Abstract
In this study, we have investigated the effect of specific mutations in human immunodeficiency virus type 1 (HIV-1) envelope (Env) on antibody production in an effort to improve humoral immune responses to this glycoprotein by DNA vaccination. Mice were injected with plasmid expression vectors encoding HIV Env with modifications in regions that might affect this response. Elimination of conserved glycosylation sites did not substantially enhance humoral or cytotoxic-T-lymphocyte (CTL) immunity. In contrast, a modified gp140 with different COOH-terminal mutations intended to mimic a fusion intermediate and stabilize trimer formation enhanced humoral immunity without reducing the efficacy of the CTL response. This mutant, with deletions in the cleavage site, fusogenic domain, and spacing of heptad repeats 1 and 2, retained native antigenic conformational determinants as defined by binding to known monoclonal antibodies or CD4, oligomer formation, and virus neutralization in vitro. Importantly, this modified Env, gp140ΔCFI, stimulated the antibody response to native gp160 while it retained its ability to induce a CTL response, a desirable feature for an AIDS vaccine.
Plasmid DNA vaccination has been a useful technology for the development and analysis of immunogens. This method of vaccination allows relevant posttranslational modifications, appropriate intracellular trafficking, and antigen presentation. Direct injection of naked DNA either intramuscularly or intradermally readily induces protective immune responses in animal models. Though DNA vaccines readily elicit cell-mediated immune responses, their ability to induce high-titer antibody responses has been limited, particularly to human immunodeficiency virus type 1 (HIV-1) envelope (Env). However, plasmid expression vectors can be readily modified to express different forms of HIV envelope proteins, enabling rapid and systematic testing of alternative vaccine immunogens.
To improve the immune response to native gp160 and to expose the core protein for optimal antigen presentation and recognition, we have analyzed the immune response to modified forms of the protein. The conserved N-linked glycosylation sites previously suggested to limit the antibody response (39) were comprehensively analyzed. In addition, the important coiled-coil hairpin region involved with formation of fusion intermediates has been studied. Expression vectors with deletions in the cleavage site (C), the fusion peptide (F), and the interspace (I) between the two heptad repeats, termed ΔCFI deletions, were prepared. In this report, the immune response to Env candidates expressed in plasmids with codons modified to improve gene expression has been analyzed. Both antibody and cytotoxic-T-lymphocyte (CTL) responses were evaluated after injection of plasmid DNA into muscle. A modified gp140 DNA has been identified that better elicits antibody responses at the same time that it retains its capacity to induce CTL responses to HIV Env. This prototype may facilitate the identification of immunogens which can elicit broadly neutralizing antibody responses to HIV by gene-based vaccination.
MATERIALS AND METHODS
Immunogens.
Plasmids expressing the CXCR4-tropic HIV-1 HXB2 Env were made synthetically with sequences designed to disrupt viral RNA structures that limit protein expression by using codons typically found in human cells (1, 37, 38, 41, 42, 47). Briefly, the synthetic env gene of HXB2 (GenBank accession number K03455) was generated in three fragments by assembling the overlapping synthetic oligonucleotides using PCR amplification. Glycosylation mutants were generated by site-directed mutagenesis to replace asparagine with glutamic acid residues in a block of either 11 or 17 conserved glycosylation sites between amino acids 88 and 448 (Fig. 1). To produce a CCR5-tropic version of the HIV-1 envelope, the most divergent region encoding amino acids 275 to 361 of HXB2 (CXCR4-tropic) gp160 from Bal was replaced with CCR5-tropic HIV-1 BaL sequence (GenBank accession number M68893), which includes the V3 loop. To express truncated mutant Env proteins, stop codons were introduced after positions 752, 704, 680, or 592 to produce gp150, gp145, gp140, or gp128, respectively. The Env protein was further changed by deleting amino acids 503 to 537 and amino acids 593 to 619, which removes the cleavage site sequence, the fusion domain, and a part of the spacer between the two heptad repeats. All of these mutations were confirmed by sequencing of both strands of the cDNAs. The structures of the synthetic HIV envelope genes are shown (Fig. 1). The cDNAs were cloned in the expression vector pVR1012 (56) under the control of the cytomegalovirus immediate-early enhancer, promoter, and first intron. Sequence analysis indicated that the codon-optimized envelope contained the following minor point substitutions: F53L, N94D, K192S, I215N, A224T, A346D, P470L, T723I, and S745T.
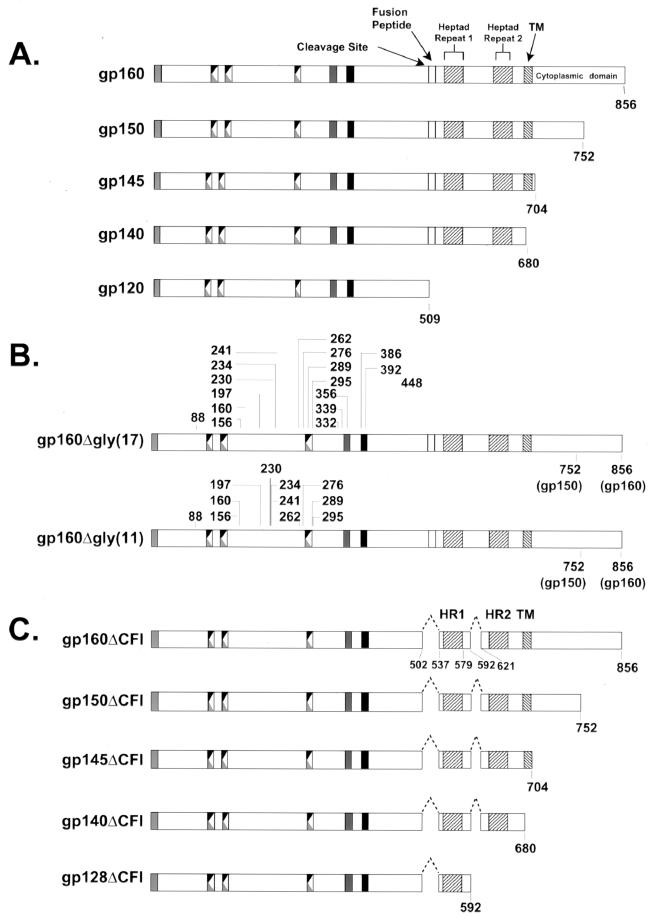
FIG. 1.
