Abstract
We have studied the replication of plasmids composed of bovine papillomavirus type 1 (BPV1) origin of replication and expression cartridges for viral proteins E1 and E2 in hamster and mouse cells. We found that the replication mode changed dramatically at different expression levels of the E1 protein. At high levels of the E1 protein, overreplication of the origin region of the plasmid was observed. Analysis of the replication products by one-dimensional and two-dimensional gel electrophoresis suggested that initially “onion skin”-type replication intermediates were generated, presumably resulting from initiation of the new replication forks before the leading fork completed the synthesis of the DNA on the episomal plasmid. These replication intermediates served as templates for generation of a heterogeneous set of origin region-containing linear fragments by displacement synthesis at the partially replicated plasmid. Additionally, the linear fragments may have been generated by DNA break-up of the onion skin-type intermediates. Analysis of replication products indicated that generated linear fragments recombined and formed concatemers or circular molecules, which presumably were able to replicate in an E1- and E2-dependent fashion. At moderate and low levels of E1, generated by transcription of the E1 open reading frame using weaker promoters, DNA replication was initiated at much lower levels, which allowed elongation of the replication fork starting from the origin to be more balanced and resulted in the generation of full-sized replication products.
Viruses express replication proteins and virus-encoded polymerases in a precisely regulated fashion. Some of the best examples of viruses capable of such complex regulation are human immunodeficiency virus type 1 (23) and other retroviruses, repetitive element LINE1 from human and rat (21, 37), human hepatitis A virus (18), human hepatitis B virus (14), human hepatitis C virus (63), duck hepatitis B virus (8), cauliflower mosaic virus (56), and many others. Tissue type-specific transcription as well as unusual initiation or elongation of translation of the replication proteins or DNA polymerases is often used by the DNA viruses to achieve precise regulation of their DNA replication. For example, the tissue tropism of papillomaviruses is due in part to epithelial cell-specific expression of E1 and E2 proteins, which allows replication of the virus genome in the cells of this tissue. Human papillomavirus type 18 (HPV18) expresses replication protein E1 from polycistronic mRNA containing E6, E7, and E1 open reading frames (ORFs), where the E1 coding region would be the last to be translated (49).
One of the obvious reasons for a low and strictly regulated level of expression of papillomavirus replication proteins could be the need to hide virus-infected cells from the cellular immune response. This conclusion is supported by the fact that in addition to the down-regulation of viral gene expression, viruses express proteins which actively interfere with processing or presentation of the antigens by the major histocompatibility complex class I pathway. It has been also clearly demonstrated in the case of cottontail rabbit papillomavirus, used as a model system for papillomavirus pathogenesis, that cellular immune responses against early viral proteins, particularly E1, E2 and E6, E7 proteins, are responsible for the regression of the cottontail rabbit papillomavirus-induced papillomas (19, 58) and that infiltration of the papillomas with CD8+ cells leads to their regression (59).
Another reason for the low, well-regulated level of expression of replication proteins could be the ability of these proteins to interact with the cellular regulatory proteins, as it has been shown for many papovavirus proteins (1, 27, 29, 35, 36, 38, 41, 46, 48, 69). Such interactions at high concentrations of respective replication proteins might have a deleterious effect on the cell cycle regulation, which may lead to the premature death of the host cell.
A third possible reason for the low level of expression of the replication proteins in the latently infected cells could be their ability to actively interfere with viral gene expression or with viral DNA replication and lead to abortive replication (4, 11, 24, 31, 53).
The replication cycle of the papillomaviruses is strictly dependent on differentiation of the epithelial tissue (2, 13, 39, 42). Two viral factors encoded by the E1 and E2 ORFs together with the host replication apparatus are necessary and sufficient for the initiation of viral DNA replication during the first amplificational phase and the latent replication phase of the replication cycle. Our studies have indicated that the papillomaviruses use cellular p53 protein for the control of overreplication of the viral genome (32; I. Ilves, M. Kadaja, and M. Ustav, unpublished data). Plasmids carrying the minimal replication origin of bovine papillomavirus type 1 (BPV1) (65) combined with the minichromosome maintenance element (MME), which is composed of the E2 protein multimeric binding sites (47), could be maintained at a stable copy number for prolonged periods in the proliferating cells expressing BPV1 replication proteins E1 and E2. Replication and nuclear retention or segregation-partitioning of the viral genome are required for stable replication of the viral genome in the proliferating cells (22, 30, 60).
We constructed a set of episomally replicating plasmid vectors carrying the BPV1 origin and E1 and E2 expression cartridges. We used different expression cassettes in which the expression of E1 and E2 ORFs was controlled by promoters of different strengths. We found that the viral protein E1 appears to play a major role in modulating the intensity of initiation of replication and formation of replication intermediates. At very high levels of E1 initiator, due to “onion skin”-type replication, a large fraction of the replicated plasmid DNA is present in other than unit-length forms. Our data suggest that such overreplication of the origin region and generation of the aberrant replicational intermediates may have a role in the pathogenic features of high-risk papillomaviruses or in the late phase of the viral life cycle.
Constructs
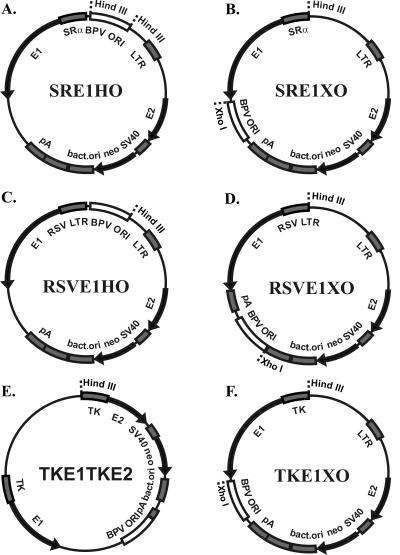
As the basic backbone, we started with the plasmid pBabeNeo (40), into which we inserted BPV1 components at different positions, in different orientations, and of different compositions; the expression cartridges for proteins E1 and E2; and the origin-containing upstream regulatory region (URR) of the BPV1, which was incorporated into an approximately 1-kb fragment (nucleotides [nt] 6959 to 40) (47). The schematic maps of the plasmids are presented in Fig. 1. We designed plasmids so that the location of the origin differed from that in the constructs—in the HindIII site, between promoters driving E1 and E2 expression (Fig. 1A and C) or in an opposite position in the XhoI site downstream of the coding sequences for viral proteins (Fig. 1B, D, and F). The constructs also carried the bacterial origin for propagation in Escherichia coli and the dominant genetic selection marker kanamycin or G418 for bacterial or eukaryotic cells, respectively. For control of the expression of the E1 and E2 proteins, promoters of different strengths—SRα (composed of the simian virus 40 [SV40] early promoter and the R-U5 segment of the human T-cell leukemia virus type 1 long terminal repeat), Rous sarcoma virus 5′ long terminal repeat (RSV LTR), Moloney murine leukemia virus (MoMuLV) LTR, and herpes simplex virus thymidine kinase (TK) promoter—were used. All sequences of plasmids are available upon request.
