin1 were not investigated in the latter report and the gD binding experiment was carried out at high receptor concentrations (5 μM), the results are not readily compared with ours and the reasons for the discrepancy are unclear at the moment.
Notwithstanding the differences in gD binding activity between murine and human nectin1, the pathway of entry into the cell is gD dependent with either receptor, implying that a high-affinity interaction with gD is not required in order for nectin1 to act as an entry receptor (25). This notion is supported by the finding that HSV, pseudorabies virus (PrV), and bovine herpesvirus 1 utilize a gD-dependent pathway of entry, yet the affinities of their respective gDs to human nectin1 differ by at least three logs of magnitude (11).
The objective of the study reported here was to identify residues on human nectin1 responsible for high-affinity binding to gD. The 11 divergent residues in murine V were replaced singly or in various combinations with the corresponding amino acids of human nectin1 with a gain-of-function approach, i.e., the acquisition of high-affinity gD binding. We also finely mapped the residues on human nectin1 responsible for reactivity to monoclonal antibody (MAb) R1.302, which is specific for the human isoform. Inasmuch as this antibody blocks HSV infection, it represents a powerful tool for the elucidation of the role of nectin1 in the infection of human cells, and its epitope, located in the V domain, must overlap at least in part with the HSV entry site on human nectin1 (8).
MATERIALS AND METHODS
Cells and viruses.
Mammalian cells were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum. The J1.1-2 cell line (J cells), a derivative of BHKtk− cells with high resistance to HSV infection, was described previously (10). The HSV type 1 (HSV-1) recombinant R8102 was constructed by N. Markowitz and B. Roizman (University of Chicago, Chicago, Ill.) by inserting a cassette containing a lacZ gene under the control of the α27 promoter (325-bp BamHI-HinfI fragment from the left end of the HSV-1 BamHI B fragment) at the UL3-UL4 boundary. The virus was derived by homologous recombination in the 3975 recombinant, which carries the thymidine kinase gene inserted at the BamHI site between UL3 and UL4 (2). The PrV recombinant carrying LacZ in place of gH was described previously (1). Virus infectivity was detected as β-galactosidase (β-Gal) activity by light microscopy observation of β-Gal-expressing cells or by reading the optical density at 405 nm after the addition of ONPG (o-nitrophenyl-β-d-galactopyranoside) (10, 30). Sf9 insect cells were cultured in TNM-FH medium (Pharmingen). SF900 II serum-free medium (Life Technologies) was used for heterologous protein expression following recombinant baculovirus infection.
Production of gD(Δ290-299t).
A truncated soluble form of HSV-1 gD, with the same amino acid sequence as gD(Δ290-299t) (31), was engineered by mutagenesis and produced by means of a baculovirus-based expression system. The gD sequence was PCR amplified with the primers sgDbacB5/2 (CCG CAG CAA GGA TCC CTT GGT GGA TGC CTC) and gD290Bg (GGG AGA TCT TAA GGC GTC GCG GCG TCC TGA GGG AAT ATC TTT CCT TGC GGC GCC ACC GTC CCC) by using HSV-1(F) purified DNA as a template and cloned in the BamHI and BglII sites of the baculovirus transfer vector pAcGP67A (Pharmingen), yielding psgDΔ. Sf9 insect cells were cotransfected with psgDΔ and BaculoGold linearized DNA (Pharmingen) according to manufacturer's instructions to obtain a recombinant baculovirus expressing soluble gD(Δ290-299t) under the control of the polyhedrin promoter. Sf9 insect cells seeded in T175 flasks were infected at a multiplicity of infection of 4 PFU/cell and grown in serum-free SF900 II medium (Life Technologies). The supernatant was harvested at 4 days postinfection, and soluble gD was purified by affinity chromatography on a column of MAb30 (5) conjugated to CNBr-Sepharose. Fractions were eluted with 3 M KSCN-10 mM Tris-HCl (pH 7.5)-0.5 M NaCl, dialyzed against phosphate-buffered saline (PBS), and concentrated by using YM3 ultrafiltration membranes with an Amicon ultrafiltration device (Millipore).
Optical biosensor analysis.
Biosensor experiments were performed on an IAsys optical biosensor (Affinity Sensors, Cambridge, United Kingdom) at 25°C. The soluble recombinant forms of murine and human nectin1 fused with human IgG Fc, designated murine nectin1-Fc and human nectin1-Fc, were anchored in two different experiments to the dextran matrix of an IAsys cuvette. The carboxyl groups of the dextran matrix were converted to active N-hydroxysuccinimide esters. Subsequently, the covalent bond between the amine moiety of the protein and the activated matrix was obtained by adding the receptors to 10 mM acetate buffer solution (pH 5.1). The coupling reaction was performed in parallel on the reference cell of the dual-well cuvette without the addition of the soluble receptor. The gD(Δ290-299t) binding experiments were carried out with PBS as a running buffer and were done in parallel in both the sample and reference cells to monitor nonspecific binding to the dextran matrix and the changes of refractive index upon gD(Δ290-299t) addition. The association of gD(Δ290-299t) to the cuvette-immobilized receptor was monitored for 5 min. An anti-human Fc antibody (Dako Laboratories) was used as a positive control for binding to the cell-coupled murine or human nectin1-Fc. The data were collected every 0.3 s and were analyzed by using a global fitting routine with Fast Plot version 3.1.0.17 (Affinity Sensors) and Graph Fit version 5.0.0.36 (Erithacus Software Ltd.) software. The sensorgrams were corrected by subtracting the contribution to the observed signal that arose from nonspecific binding.