Schematic representation of functional domains and mutations in HIV-1 Env glycoproteins. Full-length envelope polyprotein, gp160, with the indicated features based on the amino acid residues of HXB2 is shown (top). Functional domains include the gp120/gp41 cleavage site (residues 510 and 511), the fusion domain (residues 512 to 527), the two heptad repeats (residues 546 to 579 and residues 628 to 655), the transmembrane domain (residues 684 to 705), and the cytoplasmic domain (residues 706 to 856). The mutant forms of the envelope proteins are shown below the structure of gp160. COOH deletions were introduced that terminate the envelope protein at positions 752, 704, or 680 to produce gp150, gp145, or gp140, respectively. Two internal deletions that removed the cleavage site, the fusion domain, and the region between the two heptad repeats were introduced into gp160, gp150, gp145, and gp140. A further deletion in the COOH-terminal region at position 592 removed the second heptad repeat and the transmembrane domain to produce gp128ΔCFI. To disrupt potential glycosylation sites, asparagine (N) residues at 11 positions (88, 156, 160, 197, 230, 234, 241, 262, 276, 289, and 295) were replaced with aspartic acid (D) residues in both gp160 and gp150. Versions of both gp160 and gp150 were created with a total of 17 mutated glycosylation sites by including six additional N-to-D substitutions at positions 332, 339, 356, 386, 392, and 448.
Expression of envelope proteins in transfected cells.
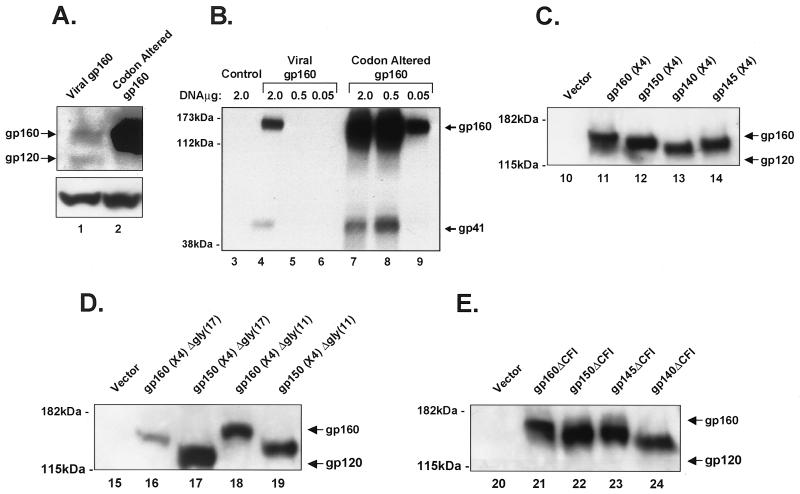
293 cells (106) were plated in 60-mm-diameter dishes. Cells were transfected on the following day with 2 μg of plasmid using calcium phosphate (16, 56). Cells were harvested 48 h after transfection using phosphate-buffered saline (PBS) containing 2 mM EDTA (PBS-EDTA). Cells were lysed in buffer containing 50 mM HEPES (pH 7.0), 250 mM NaCl, and 0.5% NP-40. The protein concentration in the lysates was determined using the Bradford assay (Bio-Rad, Hercules, Calif.), and the protein (25 μg) was analyzed by separation on 7.5% sodium dodecyl sulfate (SDS)-7.5% polyacrylamide gel and transfer to Immobilon-P membrane (Millipore, Bedford, Mass.). The antibody response to mutant Env was detected by immunoprecipitation with sera from mice followed by Western blotting (Fig. 2A, C, D, and E) with polyclonal antibody against gp160 (Intracel, Rockville, Md.). β-Actin was used as an internal control with an antibody to this gene product (Sigma, St. Louis, Mo.). As shown in Fig. 2B, 293 cells were transfected either with 2 μg of control vector or various amounts (2.0, 0.5, and 0.05 μg) of vector expressing either viral gp160 or codon-altered gp160 using the protocol above. Cells were harvested 48 h after transfection and lysed in buffer containing 50 mM HEPES (pH 7.0), 250 mM NaCl, and 0.5% NP-40. Processing of Env was detected as described above by Western blotting using supernatant from a hybridoma producing monoclonal antibody against gp41 (HIV-1 IIIB gp41 hybridoma [chessie 8]; NIH AIDS Research and Reference Reagent Program [kindly provided by George Lewis]).
FIG. 2.
Comparison of the expression of the HIV-1 gp160 with codon-optimized gp160. (A and B) Expression of plasmids encoding Rev-dependent and Rev-independent codon-modified gp160 (lanes 1 and 2). (A) The upper panel shows expression of Rev-dependent viral gp160 (left) and codon-modified gp160 (right) in transfected 293 cells. The lower panel shows comparable expression of β-actin in these transfected cells. (B) Processing of gp160 was detected with a monoclonal antibody to gp41with viral or codon-altered gp160, as indicated (lanes 3 to 9). (C) Expression of mutant CXCR4-tropic HIV Env glycoproteins with COOH-terminal truncations is shown. (D and E) CXCR4-tropic envelope proteins containing mutant glycosylation sites and mutant functional domains are shown. The indicated proteins were detected by immunoblotting as described above. Cell lysates produced by transfection with vector containing no insert were used as controls (vector, first lane in each panel).
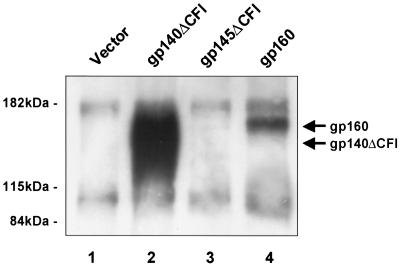
Expression of soluble ΔCFI HIV-1 envelope variants.
293 cells were transfected either with a control vector with no insert or the same vector expressing gp160, gp145ΔCFI, or gp140ΔCFI as described above. Cell-free media were collected 48 h after transfection and clarified by centrifugation at 16,000 × g at 4°C. Equal volumes of cell-free media were used for immunoprecipitation with 1 μg of HIV-1 immunoglobulin G (IgG). The precipitated proteins were separated by SDS-7.5% polyacrylamide gel electrophoresis (PAGE), and Env was detected by Western blotting using a polyclonal antiserum against gp160 (Intracel).
Cell surface expression of envelope by fluorescence-activated cell sorting (FACS) analysis.
293 cells were harvested 48 h after transfection using calcium phosphate (∼90% transfection efficiency), as in the previous section, and washed twice with PBS containing 1% bovine serum albumin and incubated for 30 min on ice with either control mouse or human IgG or monoclonal 2F5 or 2G12 or polyclonal Ig from an HIV-1-infected patient. The secondary antibody against the relevant human or mouse IgG conjugated with fluorescein isothiocyanate (Jackson Immuno Research) was added, incubated for 30 min on ice, washed three times with PBS, and analyzed by flow cytometry (FACScan). The median fluorescent intensity values were derived using Cell Quant software.
CD4 binding assay.
The binding assay with soluble CD4 (sCD4) was performed essentially as described previously (20). ELISA plates were coated with (per well) 200 ng of human sCD4 in 100 μl of PBS, pH 7.4, instead of carbonate/ bicarbonate buffer. Polyclonal Ig from an HIV-1-infected patient was used as a primary antibody instead of polyclonal (rabbit) antisera. As a negative control, total protein from extracts of 293 cells transfected with empty vector was used. Competitive inhibition of binding with sCD4 was assessed by serial dilution of sCD4 (1:3, 1:9, 1:27) from 500 ng/well compared to no sCD4 in a 96-well plate that was previously coated with CD4, followed by incubation with either gp140ΔCFI or r-gp160 (Intracel).