FIG. 1.
Schematic representation of designed plasmids. The arrows represent the location and direction of expression of genes (E1 gene, E2 gene, neomycin-kanamycin resistance marker gene neo), and the shaded boxes represent transcriptional elements like promoters, polyadenylation sites (pA), and bacterial origin. The open boxes represent the origin regions (BPV URR), which comprise the minimal origins together with the MME. Promoters of different strengths are driving the transcription of E1 and E2 in the constructs SRα, RSV LTR, TK, and MoMuLV LTR.
Expression levels of viral replication proteins
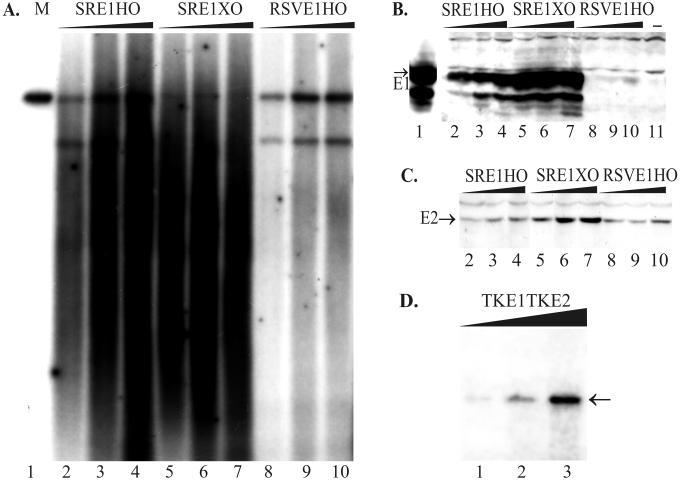
Papillomaviruses seem to use complicated mechanisms for regulation of expression of their replication proteins at transcriptional and posttranscriptional levels. Only low levels of E1 expression have been reported from the steady-state autonomously replicating BPV1 genomes (62). The viral “runaway” DNA amplification and late promoter induction are epithelial cell differentiation-dependent viral functions (3, 15, 16, 24, 50) and are coupled to expression of E1/E4, the major viral late gene product (12). In raft cultures the E2 mRNA levels of HPV31b remain relatively constant, whereas E1 RNA level is up-regulated during the maximal amplification of viral genomes, indicating that E1 may be the major regulator of viral genome amplification (43, 44). Possible regulatory effects of the viral protein E2 can add complexity to the expression pattern of viral genes. The interaction between E1 and E2 is essential in order for E2 to stimulate E1-dependent replication (57). In our transient system, we used Western blotting to analyze the expression levels of the E1 and E2 proteins in different constructs with different promoter strengths in the Chinese hamster ovary cell line CHO (European Collection of Cell Cultures, reference no. 85050302) (Fig. 2B, 2C, and 4C) and in the mouse polyomavirus-transformed C127 cell line COP5. We used either E1-specific monoclonal antibodies (kindly provided by A. Stenlund) or E2-specific monoclonal antibodies (26) for detection of expression of replication proteins, respectively. The signals of E1 and E2 proteins were visualized using peroxidase-conjugated secondary antibody and an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech). High levels of E1 accumulation were achieved in all cases in which the transcription of E1 was under the control of the SRα promoter, irrespective of the location of origin (Fig. 2B, lanes 2 to 7). Considerably lower levels of E1 were detected when the constructs included the RSV promoter (Fig. 2B, lanes 8 to 10), and reduced levels of E1 were detected when constructs included the TK promoter (Fig. 4C, lanes 1 and 5). E2 protein expression was kept under the control of the MoMuLV LTR in most of the constructs (Fig. 1A to D and F) and did not vary much (Fig. 2C). When E2 expression was directed by the TK promoter (Fig. 1E), the protein level was barely detectable (see Fig. 4C, lanes 1 and 5).
FIG. 2.
The replication properties of the designed plasmids are dependent on E1 protein expression. (A and D) A transient-replication assay was carried out with plasmids with promoters of different strengths for E1 in CHO cells. Increasing amounts of plasmid DNA (1, 2, and 5 μg) were transfected by electroporation into CHO cells, and episomal DNA was extracted 72 h later. (A) Purified episomal DNA was digested using linearizing enzymes EcoRI (lanes 2 to 4) or HindIII (lanes 5 to 10 [also panel D]) together with DpnI. Two hundred picograms of linear DNA (SRE1HO) was used as a marker on the blot (lane 1). X-ray film was exposed for 24 h (A) or 48 h (D). The position of unit-sized plasmid is indicated by the arrow (D). (B) Western blot analysis for E1 protein expression from transfected CHO cells. In this experiment, the strong SRα promoter was driving E1 expression in the case of plasmids SRE1HO and SRE1XO, which differ from each other in ori location in the plasmid (lanes 2 to 4 and 5 to 7, respectively). The weaker RSV promoter was driving E1 expression in the plasmid RSVE1HO (lanes 8 to 10). As a marker, the lysate from the COS7 cells, transfected with 0.5 μg of E1 expression construct pCGEag, was used as a positive control (lane 1), and only carrier DNA-transfected CHO cells were used as the negative control (lane 11). (C) Western blot analysis for E2 expression directed by the MoMuLV LTR from the plasmids SRE1HO (lanes 2 to 4), SRE1XO (lanes 5 to 7), and RSVE1HO (lanes 8 to 10).
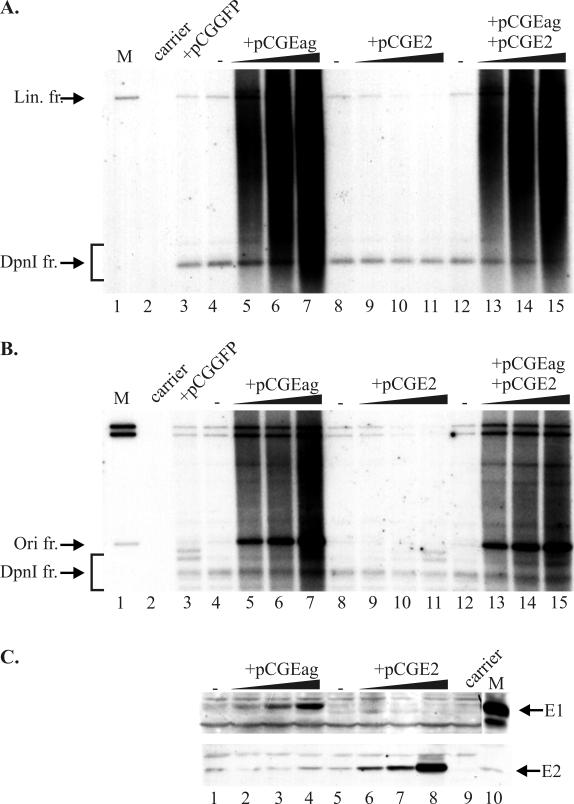
FIG. 4.