Mutagenesis.
Mutagenesis of murine nectin1 was performed with the following primers designed on the murine nectin1 coding sequence: mut_70-71_BamHI (GGT CAC ATG GCA GAA GTC CAC CAA TGG ATC CAA GCA GAA C), mut_78-84_NcoI (CCA AAC AGA ACG TGG CCA TCT ACA ACC CGT CCA TGG GTG TGT CC), mut_91-95_NheI (GTG TGT CCG TGC TAG CTC CCT ACC GCG AGC GAG TGG AGT TCC TG), mut_105-113_Asp718 (CCT CCT TCA CCG ACG GTA CCA TTC GCC TCT CCC GCC TGG AGC TGG), mut_121_NdeI (GGA GGA CGA GGG CGT CTA CAT ATG TGA ATT TGC C), and mut_138_PvuII (GCA ACC GTG AAA GCC AGC TGA ATC TCA CTG TGA TGG). The cDNA of the C3H allelic form of murine nectin1 cloned in the pcDNA3.1(−) vector (Invitrogen) was employed throughout the study (26). The primer L138P_NruI (CGG GCA ATC GCG AAA GCC AGC CCA ATC TCA CGG) was designed on the human nectin1 coding sequence in order to introduce the L138P substitution and was used on human nectin1α cDNA cloned in pcDNA3.1(−). For easiness of screening, all the primers were designed with silent mutations that yielded the recognition sequence for the indicated restriction endonucleases. Mutagenesis was performed with T4 DNA polymerase and T4 DNA ligase followed by digestion of the methylated parental DNA with DpnI and transformation in Escherichia coli mutS strain BMH71-18 (Promega). The mutagenized constructs identified by restriction nuclease pattern were retransformed in E. coli DH5α to avoid rearrangements or random mutations, and V was sequenced for accuracy.
Expression of mutant receptors.
Expression of mutant receptors at the plasma membrane and proper folding of mutant molecules were determined by two assays, reactivity to appropriate antibodies and an infectivity assay. The J cells (10) were transfected with plasmids carrying mutant receptors by using Lipofectamine reagent (Life Technologies) and used 30 h later. Alternatively, cell lines stably expressing the mutant receptors were derived by neomycin G418 selection (1.6 mg of active compound/ml) of transfected J cells for 14 days. For cell surface determination of mutant receptors, stable cell lines were stained with anti-murine nectin1 rat MAb 48-14 (undiluted hybridoma supernatant) followed by fluorescein isothiocyanate (FITC)-conjugated rabbit anti-rat IgG (Sigma) at 1:300 or with MAb CK35 directed to human nectin1 (ascites, 1:100) followed by FITC-conjugated goat anti-mouse antibody (Jackson Immunoresearch Laboratories) at 1:100; the cell lines were then read in a Beckman Coulter FACScan flow cytometer. All MAb dilutions were done in PBS containing 5% fetal bovine serum. For infectivity determinations, transient expressing cells were infected with R8102 or PrV (10 PFU/cell). After 16 h, virus infectivity was detected as β-Gal activity after staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (10, 30).
FACS and fluorescence.
For fluorescence-activated cell sorter (FACS) analysis of gD binding and immunoreactivity to MAb R1.302, stable cell lines expressing the mutant receptors were fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and incubated with 1 μg of affinity-purified recombinant gD(Δ290-299t)/ml or 10 μg of affinity-purified MAb R1.302 IgGs/ml (10, 22). gD binding was revealed by means of MAb H170 (Goodwin Cancer Research Institute, Plantation, Fla.) (1:1,000) followed by a 1:100-diluted FITC-conjugated goat anti-mouse antibody (Jackson Immunoresearch Laboratories). All steps were done in 5% fetal calf serum in PBS for 1 h at 4°C. Cells were subsequently analyzed in a FACScan (Becton-Dickinson) flow cytometer. In each experiment, a gate set on cells transfected with the pcDNA3.1(−) empty vector defined the negative cells and was kept constant for all subsequent samples. The gate containing the positive cells was also set at the beginning of each experiment and kept constant throughout the assays. For each construct, a negative control was run each time, consisting of cells reacted with MAb H170 and secondary antibody but lacking gD or cells reacted with secondary antibody but lacking MAb R1.302. For all cultures, the mean fluorescence ranged from 101 to 102 fluorescence 1 height (FL1-H). The results were expressed as the percentage of positive cells relative to the total number of scored cells (104).
Cell ELISA.
Stable cell lines expressing the mutant receptors grown in 96-well trays were washed twice with PBS and fixed with 4% paraformaldehyde in PBS supplemented with 1% H2O2 for 10 min at room temperature. Unspecific binding was blocked by incubation with 5% fetal calf serum in PBS for 1 h at 4°C. The cells were reacted with increasing concentrations of affinity-purified recombinant gD(Δ290-299t) that was biotinylated as described previously (10). Biotinylated fetuin was included as a control. The binding of biotinylated proteins was revealed by adding ExtrAvidin peroxidase (Sigma) at 1:1,000 in 5% fetal calf serum in PBS followed by o-phenylenediamine as a substrate and monitoring the optical density at 490 nm.