DNA injections in mice.
Six-week-old, female BALB/c mice were injected intramuscularly with 100 μg of purified plasmid DNA suspended in 200 μl of normal saline. For each plasmid DNA, a group of four mice was injected three times at intervals of 2 weeks. The mice were bled 2 weeks after the last injection, and sera were collected and stored at 4°C.
Immunization of guinea pigs.
Six-week-old, female Huntley guinea pigs were injected intramuscularly with 500 μg of purified plasmid DNA encoding the CXCR4-tropic HXB2 gp145ΔCFI suspended in 400 μl of normal saline. For each plasmid DNA, a group of four guinea pigs was injected three times at intervals of 2 weeks. The guinea pigs were bled 2 weeks after the last injection, and sera were collected and stored at 4°C. The guinea pigs received a boost with replication-defective recombinant adenovirus (ADV) expressing CXCR4-tropic HXB2 gp140ΔCFI as described previously (50, 54, 56) and were bled 2 weeks after ADV injection.
Quantitation of antibody response.
Immunoprecipitation and Western blotting were used to detect the antibodies that bind to native envelope proteins. Sera from immunized mice were used to immunoprecipitate gp160 from cell lysates of gp160-transfected 293 cells and were compared with well-characterized monoclonal antibodies 1726 and 1727 (NIH AIDS Research and Reference Reagent Program) against the V3 loop of HIV-1 IIIb. Indicated dilutions were used to immunoprecipitate gp160 from the cell lysate (400 μg). Immunocomplexes were separated by SDS-7.5% PAGE and analyzed by immunoblotting using polyclonal antibody against gp160.
Reactivity of gp140ΔCFI with monoclonal antibodies.
The binding ability of several monoclonal antibodies—2F5, 2G12, F105, and b12—to gp140ΔCFI was determined as above. Antibody (5 μg) was used to immunoprecipitate gp140ΔCFI from 100 μl of membrane-free supernatant from 293 cells transfected with the expression vector expressing gp140ΔCFI. The same volume of supernatant from cells transfected with empty vector was used as a control. Antibodies were obtained from the AIDS Research and Reference Reagent Program. The [35S]methionine-labeled supernatant was incubated with progressively diluted (5, 1, and 0.1 μg) monoclonal antibodies 2F5 and 2G12 along with HIV-1 IgG, and the intensity of the gp140ΔCFI band was determined by quantitative phosphorimaging.
Sucrose density gradient centrifugation.
The oligomerization of gp140ΔCFI was investigated by sucrose gradient centrifugation followed by Western blot analysis. Supernatants from cells transfected with the gp140ΔCFI expression vector were centrifuged at 16,000 × g at 4°C to remove debris. The supernatants were further centrifuged at 100,000 × g to remove vesicles and membrane fragments. The clarified supernatants were concentrated 30-fold with a UFV2GC10 filter (Millipore). The concentrated supernatants (750 μl) were loaded onto a 10%-to-40% continuous sucrose gradient and centrifuged in a Sorvall rotor (model TH641) for 20 h at 40,000 rpm at 4°C. Fractions of 1 ml were collected manually and analyzed by both nonreducing and reducing SDS-5% PAGE. Equal volumes from each fraction were used to immunoprecipitate gp140ΔCFI by HIV-1 IgG, and the separated proteins were detected by Western blotting with a rabbit polyclonal antibody to gp160. Cross-linked phosphorylase b (Sigma) was used as a high-molecular-weight marker.
Molecular exclusion chromatography.
Gel filtration using Superdex 200 gel filtration column (Amersham Pharmacia, Piscataway, N.J.) was calibrated with markers with molecular masses of 669, 440, 232, and 158 kDa individually and together. The proteins were chromatographed using membrane-free supernatant containing gp140ΔCFI prepared as described for sucrose gradients. Proteins from relevant fractions were separated by SDS-10% PAGE and transferred to Immobilon-P membrane (Millipore). gp140ΔCFI was detected by immunoprecipitation followed by Western blotting as described above, and band intensity was determined by densitometry using the ImageQuant program. The molecular weight was determined from the curve of Kaverage (gel phase distribution coefficient) versus log (molecular weight) of the standards.
Analysis of CTL response.
Spleens were removed aseptically, gently homogenized to a single-cell suspension, washed, and resuspended to a final concentration of 5 × 107 cells/ml. Cells were incubated for 7 days in the presence of interleukin 2 (10 U/ml) and either irradiated peptide-pulsed splenocytes from naive mice or an irradiated stable cell line expressing the full-length gp160 BC-env/rev (18). Three types of target cells were used: peptide-pulsed P815 cells (ATCC TIB64), BC10ME cells stably expressing gp160, and BC10ME cells (11) pulsed with peptides derived from gp160 sequence. Target cells were labeled with 51Cr for 90 min and washed three times with RPMI 1640 medium with 10% fetal bovine serum, 2 mM glutamine, 5 × 10−5 M β-mercaptoethanol and amphotericin (Fungizone) (250 U/ml), and resuspended in this medium. Cytolytic activity was determined in triplicate samples using all different target cell dilutions in a 5-h 51Cr release assay (34).
Neutralization assay.
Antibody-mediated neutralization was assessed in MT-2 cells with HIV-1 IIIB. Neutralizing antibody titers in the MT-2 assay were defined as the reciprocal serum dilution at which 50% of cells were protected from virus-induced killing as measured by neutral red uptake (26). Assay stocks of HIV-1 IIIB were produced in H9 cells.
RESULTS
Development of HIV Env vectors.
To develop Env glycoprotein variants that might better induce humoral immunity after DNA immunization, a series of plasmid expression vectors was generated (Fig. 1). Full-length HIV Env (gp160) was highly expressed in the absence of HIV accessory proteins at levels ≥10-fold higher than Rev-dependent viral gp160 in transfected 293 cells as seen by Western blot analysis (Fig. 2A and B), and the relevant mutant proteins were detected at the expected apparent molecular weights (Fig. 2C to E). As might be anticipated, gp160 expressed from the synthetic gene was not efficiently processed in transfected 293 cells, similar to previously published studies (3, 57), presumably because the high levels of gp160 saturate the cellular proteases responsible for cleavage, as overexpression of furin can overcome this problem (3); nonetheless, processed gp160 was detected with a monoclonal antibody to gp41 (Fig. 2B, lanes 3 to 9). This result is consistent with previous studies of viral gp160 in transfected cells, in which 75% may remain unprocessed (29).