Admixture of replication proteins together with TKE1TKE2 vector DNA into CHO cells. (A and B) One microgram of the vector was transfected alone (lanes 4, 8, and 12) or with 0.1, 0.25, or 0.75 μg of the appropriate expression vector—pCGEag (for E1 expression) (lanes 5 to 7) or pCGE2 (for E2 expression) (lanes 9 to 11)—or with two expression vectors together (lanes 13 to 15). Mock-transfected cells (lane 2) and cotransfection with green fluorescent protein expression vector pCGGFP (0.75 μg) (lane 3) were used as negative controls. Cells were harvested 48 h after transfection, and episomal DNA was analyzed by Southern blotting with appropriate size markers (lane 1). Full-length vector DNA (B) and the 1-kb origin fragment (A) were used as probes, respectively. DpnI-resistant replicated DNA fragments as well nonreplicated DpnI-digested material are indicated on both blots. (A) The linearizing enzyme HindIII was used together with DpnI. (B) Enzyme KspAI (cleaves out an ∼1.0-kbp origin fragment) together with DpnI was used for restriction of the replication products. (C) Western blot analyses of expression levels of replication proteins E1 and E2 in samples at 48 h after incubation. Purified E1 and E2 proteins were loaded as positive controls (indicated by arrows, respectively). Abbreviations: Lin. fr., linear fragment; Ori fr., origin sequence-containing fragment.
Replication properties of the constructed plasmids
To determine how the different levels of viral proteins affect the replication of plasmid DNA, we analyzed the replication products in a short-term replication assay of hamster (CHO) and mouse (COP5) cell lines. Different amounts of plasmid DNA were transfected into the cells by electroporation (64) and transfection efficiencies were evaluated in parallel by in situ staining of cells transfected with β-galactosidase-expressing constructs. The analysis of replication products was carried out by Southern blotting 48, 72 or 96 h after transfection. Episomal DNA was harvested by the Hirt procedure (20), and low-molecular-weight DNA was purified and digested with DpnI, to remove dam-methylated input DNA, and with the linearizing enzyme. In the plasmids with the SRα promoter in front of E1, we detected very strong replication signals for both constructs—SRE1HO and SRE1XO (Fig. 2A, lanes 2 to 7). However, there were no unit-sized replication products; products extended from relatively small fragments to products longer than full length. The intensity of the replication signal was considerably lower in plasmids with RSV-directed E1 expression: the smear was much less intensive, and the unit-sized linear fragments were clearly detectable (Fig. 2A, lanes 8 to 10). In the cases in which the TK promoter was driving E1, the replication activity of the vector was much lower (Fig. 2D, lanes 1 to 3; also see Fig. 4A and B), however, the apparent replication signal showed the unit-sized de novo-synthesized DNA. Similar results were obtained with the mouse COP5 cells (data not shown). In some cases, as seen in Fig. 2A, the recombination events have given rise to smaller, distinctively sized products, which replicated nearly as efficiently as full-size plasmids.
The intensity of replication signal correlated with the E1 protein expression level. As seen in Fig. 2B, the increasing amounts of transfected plasmid DNA induced very high levels of the E1 protein in the cases of SRE1HO and SRE1XO, compared to RSVE1HO, while expression of E2 varied much less between the different constructs (Fig. 2C).
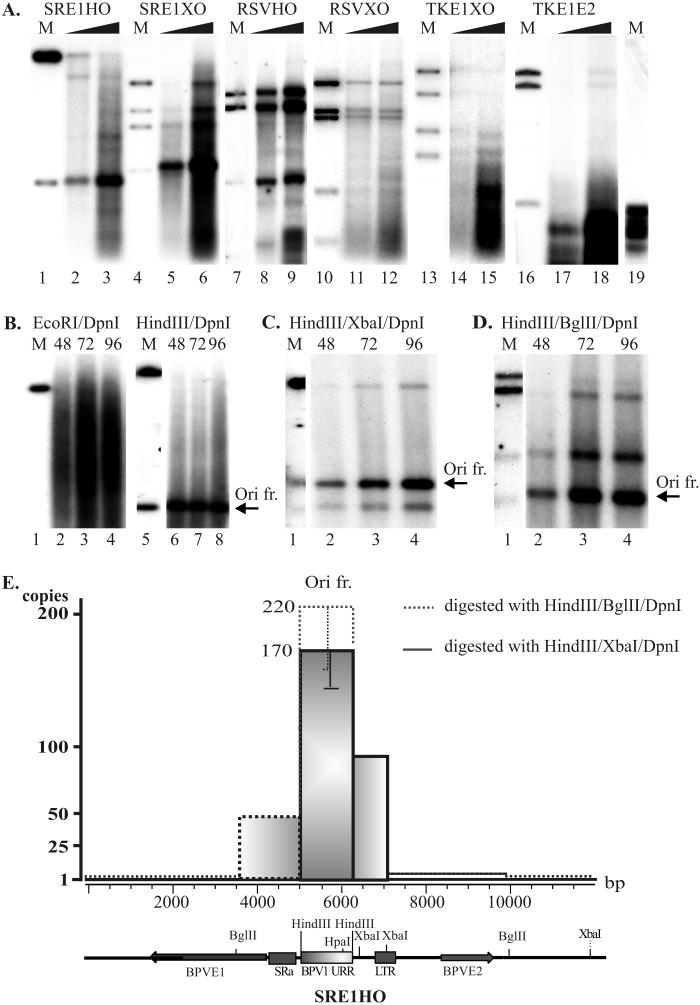
In the CHO cells transfected with plasmid in which the E1 expression was driven by the SRα promoter (SRE1HO), the increasing smear was detected 48, 72, and 96 h after transfection (Fig. 3B). In order to understand the nature of the heterogeneous replication products generated in the cells, we used different combinations of endonucleases to excise the specific fragments from the replicated plasmid to identify which regions of the plasmid were over- or underrepresented in the replicated plasmid. Digestion of the episomal replication products with a linearizing enzyme (EcoRI) generated a family of fragments which became more abundant at later time points (Fig. 3B, lanes 2 to 4). Cleavage with HindIII generated a strong, unit-sized BPV1 origin-containing fragment, which was clearly detectable from the background smear (the arrow points to the HindIII-HindIII origin fragment in Fig. 3B, lanes 6 to 8; also Fig. 3A, lanes 2 and 3). Comparing the approximately 10-kb backbone band to this short ∼1-kb origin band (compare also the ratio for marker bands in Fig. 3B, lane 5), showed that the replication of plasmid has initiated from the BPV1 origin and that a considerable part of the signal was within the excised origin fragment. Using the plasmid in which the origin fragment was transferred to the opposite position in the plasmid, we observed similar amplification of the BPV1 origin area (Fig. 3A, lanes 5 and 6). These data indicate that in the case of both plasmids, assuring a high level of E1 expression resulted in amplificational replication of the BPV1 origin region. Expression of the E1 protein at lower levels induced less-intense amplification (Fig. 3A, lanes 8, 9, 11, 12, 14, 15, 17, and 18).