Molecular modeling.
For global sequence alignment, we chose as a model sequence the myelin membrane adhesion molecule P0 (Protein Database [PDB] identification code 1NEU) for two reasons. First, among the molecules whose three-dimensional (3D) structure has been resolved at high resolution (3 Å), P0 is the molecule with the highest sequence homology to the target sequence (36). Second, the 3D structure of PVR bound to poliovirus present in the PDB is known only at 22 Å (backbone level), and furthermore, PVR structure itself was modeled on the P0 structure (3). Modeling was performed essentially according to the following protocol: (i) sequence alignment of the target sequence with the database in order to find the template, (ii) selection of the template with the highest sequence homology to the target, (iii) alignment of the target and the template with LALIGN with the similarity matrix BLOSUM 62 without end gap penalty (http://www.ch.embnet.org/software/LALIGN_form.html [27]) and CLUSTAL W (1.81) with the similarity matrix BLOSUM (http://www2.ebi.ac.uk/clustalw [40]), (iv) building of the 3D structure with MODELLER (34), and (v) evaluation of the goodness of the model with PROCHECK. The secondary structure and the bonding state of the cysteine residues were predicted with SECPRED (http://www.biocomp.unibo.it [15]) and CYSPRED (http://www.biocomp.unibo.it [12]), respectively. The solvent exposure of residues was evaluated with the DSSP program (16). In the comparative model building, for a given alignment, 10 model structures were built and evaluated with the PROCHECK suite of programs (19). Only the best model evaluated was retained after the analysis. RASMOL (35) was used for visualization. Hydrogen bonds were estimated with HBPLUS (24). Salt bridges were computed with the WHAT IF program (http://www.cmbi.kun.nl).
RESULTS
Affinity of murine and human nectin1 to gD.
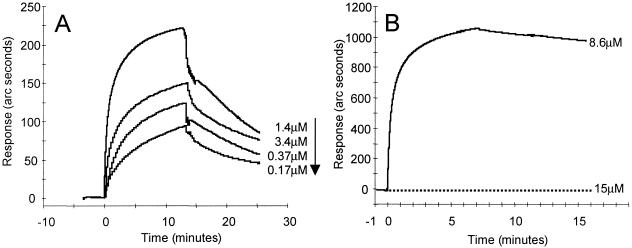
Optical biosensor technology, which measures real-time interaction, was used to quantify the differences in gD binding affinity between murine and human nectin1 receptors. Soluble recombinant forms of murine nectin1 and human nectin1 fused to human IgG Fc, designated murine nectin1-Fc and human nectin1-Fc, respectively, were immobilized to IAsys dual-well carboxymethyl dextran matrix cuvettes. gD(Δ290-299t) was added onto the chips at concentrations ranging from 0.025 to 1.4 μM. gD(Δ290-299t) had a 2.4 × 10−7 M equilibrium dissociation constant (Kd) for its binding to human nectin1-Fc. Under the same conditions, and even at 10-fold higher ligate concentrations, murine nectin1-Fc did not show any significant binding to gD(Δ290-299t) above the level observed in the reference channel devoid of murine nectin1-Fc. This behavior prevented the determination of Kd. The quality control of the immobilized murine nectin1-Fc was performed by the use of anti-human Fc antibody (Dako Laboratories), which displayed a very strong binding. In Fig. 1A, the overlaid sensorgrams show the response plotted as a function of time for gD(Δ290-299t) added at different concentrations (0.17 to 1.4 μM) to the human nectin1-Fc-based cuvette. The response obtained by adding a 15 μM gD(Δ290-299t) solution on murine nectin1-Fc is reported in Fig. 1B together with the sensorgram obtained by binding the control anti-human Fc antibody (8.6 μM). These results are in accordance with those of previous reports in which gD binding was either undetectable or barely detectable, depending on the sensitivity of the assay (25, 26). The Kd of human nectin1-Fc binding to gD is consistent with determinations obtained at another laboratory with a somewhat different construct, i.e., a truncated histidine-tagged human nectin1 (41). A detailed description of the binding properties of human nectin1-Fc, with gD(Δ290-299t) and gD305t as the ligates, is forthcoming (S. Cimitan, L. Menotti, G. Campadelli-Fiume, and C. Bertucci, unpublished data).
FIG. 1.
Real-time analysis of the gD(Δ290-299t) binding to the receptors. (A) Overlaid sensorgrams showing the interaction of decreasing concentrations of gD(Δ290-299t) with human nectin1-Fc; (B) overlaid sensorgrams showing the interaction of gD(Δ290-299t) (dotted line) and the control anti-human Fc antibody (solid line) with murine nectin1-Fc. The graphs represent data from which nonspecific binding was subtracted.
Mutagenesis design.