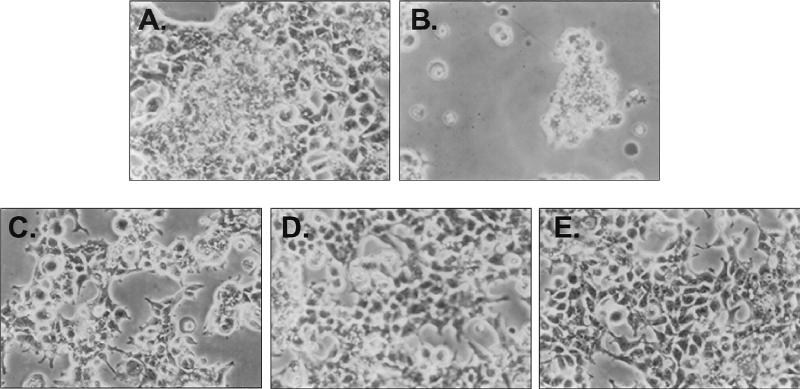
Synthetic HIV gp160 induced toxicity in transfected cells, with cell rounding and detachment evident within 48 h (compare Fig. 3A and B). This cytotoxicity was reduced by elimination of the COOH-terminal cytoplasmic domain. Env protein that terminated at amino acid 752 (gp150) was less cytotoxic than gp160, while the shorter proteins (gp145 and gp140) produced little or no effect (Fig. 3C, D, and E, respectively).
FIG. 3.
Cytotoxicity of full-length gp160 is eliminated by deletion of the COOH-terminal cytoplasmic domain. Cell rounding and detachment were not observed in control-transfected 293 cells (A), in contrast to full-length gp160 (B), and were observed to a lesser extent in cells transfected with gp150 (C), in contrast to gp145 (D) or gp140 (E).
To alter Env immunogenicity, two different approaches were explored. First the effects of glycosylation were evaluated. Two sets of mutations were introduced into both gp160 and gp150 (Fig. 1). The first set included 11 potential N-linked glycosylation sites (Δgly11), and the second set included an additional six sites downstream (Δgly17) that were eliminated by site-directed mutagenesis. Expression studies showed that the glycosylation mutants were efficiently expressed, and the glycosylation mutant protein was appropriately reduced in size compared to wild-type gp160 or gp150, consistent with reduced N-linked glycosylation (Fig. 2D). These proteins were also readily detected on the cell surface by flow cytometry (data not shown). As noted below, these glycosylation mutants had little effect on the generation of antibodies to Env, so efforts were therefore directed to a different set of mutants.
The second approach involved a series of internal deletions designed to stabilize and expose functional domains of the protein that might be present in an extended helical structure prior to the formation of the six-member coiled-coil structure in the hairpin intermediate (10, 52). To generate this putative prehairpin structure, the cleavage site was removed to prevent the proteolytic processing of the envelope and stabilize the protein by linking it covalently to the gp41 extracellular and/or transmembrane domain. To reduce toxicity and enhance stability, the fusion peptide domain was deleted. The heptad repeats in the envelope protein are important tertiary structure domains involved in trimer formation (23). The sequence between the heptad repeats was removed to stabilize the formation of trimers and eliminate formation of the hairpin intermediate. These ΔCFI deletions were introduced into full-length gp160 and COOH-terminal truncation mutants. Cells transfected with vectors encoding gp140ΔCFI, gp145ΔCFI, and gp160 readily expressed these proteins (Fig. 2E). gp140ΔCFI, which lacks the transmembrane domain, was easily detected in the supernatant (Fig. 4), indicating that it can give rise to soluble antigen. Some gp160 was also noted in media from cells transfected with wild-type gp160, likely reflecting some membrane contamination of the supernatant fraction or release of gp160 after cell lysis due to toxicity induced by gp160.
FIG. 4.
Expression of soluble ΔCFI HIV-1 envelope variants. Results of immunoprecipitation and Western blot analysis of supernatants from the indicated transfected cells are shown.
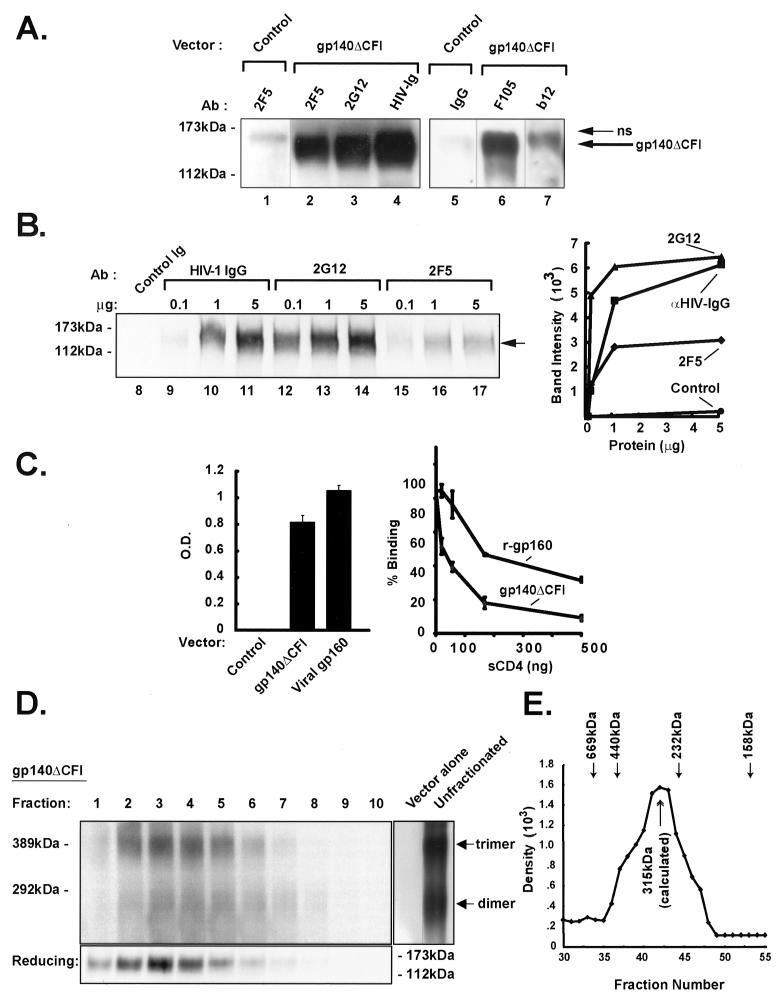
The antigenic structure of gp140ΔCFI was analyzed using different conformation-dependent monoclonal antibodies and by biochemical analysis. At least four well-defined monoclonal antibodies—2F5, 2G12, F105, and b12—bound specifically to gp140ΔCFI (Fig. 5A). The specificity of binding by 2F5 and 2G12, in comparison to anti-HIV-1 IgG, was confirmed further by performing a quantitative dose-response analysis (Fig. 5B). Similarly, gp140ΔCFI retained its ability to interact specifically with its natural receptor, CD4, and its binding was competitively inhibited by sCD4 (Fig. 5C). As a negative control, total protein from 293 cells transfected with empty vector was used, in which the optical density was ≤0.1, confirming the specificity of binding with this assay. Finally, the ability of gp140ΔCFI to oligomerize was tested biochemically. By sucrose density gradient analysis and Western blot analysis, gp140ΔCFI predominantly formed trimeric and dimeric oligomers (Fig. 5D). The patterns that have been observed are typical of other envelope proteins, with monomers, dimers, trimers, and multimers. Notably, there was a significant component of trimer in the equilibrium gradient, showing that the protein is capable of forming the appropriate oligomer. It is also likely that some aggregation occurs, and additional high-molecular-mass (>500-kDa) multimers were observed in fraction 1 (data not shown), typical of such analyses (55). The proteins in all fractions migrated at the position of the monomer under denatured reducing conditions (Fig. 5D, lower panel). Membrane-free supernatants were analyzed by molecular exclusion chromatography using Superdex 200. The major peak was found at the calculated size of ∼315 kDa (Fig. 5E), consistent with the predicted size of the trimeric complex. Flow cytometric analysis of cell surface envelope glycoproteins indicated that both gp140ΔCFI and gp145ΔCFI were synthesized and processed within cells. Both proteins were expressed on the cell surface after transfection, as recognized by flow cytometry with HIV Ig or two different monoclonal antibodies, 2F5 and 2G12 (Fig. 6). Though gp140ΔCFI was secreted from cells, it was also readily detected on the cell surface, likely due to its high level of synthesis in transfected cells, as has been described for other secreted and highly expressed proteins previously (24, 45).