FIG. 3.
Restriction analysis of replication products. (A) Transient-replication assay of vector constructs with promoters of different strengths driving E1 expression (SRα, RSV, TK) and with two ori locations (in HindIII or in XhoI sites). After transfection of 1 and 2 μg of vector DNA into CHO cells, episomal replication products were isolated, purified, and analyzed 72 h posttransfection using DpnI and different enzyme combinations: HindIII (lanes 2 and 3), MunI/Eco47III (lanes 5, 6, 11, 12, 14, 15, 17, and 18), and MunI/Eco47III/HindIII (lanes 8 and 9). Lanes 1, 4, 7, 10, 13, and 16 contain markers for the input plasmids, cleaved with respective enzymes. (B to D) Analysis of replication products in plasmids with high-level E1 expression (construct SRE1HO) in the CHO cell line, using 1.0 μg of transfected DNA. The cells were harvested 48, 72, and 96 h after transfection, and low-molecular-weight DNA was analyzed on 0.8% agarose gel by Southern blotting. DNA was digested with DpnI and additionally with the linearizing enzyme EcoRI (panel B, lanes 2 to 4); with enzyme HindIII (panel B, lanes 6 to 8), which cleaves out the origin-containing fragment (arrow); or with the enzyme combinations HindIII/XbaI (C) and HindIII/BglII (D). ori-sequence containing bands are shown by arrows. Marker DNAs (400 pg), digested with respective enzymes, are also shown. The blots were exposed for 24 h. (E) Schematic representation of amplification of SRE1HO vector. 32P-labeled dCTP incorporation was normalized to that based on the length of restriction fragments. The number of copies of origin sequence-containing fragments synthesized was calculated by comparing the incorporation of labeled nucleotides in the origin sequence-containing fragment to the incorporation of label into other fragments generated by the enzyme cleavage. A schematic representation of the linearized SRE1HO construct is shown below the graph, where viral origin, coding sequences, promoters, and used restriction enzyme cleavage sites are presented. Ori fr., origin sequence-containing fragment.
Papillomavirus DNA replication in the presence of E1 and E2 has been shown to be bidirectional, starting from the BPV1 origin sequences (65), both in vivo and in vitro. The data provided here once again confirm that fact. We characterized the replication fork movement through the whole plasmid sequence by measuring the average content of replicated material corresponding to different combinations of synthesized fragments using restriction enzymes, moving from the origin in both directions. At high concentrations of the E1 protein, the digestion and analysis of the replicated DNA allowed us to demonstrate that the sequences around the replication origin were considerably amplified (Fig. 3A [lanes 2, 3, 5, and 6] to D). In all cases the strongest signal was picked up in the region of the BPV1 origin fragment (arrows in Fig. 3B to D). Measuring the intensity of replication signals in the appropriate bands and normalizing them to the size of the plasmid fragments allowed us to visualize the abundance of replicated DNA distribution along the plasmids (Fig. 3E). The presented data indicate that DNA de novo synthesis was initiated at the origin of BPV1, irrespective of its location in the plasmid. However, replication is not processive and tends to stall (Fig. 3E). A theta mode of replication in a bidirectional manner requires initiation at every round of replication, and the data presented above clearly show that initiation had been very intensive and depends exclusively on the presence of E1 and E2 proteins. The presented results do not indicate that unspecific initiation of replication outside of the origin region from the transfected plasmids could be detected at high E1 concentrations. Elimination of the E2 protein expression by frameshift abolished any replication initiation from the plasmid, although E1 was expressed at the same levels in the cells (data not shown). At high levels of E1 protein, the origin region may be amplified up to 200 times compared to the sequences located opposite to the origin in the plasmid (Fig. 3E), and the same enhancement (up to 200 copies) could be observed in both origin locations in the plasmid configuration (Fig. 3A, lanes 2, 3, 5, and 6). Our data suggest that new rounds of initiation of DNA replication had started before the previous replication fork had finished synthesis of the leading and lagging strands.
Analysis of replication products by one-dimensional electrophoresis
The data presented above suggest that overreplicated origin-containing fragments were generated as a result of multiple initiations that took place at the same BPV1 replication origin before previous initiations had completed the synthesis. This led to disproportional amplification of certain origin-containing plasmid regions. Many general mechanisms have been described for gene amplification in eukaryotic cells (reviewed in reference 54), and one of them is a disproportional replication, in which a portion of the genome is replicated more than once during a single cell cycle. Consequently, multiple initiations of replication are likely in order to generate free strands of DNA within the replication bubble, a version of which is the onion skin replication model of Botchan et al. (6), described for integrated SV40 genomes. Similar quantitative changes in genome copy number can also be obtained near the origin region in our system at a high level of E1 expression.
Reinitiation of DNA replication was not observed in soluble cell-free systems in an in vitro BPV1 replication assay when the level of DNA synthesis was low (68). At high E1 concentrations the reinitiation could be achieved using COS cell extract, therefore indicating that E1 functions as an initiator of lytic replication (5). Liu et al. (33) have shown that the elongation stage of HPV11 in vitro DNA replication requires E1 and that E2 appears not to be essential for elongation. HPV31 E1 is shown to stimulate replication; in contrast, increased expression of E2 decreased replication (61). Our data show clearly that amplification of origin region is reduced by lowering the expression level of E1. At considerably lower levels of protein from weaker RSV promoters, the initiation of replication was much less efficient, and the elongation of the replication fork resulted in longer replication products, indicating more-balanced initiation and elongation of the replication forks (Fig. 3A, lanes 8, 9, 11, and 12). In the case of plasmids with the E1 expressed from the TK promoter, replication activity of the vector was relatively low, and a single-unit de novo-synthesized plasmid DNA was generated (Fig. 2D; Fig. 3A, lanes 14, 15, 17, and 18; Fig. 4A and B, lanes 4, 8, and 12). However, when a construct such as TKE1TKE2 was cotransfected with increasing amounts of the E1 expression vector pCGEag (65), the amplification of the origin region was induced from the BPV1 origin in this plasmid (Fig. 4A and B, lanes 5 to 7). The same amount of addition of expression vector pCGE2 had no effect, or even weak repression was seen (Fig. 4A and B, lanes 9 to 11). When cotransfected with both expression vectors, the origin plasmid had an identical effect with only E1 added (Fig. 4A and B, lanes 13 to 15). Increasing E1 expression in this experiment amplified the origin region by up to 15, 30, and 80 copies, respectively (Fig. 4B, lanes 5, 6, and 7).