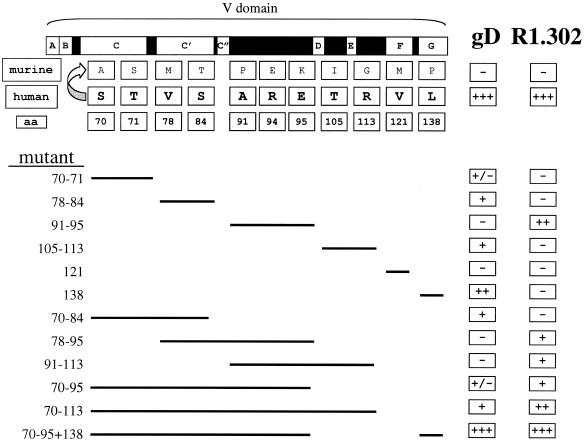
Immunoglobulin domains are rigid structures essentially made of two β-sheets and intervening loops. Their mutagenesis may present special problems, since substitutions of residues located in β-strands can cause loss of function, not because the key residues that mediate the function under study have been modified, but because the structure has been altered. To ensure that our results were not dependent on inadvertent alteration of the V structure, we applied a gain-of-function approach based on the exchange of murine nectin1 residues with the corresponding residues from human nectin1, the acquisition of high-affinity binding to gD, and MAb R1.302 reactivity. The experiment was designed to simultaneously substitute proximal amino acids. The 11 substitutions were thus grouped into six sets, which included the residues 70 and 71; 78 and 84; 91, 94, and 95; 105 and 113; 121; and 138 (Fig. 2). In order to test the effect of multiple substitutions, once a set of residues was exchanged, the mutant was employed as the template for a second round of mutagenesis to yield a second-generation mutant. This incremental approach was applied repeatedly. Figure 1 illustrates the residues which differ between murine and human nectin1, the way in which they were grouped into sets for mutagenesis, and the spectrum of mutants which were generated. The assigned numbers indicate the coordinates of the exchanged residues. The initial murine nectin1 cDNA and, consequently, all mutants were cloned in the pcDNA3.1(−) vector for expression in mammalian cells.
FIG. 2.
Schematic representation of the mutagenesis design and summary of results. The top row shows the schematic linear diagram of nectin1 V. Empty boxes with letters indicate β-strands A to G. Black boxes indicate the intervening loops. The murine and human rows show the divergent residues between murine and human nectin1 V as well as their coordinates and predicted positions. The lines below indicate how proximal substitutions were grouped into sets and show the mutants that were created. The figures in the left column indicate the coordinates of the human residues that were introduced in the murine nectin1. For each mutant, the results are summarized in the two rightmost columns, which report gD binding and MAb R1.302 reactivity. +, −, +/− denote reactivity to different degrees, lack of reactivity, and very weak reactivity, respectively. aa, amino acid.
gD binding activity of mutant receptors.
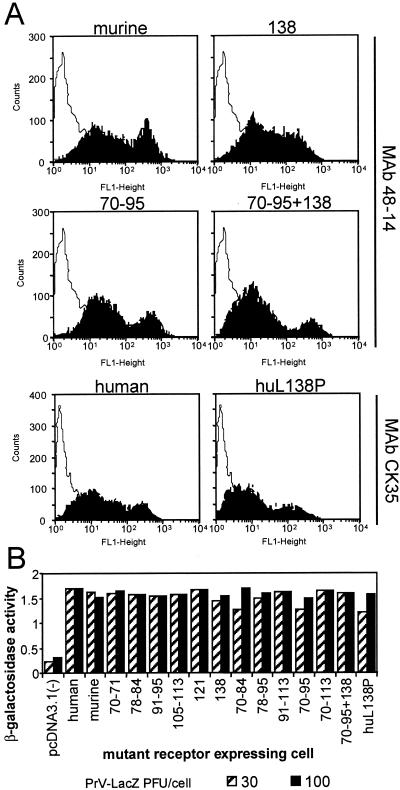
J cells, which are negative for expression of HSV receptors and resistant to HSV entry, were transfected with the mutant plasmids and used in transient expression assays or following selection with neomycin G418 (stable transformants). Preliminarily, we ascertained that all mutant receptors were expressed, appropriately folded, and transported to the plasma membrane in comparable amounts by two approaches. In the first, we measured the cell surface expression of the mutant receptors in the transformants by immunoreactivity to the rat MAb 48-14 directed to murine nectin1 and, for the human nectin1, by immunoreactivity to CK35, a monoclonal antibody directed to the first C2-type domain of human nectin1 (17, 29). The FACS analysis (Fig. 3A) shown for the most representative constructs revealed no major defect in cell surface expression. Specifically, the 138 and the 70-to-95-plus-138 mutants, which show the highest difference in gD binding activity relative to that of the parental murine nectin1 (Fig. 4), show levels of cell surface expression comparable to that of murine nectin1. Similarly, cells expressing the human nectin1 or the L138P mutant thereof show similar levels of cell surface expression as measured with CK35 MAb (Fig. 3A). The second approach was a functional assay and measured the ability of cells expressing the mutant receptors to be infected by HSV or PrV recombinants that carry a LacZ reporter gene (1). Virus entry was monitored as β-Gal activity. Cells expressing each mutant were found to be infectible at comparable levels (results are shown for PrV in Fig. 3B). Cumulatively, the two assays provided evidence that the cells expressing the mutant receptors carried appropriately folded molecules and that there was no major defect in cell surface transport.
FIG. 3.