FIG. 5.
Interaction of gp140ΔCFI with defined monoclonal antibodies or CD4 and biochemical analysis for oligomerization of gp140ΔCFI. (A) Analysis of the antigenic structure of soluble gp140ΔCFI with monoclonal antibodies. The envelope glycoproteins from the supernatants of the 293 cells transfected with the vector expressing gp140ΔCFI were immunoprecipitated with either 5 μg of monoclonal antibodies 2F5, 2G12, F105, and b12 or with 5 μg of HIV-1 Ig. The proteins were analyzed by SDS-PAGE and detected by Western blotting using the polyclonal antibodies against gp160. The arrow indicates the position of gp140ΔCFI. A nonspecific (ns) band that cross-reacted with the antibody is indicated. (B) Quantification of 2F5 and 2G12 binding to gp140ΔCFI using the indicated concentrations of each antibody as shown compared to a nonreactive Ig isotype (control). The intensity of the gp140ΔCFI band was determined by quantitative phosphorimaging. The arrow indicates the position of gp140ΔCFI. (C) Interaction of soluble gp140ΔCFI protein with CD4. Binding of gp140ΔCFI and gp160, compared to that of controls transfected with vector alone, in an ELISA with CD4 is shown (left panel). The values represent the mean and standard deviation (error bars) for each point. The ability of sCD4 to compete with binding to these envelopes is shown (right panel). (D) Biochemical analysis of soluble gp140ΔCFI oligomerization. Western blot analysis to detect the gp140ΔCFI in different fractions, fractions 1 to 10, after sucrose density gradient. Fraction 1 represents the greatest density, and fraction 10 represents the least density. Equal volumes of the samples from each fraction were analyzed in an SDS-polyacrylamide gel under nonreducing conditions, except the molecular weight marker. The proteins were detected by Western blotting using polyclonal antibody against gp160. The positions of dimer, trimer, and aggregates are shown. The lower panel shows the presence of monomer in these fractions run under reducing conditions. (E) Molecular-exclusion chromatography of soluble of gp140ΔCFI. Membrane-free supernatant containing gp140ΔCFI was analyzed on a Superdex 200 column and compared with a mixture of molecular weight standards; the position of each marker is indicated by arrows. Fractions were analyzed for gp140ΔCFI by immunoprecipitation followed by Western blotting and were quantitated by densitometry.
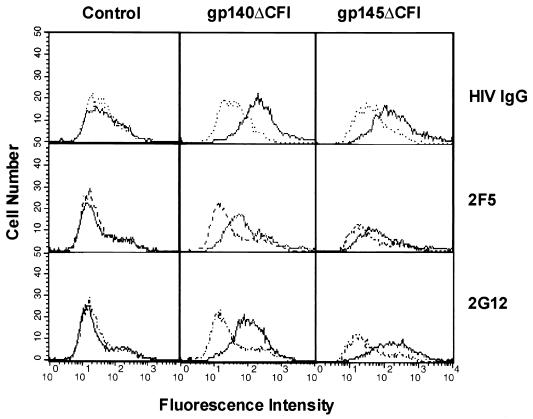
FIG. 6.
Confirmation of cell surface expression of gp140ΔCFI and gp145ΔCFI by flow cytometry. 293 cells were transfected with either a control vector or vectors expressing gp140ΔCFI or gp145ΔCFI, as indicated. The transfection efficiency (∼90%) was calculated based on staining after transfection of the same number of cells under identical conditions with an equal amount of DNA expressing β-galactosidase. Cell surface expression was detected by FACS analysis using Ig purified from the sera of HIV-1-infected individuals (HIV Ig) or different monoclonal antibodies, 2F5 and 2G12. The transfected cells are labeled at the top, and the different antibodies are indicated at right. Light dotted lines indicate FACS analysis with control human Ig, and dark dotted lines indicate FACS analysis with human monoclonal antibodies against HIV-1.