Analysis of replication products by 2D electrophoresis
Appearance of the amplified origin region within replicated DNA by the onion skin-type replication is possible; however, other mechanisms have been suggested to generate amplification products similar to those of certain regions in the genome. Several mechanisms were proposed to explain the various phenomena of gene amplification, and considering the diversity in size and molecular configuration of the amplified sequences, many mechanisms probably could exist in parallel (for reviews, see references 55 and 67). DNA breaks and the following ligation and/or recombination could explain the formation of extrachromosomal circles associated with genomic instability (66). For example, it was demonstrated by two-dimensional (2D) gel electrophoresis that in N-methyl-N′-nitro-N-nitrosoguanidine-treated SV40-transformed CHO cells the viral origin sequences are amplified as circular molecules of various sizes, containing inverted repeats which were suggested to be early amplification products (9). The neutral-neutral 2D electrophoretic analysis, developed by Brewer and Fangman (7) has been shown to be useful for identification of heterogeneous populations of DNA molecules (9), because it separates molecules according to size and topology.
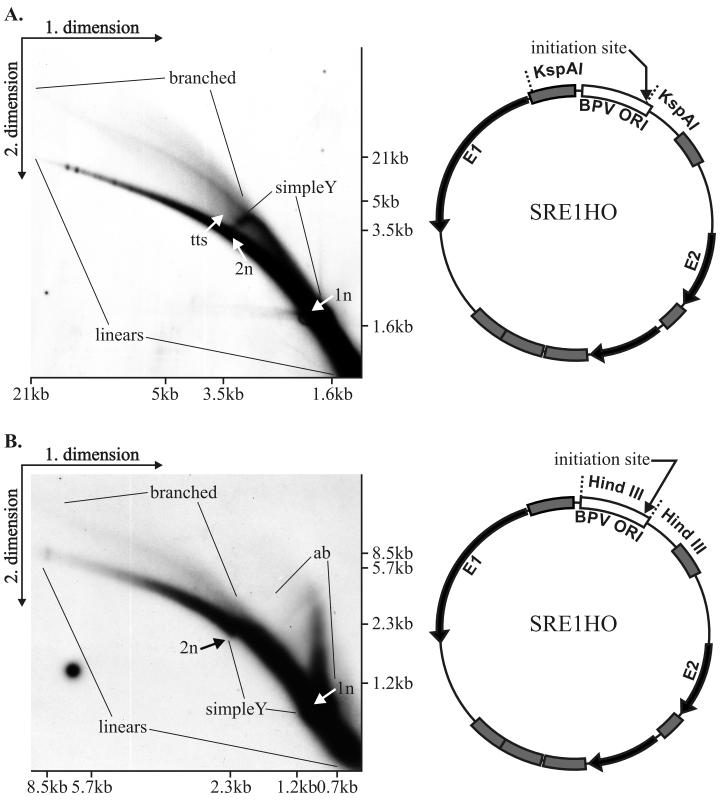
We analyzed the amplified replication products in our system using a similar neutral-neutral 2D gel system. One microgram of the plasmid (SRE1HO) was electroporated into CHO cells. Sixty-five hours posttransfection DNA was prepared by Hirt lysis (20) followed by digestion with KspAI. This enzyme cleaves within the minimal origin region where replication is initiated and within the SRα promoter. The first dimension was run in a 0.5% agarose gel in 1× Tris-borate-EDTA at 0.7 V/cm for 39 h. The second dimension was run in a 1.1% agarose gel in 1× Tris-borate-EDTA at 5.5 V/cm for 9 h with buffer recirculation. Ethidium bromide at a concentration of 0.3 mg/ml was added into the gel and buffer of the second dimension. The separated DNA fragments were transferred onto the membrane and probed with a 32P-labeled BPV1 ORI probe (fragment of the genome from nt 6959 to 40) (Fig. 5A). All 2D gel experiments described below were done by a similar protocol and using the same probe as specified in the figure legends.
FIG. 5.
Neutral-neutral 2D agarose gel electrophoresis analysis of replication products of SRE1HO plasmid in CHO cells. (A) Plasmid DNA (1 μg) was transfected into CHO cells by electroporation, and episomal DNA was extracted by alkaline lysis. Extracted DNA was digested with KspAI, which cleaves the plasmid at the minimal origin region and within the SRα promoter, resulting in the fragment carrying most of the URR. The digested material was separated on the 2D gel, transferred to the nylon filter, and probed with the URR region. The arc of linear molecules, along with positions of double-stranded monomers (1n) (size, 1,650 bp) and dimers (2n), is indicated. In addition, replication intermediates (simple and branched Y structures) and termination structures (tts) were clearly seen on the blot. Appropriate molecular size marker positions in both dimensions are indicated also. The schematic map of the plasmid with recognition sites for KspAI is presented to the right of the blot. The replication initiation site is indicated by an arrow. (B) Analysis of replication intermediates within the 1.1-kb URR fragment. A 2-μg aliquot of SRE1HO plasmid was transfected by electroporation into CHO cells. Episomal DNA was extracted by Hirt lysis 48 h posttransfection, digested with HindIII, and separated by 2D gel electrophoresis. Migration of the monomer (1n)- and dimer (2n)-sized double-stranded DNA fragments on the arc of linear molecules and location of the simple Y arc and branched Y fragments as well as the arc for the asymmetric bubble (ab) are indicated. Recognition sites for HindIII and E1 binding sites (initiation site) are indicated by an arrow on the schematic map of the plasmid. Both blots have been probed with a radiolabeled BPV-1 URR probe (nt 6959 to 40), identical to the BPV Ori region indicated on the vector map.