Cell surface expression of the mutant receptors. (A) Stable cell lines derived from G418 selection of transfected J cells expressing the wild-type or mutant murine nectin1 receptors were reacted with the rat 48-14 hybridoma supernatant followed by anti-rat IgG conjugated with FITC. Stable cell lines expressing human wild-type or mutant receptors were reacted with MAb CK35 followed by anti-mouse IgG conjugated with FITC. Selected examples of fluorescence analyzed in a Becton Dickinson flow cytometer are shown. (B) PrV entry activity of wild-type and mutant receptors. Stable cell lines expressing the indicated constructs were exposed to increasing amounts of PrV-LacZ. After 16 h, β-Gal activity was measured with ONPG as a substrate and absorbance was monitored at 405 nm.
FIG. 4.
Binding of gD(Δ290-299t) to cells expressing the mutant receptors. J cells expressing the indicated constructs were reacted with gD(Δ290-299t) followed by MAb H170 to gD and anti-mouse IgG conjugated with FITC. Fluorescence was analyzed in a Becton Dickinson FACS. The results were expressed as the percentage of positive cells relative to the total number of scored cells. (A) Results are plotted as the average of three independent experiments. Error bars represent standard deviations. (B) Representative examples.
The mutants were subsequently characterized with respect to gD binding activity by measuring the ability of mutant nectin1-expressing cells to bind soluble recombinant gD [gD(Δ290-299t)] (31). This approach has the advantage that the receptor is analyzed in its natural folding, degree of oligomerization, and context, i.e., the cell. Binding was quantified by FACS analysis and by cell ELISA. Results of FACS analyses are quantified in Fig. 4A and depicted in Fig. 4B. Under these conditions, the binding of gD(Δ290-299t) to human nectin1-expressing cells was readily detected, whereas the binding to murine nectin1-expressing cells was undetectable. Substitutions of the sets of residues 70 and 71, 78 and 84, 105 and 113, and 121 increased the gD binding activity to a limited extent. Various combinations of mutations, including the combination by which all of the divergent residues between 70 and 95 were exchanged, also increased gD binding to a limited extent. In some cases, combinations of substitutions decreased, rather than increased, the gD binding (compare the 78-to-84 and 78-to-95 mutants). These results were surprising in light of earlier studies which mapped the gD binding region on human nectin1 to the 64-to-94 region. The unexpected result was that the single P138L substitution conferred high gD binding, to a degree not reached by any of the single mutations or combinations tested. This substitution, when combined with the upstream 70-to-95 substitutions, which by themselves exerted limited effect, completely restored the gD binding activity typical of human nectin1. All cultures were also observed by fluorescence microscopy, which confirmed the FACS results (data not shown).
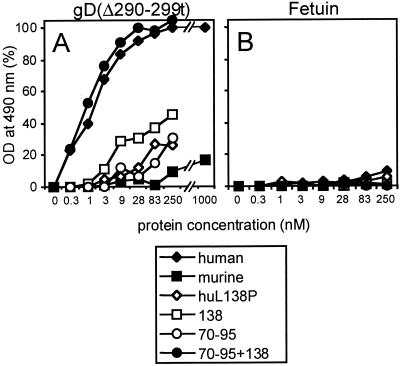
In the cell ELISA, nonpermeabilized cells were reacted with increasing concentrations of biotinylated gD(Δ290-299t) or with biotinylated fetuin as a control. The representative results reported in Fig. 5 are in agreement with the FACS analyses and show that the binding to human nectin1 cells reached a plateau at approximately 50 nM gD, whereas the binding to murine nectin1-expressing cells was barely detectable at 1 μM (Fig. 5). The P138L substitution in murine nectin1 increased the gD binding activity, and the effect was already evident at 10 nM gD. The gD binding was enhanced when both the 138 and the upstream (70-to-95) substitutions were present. The latter substitutions alone exerted a limited effect.
FIG. 5.
Effects of the substitution of the amino acid in position 138 on the binding of gD(Δ290-299t) to murine and human nectin1 receptors. Stable cell lines expressing the indicated mutant receptors seeded in 96-well trays were incubated with increasing concentrations of biotinylated gD(Δ290-299t) (A) or biotinylated fetuin (B) as a control. Binding was revealed by adding ExtrAvidin peroxidase followed by o-phenylenediamine and monitoring the optical density (OD) at 490 nm. The subtracted blank consisted of stable cell lines transfected with the pcDNA 3.1(−) vector.
In order to confirm the role of residue 138, we performed the reverse mutation, i.e., we substituted L138 with P in human nectin1. The binding to gD(Δ290-299t) showed a dramatic reduction in both the FACS analysis (Fig. 4) and the cell ELISA (Fig. 5).
MAb R1.302 epitope.
Reactivity to MAb R1.302 was measured in order to define key residues responsible for the specific reactivity of the antibody to human nectin1. Binding to cells expressing the mutant receptors was quantified by FACS analysis. The results are summarized in Fig. 6A, which also shows representative examples (Fig. 6B). In each case, the results obtained by FACS analysis were confirmed by fluorescence microscopy (data not shown). The residues that conferred MAb R1.302 reactivity on murine nectin1 were 91 to 95. Accordingly, mutants containing multiple substitutions that included the 91-to-95 region (e.g., 91-to-113 and 70-to-113) also displayed antibody reactivity. Furthermore, P138L substitution in the 70-to-95 mutant conferred the highest reactivity, rendering the mutant undistinguishable from human nectin1. Two previous reports identified part of the MAb R1.302 epitope as being located between residues 64 and 102, i.e., extending downstream of the 64-to-94 gD binding site, or between residues 70 and 104 (7, 17). Here we confirmed and extended previous results and identified A91, R94, and E95 as critical residues for reactivity of this human-nectin1-specific antibody. Residue 138 appears to affect the molecule stability or overall conformation and structure, as the highest interaction is observed when L, not P, is present at this position (see Discussion).