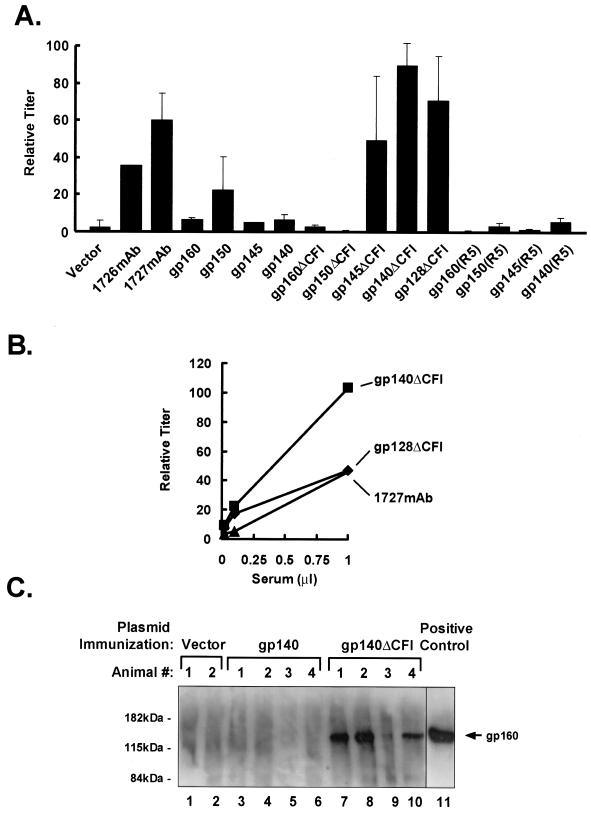
Increased antibody response after DNA vaccination with ΔCFI Env mutants. The ability of these Env proteins to elicit a humoral immune response was determined in mice by injection with these plasmid DNA expression vectors. To enhance the sensitivity of these sera for native conformational epitopes, antibody responses were monitored by their ability to immunoprecipitate wild-type gp160 from cell lysates followed by SDS-PAGE and Western blotting (Fig. 7). In some cases, antibody reactivity was also confirmed by indirect immunofluorescence (data not shown). To quantitate the antibody response, immunoprecipitation with different dilutions of immunized mouse sera was performed. The intensity of the gp160 band was determined by densitometry and standardized relative to a positive control serum used to normalize data between experiments. The approximately linear dose response of gp160 intensity with serum dilution allowed quantification of the anti-gp160 antibody response in mouse serum. Data from immunized mice showed that none of the wild-type Env proteins, neither the gp160, gp150, gp145, nor gp140 COOH-terminal truncations, generated consistently high antibody responses (Fig. 7A). Of these proteins, wild-type gp140 was somewhat more effective than gp160 in generating antibody responses, but these responses remained low and inconsistent. Immunization with vectors designed to express glycosylation-deficient envelope proteins also did not improve the humoral response (data not shown). In contrast, the ΔCFI mutants substantially increased the anti-gp160 antibody response when the transmembrane domain was truncated (Fig. 7A [gp145, gp140, and gp128 versus gp160 and gp150]). gp140ΔCFI provided more-consistent and greater increases in antibody response than gp128ΔCFI (Fig. 7B). In all cases, these vectors that encoded gp140ΔCFI, but not those that encoded wild-type gp140, induced antibodies that were reactive to native gp160 (Fig. 7C).
FIG. 7.
Antibody response against HIV-1 envelope proteins in DNA immunized mice. (A) Comparison of the antibody response in mice immunized with gp140ΔCFI or other Env plasmid expression vectors. Sera were collected 2 weeks after the last immunization and used to immunoprecipitate codon-altered gp160 from lysates of transfected 293 cells as described before. The quantitation of the immunoprecipitated gp160 was done as described for panel B. The average of the normalized data has been presented as a bar diagram. Error bars are indicated. (B) Antibody responses in mice immunized with gp140ΔCFI or gp128ΔCFI relative to a V3-specific monoclonal antibody standard (monoclonal antibody 1727). Antisera from immunized mice were diluted in immunoprecipitation buffer, and 1 μl of each diluted serum was used to immunoprecipitate codon-altered HIV-1 gp160 from lysates of transfected 293 cells as described in the legend to Fig. 3A. The gels were scanned, and the intensity of the gp160 band was determined by densitometry using the program ImageQuant and presented relative to the intensity of gp160 immunoprecipitated with positive control sera (rabbit anti-gp160), which was used to normalize data between experiments. These data are presented graphically to facilitate comparison among groups. Monoclonal antibody 1727 interacts with the V3 loop of HIV IIIb and was kindly provided by the NIH AIDS Research and Reference Reagent Program, from Jon Laman. (C) Antibody responses in mice immunized with gp140 or gp140ΔCFI were determined by immunoprecipitation and Western blotting. Animals received two booster doses (100 μg) of the same plasmid, 2 weeks apart. Sera (1 μl) collected 2 weeks after the last immunization were used to immunoprecipitate codon-optimized HIV-1 gp160 from lysates of transfected 293 cells containing 400 μg of total protein. Each lane corresponds to the serum from an animal immunized with either the control vector (lanes 1 and 2), CXCR4-tropic gp140 (lanes 3 to 6), or plasmid that expresses gp140 with the indicated mutant functional domains (lanes 7 to 10). A mouse monoclonal antibody to gp160 (HIV-1 V3 monoclonal [IIIB-V3-13]; NIH AIDS Research and Reference Reagent Program) was used as a positive control (lane 11).
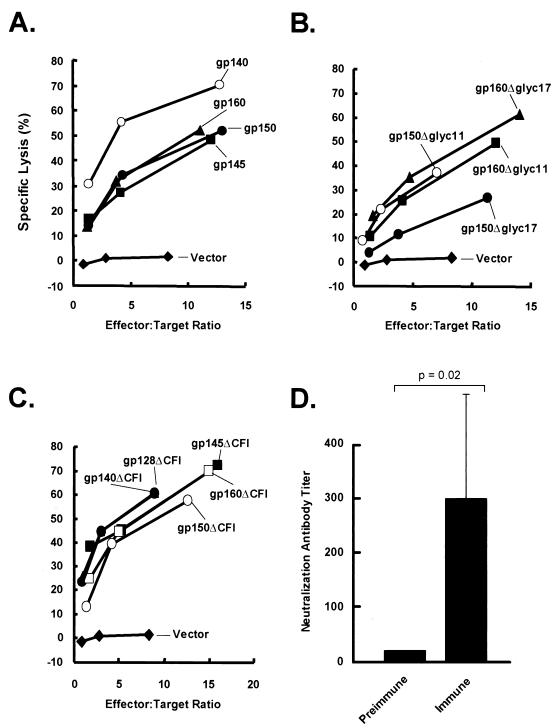
To determine whether these modifications of Env adversely affected CTL responses, spleen cells from immunized mice were tested for their ability to lyse relevant target cells. All mice immunized with wild-type codon-altered Env vectors, including COOH-terminal deletion mutations (Fig. 8A) or glycosylation mutants (Fig. 8B) elicited strong CTL responses against cell lines pulsed with HIV Env peptides. These findings were confirmed using stably transfected cells that expressed Env (data not shown). Importantly, gp140ΔCFI, which elicited increased antibody responses relative to the comparable wild-type Env, readily induced CTL responses to native Env, as did other ΔCFI mutants (Fig. 8C). Addition of anti-CD8 antibody inhibited cytolytic activity, as did depletion of CD8 cells using magnetic beads coupled with anti-CD8 antibody, thus confirming a CTL response to these immunogens after genetic immunization (data not shown). This response was detectable for at least 6 months after immunization. Finally, the ability of gp140ΔCFI to generate neutralizing antibodies was tested to determine whether this immunogen can elicit a neutralizing antibody response to functional Env on virus. This experiment was performed with guinea pigs due to the high background and variability in mouse antisera in the neutralization assay. Because these guinea pigs do not respond as efficiently to plasmid DNA alone, a DNA prime-ADV boost protocol was used, as previously described in an Ebola virus challenge model (50, 54). Four animals were immunized with gp140ΔCFI plasmid DNA as was done for mice and received boosters at week 6 with 1011 particles of ADV-gp140ΔCFI. Titers were increased from undetectable levels (<1:20) in preimmune animals to ≥1:100 after immunization (Fig. 8D) (P = 0.02), documenting that the antisera react with functional Env on virus.
FIG. 8.