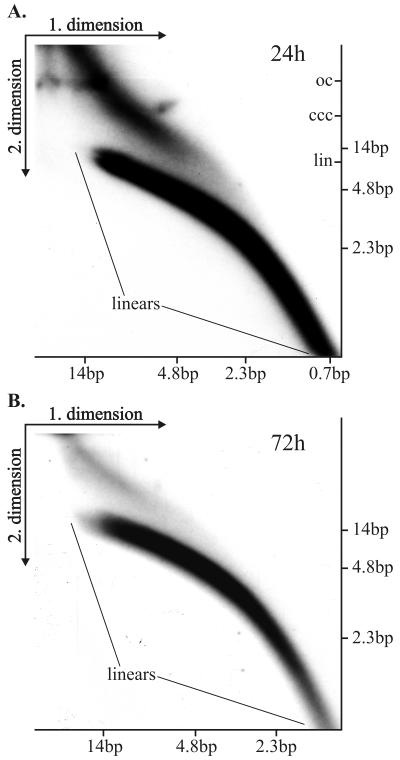
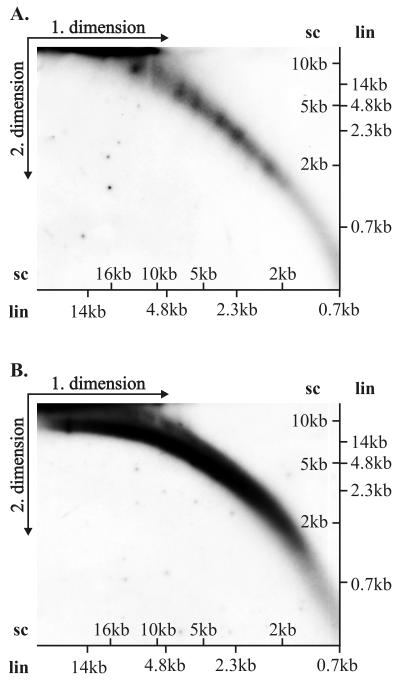
First, on the blot (Fig. 5A) the arc of linear fragments, ranging in size from a couple hundred nucleotides up to 20 kb, can be identified. We speculate that large linear fragments are generated as a result of ligation and rearrangement of linear replication products. Second, the simple arc of the replication fork Y, initiating from the 1N size of the cleaved fragment extending to 2N size, can be identified on the blot, indicating that the BPV1 origin is used actively for initiation of DNA synthesis. Third, some termination structures within this fragment can be also detected, indicating that rereplication could occur in an E1- and E2-dependent fashion, where several origin fragments have been ligated in the same molecule but only a few of them actively fire for synthesis. Fourth, the larger structures, presumably representing various branched onion skin-type replication intermediates, can easily be detected. The cleavage of replication products with HindIII analyzed on 2D gel (Fig. 5B) confirmed that initiation of replication takes place at the usual site around the HpaI site, because the simple arc of the replication forks as well as the asymmetric bubble within the linear fragment could be identified in addition to the arc of linear fragments and branched Y structures. We were surprised by the abundance and size of the linear fragments appearing on the blots. The size of the BPV1 URR-containing fragments was considerably larger than the unit size of the fragment, generated by enzyme cleavage in both cases. We varied considerably the cleavage conditions and took specific care in preparation of the episomal DNA, but the abundance and the size of the linear fragments did not change. The appearance of the very heterogeneous large URR-containing fragments suggested that at least part of these fragments are generated as a result of displacement synthesis from the onion skin-type replication intermediates, followed by ligation and recombination, resulting in the larger-than-unit-sized structures. Additionally, it is also possible that origin-containing linear fragments have been integrated into chromosomal DNA, which leads to amplification of certain genomic regions and results in the linear heterogeneous replication products of large size. To confirm the generation of the linear fragments by the displacement synthesis from the templates, generated from the onion skin replication intermediates, the Hirt procedure-extracted episomal DNA was analyzed without any enzyme treatment. The arc of linear fragments can be detected already 24 h after the transfection, ranging from very small fragments of several hundred base pairs up to the 14-kb fragments (Fig. 6A). In addition, the strong signal of the circular and presumably onion skin-type replication intermediates of heterogeneous size can be detected. Analysis of the episomal DNA extracted from the cells 72 h after transfection showed basically the same appearance of the fragments, though most of the linear fragments appeared as larger molecules (Fig. 6B and data not shown). It is clear that linear fragments can generate circular molecules which can replicate in an E1- and E2-dependent fashion as they contain the BPV1 replication origin. In order to identify the circular molecules within the replication products, we fractionated the Hirt extract prepared from the transfected cells 48 h after transfection, using conventional CsCl density gradient centrifugation in the presence of ethidium bromide (52). Gradient was formed according to the usual procedure using vertical rotor VTI80 in a Beckman ultracentrifuge (L8-M) at 50,000 rpm at 20°C for 24 h. The fraction containing linear fragments and the open circular form of the plasmid and the fraction containing covalently closed circular plasmid were isolated, ethidium bromide was removed, and the fractions were analyzed on the 2D gels. As covalently closed and nicked circles have retarded migration in 2D gel electrophoresis in comparison with linear molecules with the same mass, their migration pattern on 2D gels gives distinct arcs (9). Figure 7B shows the analysis of the fraction containing linear fragments and open circular molecules probed with the URR probe. The arc of the linear fragments and the arc of open circular molecules can be seen on this blot (Fig. 7B). In Fig. 7A, the fraction of covalently closed molecules, analyzed on the parallel 2D gel, did not show any linear fragments, but a variety of the covalently closed circular molecules of distinctive size could be found as the covalently closed circular form of the replication products. These data suggest that indeed some products can circularize and replicate efficiently in an E1- and E2-dependent fashion.
FIG. 6.
Neutral-neutral 2D electrophoresis of uncut plasmid molecules. CHO cells were transfected with 2 μg of SRE1HO plasmid DNA (size ≈ 11 kb). The uncut episomal DNA was extracted at 24 h (A) or at 72 h (B) after transfection and analyzed simultaneously by 2D gel electrophoresis with the appropriate molecular size markers for linear DNA fragments in both dimensions as indicated on the blots. (A) The arcs of the linear molecules are indicated as well the migrating positions of the covalently closed circular (ccc), linear (lin), and relaxed circular (oc) topological forms of the SRE1HO in the second dimension. The blots were hybridized with the BPV1 URR probe (nt 6959 to 40) and exposed for 48 h.
FIG. 7.
Neutral-neutral 2D gel electrophoresis of replication products fractionated by CsCl gradient centrifugation. Extrachromosomal Hirt method-extracted DNA was harvested 48 h after transfection of CHO cells with 2 μg of SRαHO plasmid DNA (∼11 kb) and fractionated by the CsCl-ethidium bromide density gradient. The fractions containing covalently closed circular form and open circular and linear DNA replication products were isolated. The fraction of supercoiled molecules (A) and the linear molecules and the relaxed circles (B) were analyzed simultaneously under the same 2D gel conditions. Positions of marker bands for linear DNA (lin) and for supercoiled DNA (sc) are indicated for both dimensions. Both blots were hybridized with the BPV1 URR probe (nt 6959 to 40).