FIG. 6.
Reactivity of cells expressing the mutant receptors to MAb R1.302. J cells expressing the indicated constructs were reacted with MAb R1.302 followed by anti-mouse IgG conjugated with FITC. Fluorescence was subjected to FACS analysis. (A) Results are plotted as the average of three independent experiments. Error bars represent standard deviations. (B) Representative examples.
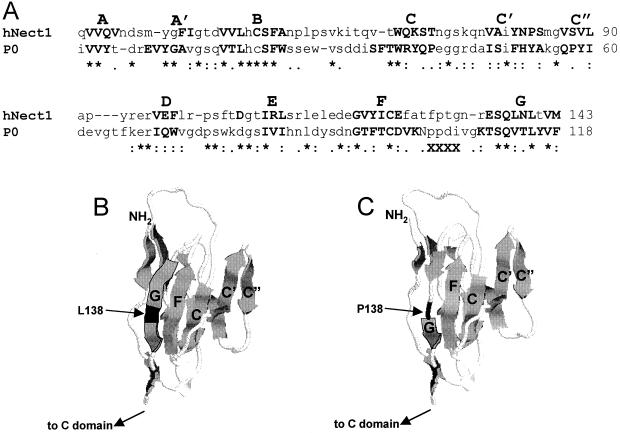
Predictions of structural modifications induced by L138P substitution.
To derive a 3D model of nectin1 V, we applied a procedure based on sequence comparison (building by homology). This same procedure was applied in earlier studies in order to generate the low-resolution structures of PVR bound to poliovirus (3, 14). The template selected on the basis of highest sequence homology was the myelin membrane adhesion molecule P0 (PDB code, 1NEU), whose crystal structure has been determined for the extracellular domain at a 3-Å resolution (36) (Fig. 7). By contrast, the PVR structure has been resolved only at 22 Å, and its predicted model has also been built by homology with P0 myelin (3, 14). Two models were generated: one with L in position 138 (L-nectin1) and the other carrying the P-for-L substitution (P-nectin1) (Fig. 7B and C). The putative low-resolution model of V substantially resembles the Ig-like β sandwich of the template structure. The site interacting with gD comprises the CC′C″ surface identified by genetic studies (7), while L138 is located in β-strand G at the opposite surface in the V model (Fig. 7B). The distance is such that no major direct influence can be foreseen between residue 138 and the CC′C″ surface. The computer model predicts that the overall V structure is conserved when the single point substitution (P for L) is considered. However, P lies outside β-strand G, and thus, β-strand G becomes shorter (Fig. 7C). The L-nectin1 and P-nectin1 models contain 73 and 57 H bonds, respectively (as evaluated with HBPLUS) (24). In addition, the solvent accessibility is lower for the L-nectin1 than for the P-nectin1 model (71 versus 73 nm2). The number of salt bridges does not differ between the two models, being three in both cases. The evaluation of the general physicochemical properties of the two models suggests that the P-for-L substitution may have an overall destabilizing effect on the V structure, as indicated also by the increase in solvent accessibility. The finding that P138L substitution increases the reactivity to MAb R1.302, which is known to recognize a conformation-dependent epitope (compare mutant 70-to-95 and mutant 70-to-95-plus-138), provides genetic support for the computer modeling conclusions that P at position 138 affects V conformation and stability.
FIG. 7.
Computer modeling of the V domain of nectin1. (A) CLUSTAL W alignment of V sequences of human nectin1 (hNect1) with V of myelin membrane adhesion molecule P0 (PDB code, 1NEU). P0 is the template structure; β-strands as taken from the PDB file are shown in boldface. The putative β-strands, as evaluated after building by homology of L nectin, are highlighted in boldface. X's in the third line mark amino acids 103 to 106, which are absent in the P0 PBD file and could not be taken into account for human nectin1 secondary-structure prediction. (B and C) 3D models of the V domain of human nectin 1 with L138 (B) and P138 (C). The effect of the P-for-L substitution in β-strand G is visible. The secondary structure was represented in cartoon form with RASMOL. β-strands are represented as large arrows.
DISCUSSION
In an earlier report, the site on human nectin1 required to mediate HSV-1 entry and exhibiting high-affinity binding to gD was mapped to the 64-to-94 region by exchanging amino acid stretches between human nectin1 and PVR (7). The region likely includes the CC′C″ β-strands and loops and was identified as the gD binding region in binding competition experiments as well (17). The HSVrid1/2 entry site on human nectin2 also maps to this region (23). Eleven amino acids are divergent between the V domains of murine and human nectin1. Six of them reside in the 64-to-94 portion, and we expected that these were the residues capable of conferring high-affinity binding to gD. Surprisingly, substitutions in this group of amino acids affected gD binding affinity to a low extent, whereas the single P138L exchange had a great effect, which was detectable at low gD concentrations. Transferring both the 138 and 70-to-95 human residues conferred on murine nectin1 a gD binding activity indistinguishable from that of human nectin1. The key role played by residue 138 was further strengthened by the reverse L138P substitution on human nectin1, which dramatically decreased gD binding. Alignment of the sequences of nectin1 homologs from human and different animal species shows that L at position 138 is highly conserved in all known human isoforms and animal species (hamster, swine, cow, and monkey), including those known to mediate HSV entry (10, 13, 28), and that only one allele of murine nectin1 carries P138.