CTL response against HIV-1 envelope proteins in DNA-immunized mice and generation of a neutralizing antibody response in guinea pigs. (A) The CTL response to CXCR4-tropic Env and indicated deletion mutants is shown. (B and C) The CTL responses to CXCR4-tropic envelope with glycosylation site and ΔCFI mutations are shown, respectively. Spleen cells were isolated from immunized mice 2 weeks after the final immunization and stimulated in vitro with irradiated cells expressing gp160 with addition of human interleukin 2 (5 U/ml) at day 4. The cytolytic activity of the restimulated spleen cells was tested after 7 days against V3 peptide-pulsed BC10ME cells. Similar findings were observed with target cells that stably express full-length Env (data not shown). (D) Preimmune sera were collected from four guinea pigs prior to immunization or after DNA priming and ADV boosting with gp140ΔCFI as described in the text. Both preimmune sera and postimmune sera were diluted, and neutralizing activity was measured by reduction of HIV-IIIB virus compared to the untreated control. Neutralizing antibody titers were analyzed as previously described (26). The data represent the dilutions at which the sera can neutralize the virus in the MT2 assay. Standard deviations are indicated.
DISCUSSION
To develop DNA vaccine candidates for HIV, we developed a series of synthetic genes designed to express HIV Env mutants in human cells. In the absence of HIV regulatory proteins, these codon-altered envelope protein genes were readily expressed in human cells. Like other DNA vaccines, immunization with these vectors elicited strong CTL responses in mice, and antibody responses were not robust in mice immunized with wild-type Env expression vectors. Mutations in highly conserved N-linked glycosylation sites did not significantly alter humoral or cellular immune response to native Env. In contrast, a mutant Env with deletions in the cleavage site, fusion domain, and a region between the heptad repeats elicited a more-potent humoral immune response and retained its ability to stimulate Env-specific CTL.
The immune response to HIV infection in long-term nonprogressors (9, 35) and HIV-exposed sex workers suggests that specific viral immunity may limit infection and the symptoms of disease, but no single characteristic correlates with protective immunity. Depletion of CTLs in chronically infected macaques enhances viremia (19, 46). In humans, higher CTL responses correlate with lower viral load and stabilization of clinical symptoms (30, 33). In animal models, passive transfer of neutralizing antibodies can also contribute to protection against virus challenge (8, 15, 28, 31, 32, 36, 43, 48). Neutralizing antibody responses can also be developed in HIV-infected individuals (8, 36, 43) and are associated with lower viral loads in long-term nonprogressors (25). Though broadly neutralizing antibody responses are uncommon, they are directed largely against the Env protein of the virus (40, 49). In early human vaccine trials, gp120 protein immunogens have yielded disappointing results: vaccine-induced antibodies have not been broadly neutralizing and have sometimes enhanced infection in vitro (5, 7, 12, 17, 51). Monomeric gp120 loses oligomer-dependent epitopes and does not include sequences in the ectodomain of the gp41 that become exposed during virus entry (6, 27, 32). It is assumed that broadly neutralizing antibodies must bind well to native gp120/gp41 on the surface of the virus (8).
Recent reports suggest that gp160 forms trimers in vivo and the domain required for trimer formation resides in the ectodomain of the gp41 (55). Such trimeric forms of HIV envelope protein are likely to present different epitopes to the immune system compared to monomeric gp120. In addition to the linear epitopes in the envelope, this trimeric structure is likely to display conformational epitopes important for B-cell triggering of a relevant antibody response. In this regard, gp140ΔCFI, which induced the greatest antibody response, is released in a soluble trimeric form (Fig. 4 and 5). This finding was confirmed both by sedimentation gradient and gel filtration analysis (Fig. 5D and E). Biochemical purification is likely to yield dimeric and monomeric forms, and another potential advantage of gene-based vaccines is the ability to make physiologically relevant complexes in vivo. In contrast to gp140ΔCFI, wild-type Env did not elicit high-titer antibody responses. The toxicity of Env in mammalian cells has been seen previously and could limit both the amount and duration of envelope protein expression in vivo, thus affecting immunogenicity. The envelope is also heavily glycosylated, and removal of partial or complete gp120 glycosylation sites has resulted in higher titers of strain-specific neutralizing antibody responses to mutant simian immunodeficiency viruses in monkeys, presumably through enhanced exposure of critical epitopes (4, 39). Though it seemed reasonable that deglycosylation would reveal epitopes otherwise masked in the native protein, we did not observe enhanced immune reactivity by DNA vaccination using different glycosylation site mutants, both in gp160 and gp150. This difference with the previous study is likely due to the fact that DNA vaccination rather than viral infection was utilized for immunization. Though glycosylation mutants are unlikely to prove helpful with the former method of immunization, it remains possible that modification of glycosylation sites may be effective with other vectors or adjuvants.
HIV-1 Env is proteolytically cleaved by a cellular proprotein convertase into gp120 and gp41. The gp41 subunit is composed of cytoplasmic, transmembrane, and ectodomain segments. The role of the ectodomain of the envelope in membrane fusion, particularly its hydrophobic glycine-rich fusion peptide, is well established. Two regions with heptad coiled-coil repeats in the ectodomain of gp41 are involved in viral fusion (13, 53). Upon fusion, these two alpha-helices, connected via a disulfide-stabilized loop (14, 21, 44), presumably undergo a transient conformational change to a fusion active state. These changes allow the formation of a six-member helical hairpin intermediate structure that presumably exposes the fusion peptide at the NH2 terminus of gp41, allowing fusion to the target cell membrane (2, 22). The ΔCFI mutation was intended to eliminate cleavage of gp140, remove the unstable hydrophobic region, and stabilize oligomer formation. Though detailed structural data are not yet available on this protein, the immunologic and biochemical analyses indicated that these mutations retain native antigenic determinants, defined by known monoclonal antibodies, and oligomeric properties, similar to native viral envelope (Fig. 5). The gp140ΔCFI mutant also elicits both humoral and cellular immune responses, probably by virtue of protein stabilization and secretion. For example, the neutralizing epitope (ELDKWAS) in the ectodomain of gp41 (31) is present in the series of deletions and truncations of the envelope, and gp140ΔCFI is reactive with the 2F5 neutralizing monoclonal antibody that binds to this epitope (Fig. 6). It is also relevant that protein misfolding is unlikely to account for enhanced antibody responses, as several vaccine candidates that induced such responses showed expected cell surface expression and/or secretion (Fig. 4, 6, and 7 [gp140ΔCFI and gp145ΔCFI]), and several immunogens that were not appropriately exposed had no effect (data not shown).
Importantly, these immunogens also induced CTL responses to Env. It is evident that DNA immunization often elicits predominant humoral or cellular immune responses that are determined by features of the immunogen that are not understood. For example, we have found that responses to Env are primarily cellular while those to Nef display enhanced humoral immunity. Though gp128ΔCFI induced slightly more potent CTL activity, gp140ΔCFI was better able to elicit an antibody response, retained the 2F5 epitope, and was readily able to elicit such responses, both to peptide-pulsed cells and stably transduced target cells. Thus, the enhanced humoral immune response introduced by this vaccine candidate did not appear to diminish the CTL response. Taken together, these results suggest that gp140ΔCFI serves as an improved immunogen that can more effectively elicit a neutralizing antibody response against the envelope by DNA vaccination while preserving its ability to induce a CTL response.