The data provided above suggest the following mechanism for generating the amplification products at high E1 protein concentrations. Initially, several consecutive replication initiation events per one plasmid molecule occur at high E1 concentrations, generating the so-called onion skin-type replication intermediates. It is logical that the first leading replication fork on the covalently closed circular plasmid has a lower elongation rate due to the torsional stress. This stress can be elevated, for example, by the lack of topoisomerase I at the replication fork due to the squelching of topoisomerase I by the excess of E1 protein (such an interaction between E1 and topoisomerase I can occur, as has been shown for large T antigen and topoisomerase I [17]). The replication forks initiated on the newly synthesized double-stranded DNA have no such constraints and can easily catch up with the leading fork. As a result, a very unstable structure is formed, especially when the next replication fork collides into the stalled previous fork. The newly replicated strands can easily break off the plasmid, resulting in replication products which can be subjects for many possible reactions, such as ligation, recombination, recircularization, and others, including being a template for further replication due to the presence of a replication origin. The inverted repeat intermediates run on a 2D gel behave in a manner similar to two-strand linear fragments. Breaks, ligations and recombinations could explain the formation of massive heterogeneous amplified products, generating the pool of linear fragments and circular molecules, detected as a family of heterogeneous replication products. Similar generations of the heterogeneous replication products have been described for the SV40 and for polyomavirus replication in the nonpermissive cells. Populations of SV40 defective genomes were shown to be supercoiled and relaxed circular DNAs of heterogenous sizes (9, 28). In some cases the formation of extrachromosomal circles with inverted repeats was proposed to be one of the initial steps of gene amplification (45, 51, 66, 67).
Clearly, random overreplication of a portion of ori plasmid—the unstably amplified state—is not beneficial for the stability of the viral genome. Therefore, it would be vital for the virus to maintain the expression of viral replication protein E1 at a low, controlled level. HPV genomes usually persist as episomal molecules in HPV-associated preneoplastic lesions, whereas they are frequently integrated into the host cell genome with random distribution in HPV-related cancer cells (34). Integration of viral genome is a rare and rate-limiting event in transformation and usually takes place in the case of high-risk papillomaviruses. Papillomavirus sequences detected from cancer cell lines or from specimens are often different from their expected full size (10, 25, 34) but, however, consistently contain transcriptionally active viral oncogenes E6 and E7. In light of the out-of-schedule firing of the origin at high E1 concentrations, we propose that onion skin structure intermediates and the conversion from onion skins to linear fragments carrying E6 and E7 coding sequences, together with the integration of such sequences into the host DNA, may generate one mechanism for stable oncogenic transformation of high-risk HPV-infected cells. To avoid such an unstably amplified state, papillomaviruses have to express E1 protein at a low, regulated level, at least in latently infected cells. However, high levels of protein may be instrumental for inducing a metastable genome to switch from the bidirectional replication to the rolling-circle replication mode. It has been suggested that a switch from a theta mode to a rolling-circle replication takes place during the vegetative phase of the high-risk papillomaviruses (13).
Further studies are needed to determine whether there are numerous or unique recombinational joints or whether there is a sequence specificity for sites of recombination. More specifically, would the cellular environment influence the respective changes? In principle, the experimental system described here could be used as a functional assay for the events leading to the generation and processing of the replication intermediates and for the study of the processes of recombination-repair.
Acknowledgments
We thank Arne Stenlund for providing BPV1 E1-specific monoclonal antibodies, Anne Kalling for excellent technical assistance in cell culture work, and undergraduate students Mihkel Allik and Olga Romantsova for their first-time and successful assistance. We are grateful to Tiina Sedman for introducing us to 2D analysis.
This study was supported by grants 4475 and 4476 from the Estonian Science Foundation, grant INTNL55000339 from the Howard Hughes Medical Institute, and grant CT96-0918 from the European Union.
REFERENCES
- 1.Amin, A. A., S. Titolo, A. Pelletier, D. Fink, M. G. Cordingley, and J. Archambault. 2000. Identification of domains of the HPV11 E1 protein required for DNA replication in vitro. Virology 272:137-150. [DOI] [PubMed] [Google Scholar]
- 2.Barksdale, S., and C. Baker. 1993. Differentiation-specific expression from the bovine papillomavirus type 1 P2443 and late promoters. J. Virol. 67:5605-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedell, M. A., J. B. Hudson, T. R. Golub, M. E. Turyk, M. Hosken, G. D. Wilbanks, and L. A. Laimins. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J. Virol. 65:2254-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belyavskyi, M., M. Westerman, L. DiMichele, and V. Wilson. 1996. Perturbation of the host cell cycle and DNA replication by the bovine papillomavirus replication protein E1. Virology 219:206-219. [DOI] [PubMed] [Google Scholar]
- 5.Bonne-Andrea, C., S. Santucci, and P. Clertant. 1995. Bovine papillomavirus E1 protein can, by itself, efficiently drive multiple rounds of DNA synthesis in vitro. J. Virol. 69:3201-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botchan, M., W. Topp, and J. Sambrook. 1979. Studies on simian virus 40 excision from cellular chromosomes. Cold Spring Harbor Symp. Quant. Biol. 43:709-719. [DOI] [PubMed] [Google Scholar]
- 7.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 8.Chang, L. J., P. Pryciak, D. Ganem, and H. E. Varmus. 1989. Biosynthesis of the reverse transcriptase of hepatitis B viruses involves de novo translational initiation not ribosomal frameshifting. Nature 337:364-368. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, S., and S. Lavi. 1996. Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol. Cell. Biol. 16:2002-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corden, S. A., L. J. Sant-Cassia, A. J. Easton, and A. G. Morris. 1999. The integration of HPV-18 DNA in cervical carcinoma. Mol. Pathol. 52:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demeret, C., S. Goyat, M. Yaniv, and F. Thierry. 1998. The human papillomavirus type 18 (HPV18) replication protein E1 is a transcriptional activator when interacting with HPV18 E2. Virology 242:378-386. [DOI] [PubMed] [Google Scholar]
- 12.Doorbar, J., R. C. Elston, S. Napthine, K. Raj, E. Medcalf, D. Jackson, N. Coleman, H. M. Griffin, P. Masterson, S. Stacey, Y. Mengistu, and J. Dunlop. 2000. The E1E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J. Virol. 74:10081-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, E., and P. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouillot, N., S. Tlouzeau, J. M. Rossignol, and O. Jean-Jean. 1993. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 67:4886-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gai, D., R. Roy, C. Wu, and D. T. Simmons. 2000. Topoisomerase I associates specifically with simian virus 40 large-T-antigen double hexamer-origin complexes. J. Virol. 74:5224-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glass, M. J., X. Y. Jia, and D. F. Summers. 1993. Identification of the hepatitis A virus internal ribosome entry site: in vivo and in vitro analysis of bicistronic RNAs containing the HAV 5′ noncoding region. Virology 193:842-852. [DOI] [PubMed] [Google Scholar]