The role played by residue 138 in gD binding to nectin1 should be interpreted in light of the V structure. This has not been solved, and it was deduced here by computer modeling by using as the template model the crystallographic structure of myelin membrane adhesion molecule P0 (PDB code, 1NEU), which has been solved at a 3-Å resolution (Fig. 7). As mentioned above, the V structure is typically formed by two interacting β-sheets, of which one is made of β-strands C, F, and G and the other of β-strands D, E, F, and A. The two sheets are joined at one side by the C′C″ β-strands and the C′-C″ loop. At the opposite side lie β-strands G and A. The deduced structure of nectin1 V predicts that L138 is located in β-strand G. This raises the issue of how residue 138 affects gD binding to nectin1, given that it is located distally from the gD-binding site. Computer modeling of the L138P substitution indicates that P is located outside β-strand G whereas L is located within β-strand G, with the net effect of reducing the dimension of β-strand G and the number of hydrogen bonds. In addition, P appears to be more exposed to solvent. It can therefore be suggested that the L138P substitution may decrease the overall stability of V and thus promote a long-range effect on the CC′C″ surface. The present data suggest that the lower stability of the molecule structure affects the interaction of gD binding to nectin1. Genetic evidence in favor of these predictions is provided here by the finding that P138L exchange in the 70-to-95 mutant increased the reactivity to MAb R1.302.
The interaction of PVR with poliovirus has been the subject of numerous studies. The crystallographic structure of poliovirus has been solved (32), and therefore, structural analyses are fairly advanced. Even in the case of PVR, the CC′C″ surface represents the major site that binds poliovirus. Similarly, the CC′ surface represents the site on CD4 that binds human immunodeficiency virus. Cumulatively, these data suggest that this part of V is highly exposed on the cell surface. It is of interest to note that mutational analysis of PVR identified residues distal from the poliovirus binding surface that affect the interaction of the virus with its receptor (4).
Altogether, the results of the present study emphasize the key role played by the overall structure of nectin1 V on its ability to bind HSV gD. This role is also indirectly supported by the finding that soluble forms of V are effective in competitively blocking HSV entry into cells whereas synthetic peptides with sequences identical to that of the 64-to-94 region fail to block HSV entry (F. Cocchi, L. Menotti, M. Lopez, and G. Campadelli-Fiume, unpublished data). The present analysis provides helpful data for the ultimate construction of a molecular model of the interaction between nectin1 and HSV gD, which is still missing.
Acknowledgments
We are indebted to Stefano Salvioli from our department for invaluable assistance in FACS analyses, Gianluca Tasco for biocomputing, Samanta Cimitan for help in biosensor analysis, and Elisabetta Romagnoli for cell cultures. We thank Y. Takai for the gift of MAb 48-14 to murine nectin1, G. Cohen and R. Eisenberg for the gift of MAb CK5, and B. Roizman and T. Mettenleiter for the gifts of R8102 and PrV-LacZ. The IAsys biosensor belongs to the CIRB core facility of the University of Bologna.
The work done at the University of Bologna was supported by Telethon (grant A141), CNR-Agenzia 2000 (grants to L.M. and G.C.-F.), Cofin-MIUR, the Target Project in Biotechnology-CNR, the University of Bologna (60%) and Giovani Ricercatori 2001, and the pluriannual plan.
Footnotes
We dedicate this article to our late friend and colleague, Franco Tatò, University of Rome, for his contributions to virology and particularly for his efforts to have virology included as a discipline in the biological sciences.