- 19.Han, R., N. M. Cladel, C. A. Reed, X. Peng, and N. D. Christensen. 1999. Protection of rabbits from viral challenge by gene gun-based intracutaneous vaccination with a combination of cottontail rabbit papillomavirus E1, E2, E6, and E7 genes. J. Virol. 73:7039-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 21.Ilves, H., O. Kahre, and M. Speek. 1992. Translation of the rat LINE bicistronic RNAs in vitro involves ribosomal reinitiation instead of frameshifting. Mol. Cell. Biol. 12:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 24.Klumpp, D. J., and L. A. Laimins. 1999. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology 257:239-246. [DOI] [PubMed] [Google Scholar]
- 25.Kook, J. K., J. H. Kim, and B. M. Min. 1998. Activity of human papillomavirus type 16 P97 promoter in immortal and tumorigenic human oral keratinocytes. Int. J. Oncol. 13:765-771. [DOI] [PubMed] [Google Scholar]
- 26.Kurg, R., J. Parik, E. Juronen, T. Sedman, A. Abroi, I. Liiv, U. Langel, and M. Ustav. 1999. Effect of bovine papillomavirus E2 protein-specific monoclonal antibodies on papillomavirus DNA replication. J. Virol. 73:4670-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, M. C., B. H. Teh, and W. Y. Tarn. 1999. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 274:11832-11841. [DOI] [PubMed] [Google Scholar]
- 28.Lavi, S., and E. Winocour. 1972. Acquisition of sequences homologous to host deoxyribonucleic acid by closed circular simian virus 40 deoxyribonucleic acid. J. Virol. 9:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, D., H. Sohn, G. V. Kalpana, and J. Choe. 1999. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature 399:487-491. [DOI] [PubMed] [Google Scholar]
- 30.Lehman, C., and M. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Moal, M., M. Yaniv, and F. Thierry. 1994. The bovine papillomavirus type 1 (BPV1) replication protein E1 modulates transcriptional activation by interacting with BPV1 E2. J. Virol. 68:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lepik, D., I. Ilves, A. Kristjuhan, T. Maimets, and M. Ustav. 1998. p53 protein is a suppressor of papillomavirus DNA amplificational replication. J. Virol. 72:6822-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J., S. Kuo, T. Broker, and L. Chow. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283-27291. [DOI] [PubMed] [Google Scholar]
- 34.Luft, F., R. Klaes, M. Nees, M. Durst, V. Heilmann, P. Melsheimer, and M. von Knebel Doeberitz. 2001. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int. J. Cancer 92:9-17. [PubMed] [Google Scholar]
- 35.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massimi, P., D. Pim, C. Bertoli, V. Bouvard, and L. Banks. 1999. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene 18:7748-7754. [DOI] [PubMed] [Google Scholar]
- 37.McMillan, J. P., and M. F. Singer. 1993. Translation of the human LINE-1 element, L1Hs. Proc. Natl. Acad. Sci. USA 90:11533-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melendy, T., J. Sedman, and A. Stenlund. 1995. Cellular factors required for papillomavirus DNA replication. J. Virol. 69:7857-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyers, C., T. Mayer, and M. Ozbun. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J. Virol. 71:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor, M. J., W. Stunkel, C. H. Koh, H. Zimmermann, and H. U. Bernard. 2000. The differentiation-specific factor CDP/Cut represses transcription and replication of human papillomaviruses through a conserved silencing element. J. Virol. 74:401-410. [PMC free article] [PubMed] [Google Scholar]
- 42.Ozbun, M., and C. Meyers. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozbun, M. A., and C. Meyers. 1998. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology 248:218-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozbun, M. A., and C. Meyers. 1998. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J. Virol. 72:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passananti, C., B. Davies, M. Ford, and M. Fried. 1987. Structure of an inverted duplication formed as a first step in a gene amplification event: implications for a model of gene amplification. EMBO J. 6:1697-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng, Y. C., D. E. Breiding, F. Sverdrup, J. Richard, and E. J. Androphy. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 74:5872-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 48.Rank, N., and P. Lambert. 1995. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 69:6323-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remm, M., A. Remm, and M. Ustav. 1999. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 73:3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruesch, M. N., F. Stubenrauch, and L. A. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz, J. C., and G. M. Wahl. 1988. Formation of an inverted duplication can be an initial step in gene amplification. Mol. Cell. Biol. 8:4302-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 53.Sandler, A., S. Vande Pol, and B. Spalholz. 1993. Repression of bovine papillomavirus type 1 transcription by the E1 replication protein. J. Virol. 67:5079-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schimke, R. T. 1984. Gene amplification in cultured animal cells. Cell 37:705-713. [DOI] [PubMed] [Google Scholar]
- 55.Schimke, R. T. 1988. Gene amplification in cultured cells. J. Biol. Chem. 263:5989-5992. [PubMed] [Google Scholar]
- 56.Schultze, M., T. Hohn, and J. Jiricny. 1990. The reverse transcriptase gene of cauliflower mosaic virus is translated separately from the capsid gene. EMBO J. 9:1177-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selvakumar, R., L. A. Borenstein, Y. L. Lin, R. Ahmed, and F. O. Wettstein. 1995. Immunization with nonstructural proteins E1 and E2 of cottontail rabbit papillomavirus stimulates regression of virus-induced papillomas. J. Virol. 69:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selvakumar, R., A. Schmitt, T. Iftner, R. Ahmed, and F. O. Wettstein. 1997. Regression of papillomas induced by cottontail rabbit papillomavirus is associated with infiltration of CD8+ cells and persistence of viral DNA after regression. J. Virol. 71:5540-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stubenrauch, F., H. B. Lim, and L. A. Laimins. 1998. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J. Virol. 72:1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun, S., L. Thorner, M. Lentz, P. MacPherson, and M. Botchan. 1990. Identification of a 68-kilodalton nuclear ATP-binding phosphoprotein encoded by bovine papillomavirus type 1. J. Virol. 64:5093-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wahl, G. M. 1989. The importance of circular DNA in mammalian gene amplification. Cancer Res. 49:1333-1340. [PubMed] [Google Scholar]
- 67.Windle, B. E., and G. M. Wahl. 1992. Molecular dissection of mammalian gene amplification: new mechanistic insights revealed by analyses of very early events. Mutat. Res. 276:199-224. [DOI] [PubMed] [Google Scholar]
- 68.Yang, L., R. Li, I. Mohr, R. Clark, and M. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 69.Yao, J. M., D. E. Breiding, and E. J. Androphy. 1998. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 72:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]