REFERENCES
- 1.Babic, N., B. G. Klupp, B. Makoschey, A. Karger, A. Flamand, and T. C. Mettenleiter. 1996. Glycoprotein gH of pseudorabies virus is essential for penetration and propagation in cell culture and in the nervous system of mice. J. Gen. Virol. 77:2277-2285. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhardt, G., J. Harber, A. Zibert, M. deCrombrugghe, and E. Wimmer. 1994. The poliovirus receptor: identification of domains and amino acid residues critical for virus binding. Virology 203:344-356. [DOI] [PubMed] [Google Scholar]
- 5.Brandimarti, R., T. Huang, B. Roizman, and G. Campadelli-Fiume. 1994. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc. Natl. Acad. Sci. USA 91:5406-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi, F., M. Lopez, P. Dubreuil, G. Campadelli-Fiume, and L. Menotti. 2001. Chimeric nectin1-poliovirus receptor molecules identify a nectin1 region functional in herpes simplex virus entry. J. Virol. 75:7987-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi, F., M. Lopez, L. Menotti, M. Aoubala, P. Dubreuil, and G. Campadelli-Fiume. 1998. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc. Natl. Acad. Sci. USA 95:15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi, F., L. Menotti, P. Dubreuil, M. Lopez, and G. Campadelli-Fiume. 2000. Cell-to-cell spread of wild type herpes simplex virus 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin1 (HveC/HIgR/PRR1) and nectin2 (PRR2). J. Virol. 74:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex viruses 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly, S. A., J. J. Whitbeck, A. H. Rux, C. Krummenacher, S. van Drunen Littel-van den Hurk, G. H. Cohen, and R. J. Eisenberg. 2001. Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC(nectin-1) with different affinities. Virology 280:7-18. [DOI] [PubMed] [Google Scholar]
- 12.Fariselli, P., P. Riccobelli, and R. Casadio. 1999. Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins 36:340-346. [PubMed] [Google Scholar]
- 13.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 14.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacoboni, I., P. L. Martelli, P. Fariselli, M. Compiani, and R. Casadio. 2000. Predictions of protein segments with the same aminoacid sequence and different secondary structure: a benchmark for predictive methods. Proteins 41:535-544. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 17.Krummenacher, C., I. Baribaud, M. Ponce De Leon, J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2000. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J. Virol. 74:10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krummenacher, C., A. H. Rux, J. C. Whitbeck, M. Ponce-de-Leon, H. Lou, I. Baribaud, W. Hou, C. Zou, R. J. Geraghty, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1999. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 73:8127-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 20.Lopez, M., F. Cocchi, E. Avitabile, A. Leclerc, J. Adelaide, G. Campadelli-Fiume, and P. Dubreuil. 2001. Novel, soluble isoform of the herpes simplex virus (HSV) receptor nectin1 (or PRR1-HIgR-HveC) modulates positively and negatively susceptibility to HSV infection. J. Virol. 75:5684-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez, M., F. Eberlé, M. G. Mattei, J. Gabert, F. Birg, F. Bardin, C. Maroc, and P. Dubreuil. 1995. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene 155:261-265. [DOI] [PubMed] [Google Scholar]
- 22.Lopez, M., F. Jordier, F. Bardin, L. Coulombel, C. Chabannon, and P. Dubreuil. 1997. CD155 workshop: identification of a new class of IgG superfamily antigens expressed in hemopoiesis, p. 1081-1083. In T. Kishimoto, H. Kikutani, A. E. G. von dem Borne, S. M. Goyert, D. Y. Mason, M. Miyasaka, A. Moretta, K. Okumura, S. Shaw, T. A. Springer, K. Sugamura, and H. Zola (ed.), Leukocyte typing VI, white cells differentiation antigens. Garland Publishing, Inc., New York, N.Y.
- 23.Martinez, W. M., and P. G. Spear. 2001. Structural features of nectin-2 (HveB) required for herpes simplex virus entry. J. Virol 75:11185-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald, I. K., and J. M. Thornton. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238:777-793. [DOI] [PubMed] [Google Scholar]
- 25.Menotti, L., E. Avitabile, P. Dubreuil, M. Lopez, and G. Campadelli-Fiume. 2001. Comparison of murine and human nectin1 binding to herpes simplex virus glycoprotein D (gD) reveals a weak interaction of murine nectin1 to gD and a gD-dependent pathway of entry. Virology 282:256-266. [DOI] [PubMed] [Google Scholar]
- 26.Menotti, L., M. Lopez, E. Avitabile, A. Stefan, F. Cocchi, J. Adelaide, E. Lecocq, P. Dubreuil, and G. Campadelli-Fiume. 2000. The murine homolog of human-Nectin1d serves as a species non-specific mediator for entry of human and animal alpha herpesviruses in a pathway independent of a detectable binding to gD. Proc. Natl. Acad. Sci. USA 97:4867-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4:11-17. [DOI] [PubMed] [Google Scholar]
- 28.Milne, R. S., S. A. Connolly, C. Krummenacher, R. J. Eisenberg, and G. H. Cohen. 2001. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 281:315-328. [DOI] [PubMed] [Google Scholar]
- 29.Mizoguchi, A., H. Nakanishi, K. Kimura, K. Matsubara, K. Ozaki-Kuroda, T. Katata, T. Honda, Y. Kiyohara, K. Heo, M. Higashi, T. Tsutsumi, S. Sonoda, C. Ide, and Y. Takai. 2002. Nectin: an adhesion molecule involved in formation of synapses. J. Cell Biol. 156:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 31.Nicola, A. V., S. H. Willis, N. N. Naidoo, R. J. Eisenberg, and G. H. Cohen. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J. Virol. 70:3815-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, et al. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 33.Sakisaka, T., T. Taniguchi, H. Nakanishi, K. Takahashi, M. Miyahara, W. Ikeda, S. Yokoyama, Y. F. Peng, K. Yamanishi, and Y. Takai. 2001. Requirement of interaction of nectin-1α/HveC with afadin for efficient cell-cell spread of herpes simplex virus type 1. J. Virol. 75:4734-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 35.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374.. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro, L., J. P. Doyle, P. Hensley, D. R. Colman, and W. A. Hendrickson. 1996. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron 17:435-449. [DOI] [PubMed] [Google Scholar]
- 37.Shukla, D., M. C. Dal Canto, C. L. Rowe, and P. G. Spear. 2000. Striking similarity of murine nectin-1alpha to human nectin-1α (HveC) in sequence and activity as a glycoprotein D receptor for alphaherpesvirus entry. J. Virol. 74:11773-11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 39.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitbeck, J. C., M. I. Muggeridge, A. H. Rux, W. Hou, C. Krummenacher, H. Lou, A. van Geelen, R. J. Eisenberg, and G. H. Cohen. 1999. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J. Virol. 73:9879-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]