Acknowledgments
We thank Nancy Barrett for preparation of the figures, Mousumi Paul and Judith Stein for advice, Nancy Sullivan for discussions and thoughtful suggestions, and Cherilyn Davis and Ati Tislerics for manuscript preparation. We thank Janet Hoff of the University of Michigan for assistance during the immunization of the animals. The following reagents were kindly provided through the AIDS Research and Reference Reagent Program: HIV-1 V3 monoclonal antibody (IIIB-V3-01 and IIIB-V3-13) from Jon Laman, b12 from Dennis Burton and Carlos Barbas, 2G12 and 2F5 from Hermann Katinger, and F105 from Marshall Posner. We thank Richard Wyatt and Peter D. Kwong for helpful discussion on analyzing the trimeric structure of the protein and Shazad Majeed for assistance in the fast-performance liquid chromatography analysis.
REFERENCES
- 1.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binley, J., and J. P. Moore. 1997. HIV-cell fusion. The viral mousetrap. Nature 387:346-348. (Erratum, 389:131.) [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley, J. M., R. Wyatt, E. Desjardins, P. D. Kwong, W. Hendrickson, J. P. Moore, and J. Sodroski. 1998. Analysis of the interaction of antibodies with a conserved enzymatically deglycosylated core of the HIV type 1 envelope glycoprotein 120. AIDS Res. Hum. Retrovir. 14:191-198. [DOI] [PubMed] [Google Scholar]
- 5.Bolognesi, D. P., and T. J. Matthews. 1998. HIV vaccines. Viral envelope fails to deliver? Nature 391:638-639. [DOI] [PubMed] [Google Scholar]
- 6.Broder, C. C., P. L. Earl, D. Long, S. T. Abedon, B. Moss, and R. W. Doms. 1994. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc. Natl. Acad. Sci. USA 91:11699-11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. M. McGrath, J. Tartaglia, P. Caudrelier, R. El Habib, M. Klein, A. Lazzarin, D. M. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87-S98. [PubMed] [Google Scholar]
- 9.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 10.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 11.Collins, J. L., P. Q. Patek, and M. Cohn. 1981. Tumorigenicity and lysis by natural killers. J. Exp. Med. 153:89-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor, R. I., B. T. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, B. D. Walker, A. U. Neumann, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalams, K. J. Kunstman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 72:1552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 14.Gallaher, W. R., J. M. Ball, R. F. Garry, M. C. Griffin, and R. C. Montelaro. 1989. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res. Hum. Retrovir. 5:431-440. [DOI] [PubMed] [Google Scholar]
- 15.Gauduin, M. C., P. W. Parren, R. Weir, C. F. Barbas, D. R. Burton, and R. A. Koup. 1997. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat. Med. 3:1389-1393. [DOI] [PubMed] [Google Scholar]
- 16.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 17.Haynes, B. F. 1996. HIV vaccines: where we are and where we are going. Lancet 348:933-937. [DOI] [PubMed] [Google Scholar]
- 18.Irwin, M. J., L. S. Laube, V. Lee, M. Austin, S. Chada, C. G. Anderson, K. Townsend, D. J. Jolly, and J. F. Warner. 1994. Direct injection of a recombinant retroviral vector induces human immunodeficiency virus-specific immune responses in mice and nonhuman primates. J. Virol. 68:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlsson, G. B., M. Halloran, D. Schenten, J. Lee, P. Racz, K. Tenner-Racz, J. Manola, R. Gelman, B. Etemad-Moghadam, E. Desjardins, R. Wyatt, N. P. Gerard, L. Marcon, D. Margolin, J. Fanton, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 1998. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J. Exp. Med. 188:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent, K. A., and J. Robinson. 1996. Antigenic determinants on HIV-1 envelope glycoproteins: a dickens of a time with oligomer twist. AIDS 10(Suppl. A):S107-S114. [PubMed] [Google Scholar]
- 22.LaCasse, R. A., K. E. Follis, M. Trahey, J. D. Scarborough, D. R. Littman, and J. H. Nunberg. 1999. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science 283:357-362. [DOI] [PubMed] [Google Scholar]
- 23.Lu, M., and P. S. Kim. 1997. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 15:465-471. [DOI] [PubMed] [Google Scholar]
- 24.Manz, R., M. Assenmacher, E. Pfluger, S. Miltenyi, and A. Radbruch. 1995. Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proc. Natl. Acad. Sci. USA 92:1921-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montefiori, D. C., G. Pantaleo, L. M. Fink, J. T. Zhou, J. Y. Zhou, M. Bilska, G. D. Miralles, and A. S. Fauci. 1996. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J. Infect. Dis 173:60-67. [DOI] [PubMed] [Google Scholar]
- 26.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, J. P. 1995. HIV vaccines. Back to primary school. Nature 376:115. [DOI] [PubMed] [Google Scholar]
- 28.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 29.Moore, J. P., P. W. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musey, L., J. Hughes, T. Schacker, T. Shea, L. Corey, and M. J. McElrath. 1997. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 337:1267-1274. [DOI] [PubMed] [Google Scholar]
- 31.Muster, T., R. Guinea, A. Trkola, M. Purtscher, A. Klima, F. Steindl, P. Palese, and H. Katinger. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68:4031-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 34.Ohno, T., Z.-Y. Yang, L. Xu, M. Jaffe, E. G. Nabel, D. Normolle, and G. J. Nabel. 1997. Combination gene transfer to potentiate tumor regression. Gene Ther. 4:361-366. [DOI] [PubMed] [Google Scholar]
- 35.Pantaleo, G., S. Menzo, M. Vaccarezza, C. Graziosi, O. J. Cohen, J. F. Demarest, D. Montefiori, J. M. Orenstein, C. Fox, and L. K. Schrager. 1995. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N. Engl. J. Med. 332:209-216. [DOI] [PubMed] [Google Scholar]
- 36.Poignard, P., P. J. Klasse, and Q. J. Sattentau. 1996. Antibody neutralization of HIV-1. Immunol. Today 17:239-246. [DOI] [PubMed] [Google Scholar]
- 37.Qiu, J. T., R. Song, M. Dettenhofer, C. Tian, T. August, B. K. Felber, G. N. Pavlakis, and X. F. Yu. 1999. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J. Virol. 73:9145-9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rana, T. M., and K. T. Jeang. 1999. Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch. Biochem. Biophys. 365:175-185. [DOI] [PubMed] [Google Scholar]
- 39.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 40.Robey, W. G., L. O. Arthur, T. J. Matthews, A. Langlois, T. D. Copeland, N. W. Lerche, S. Oroszlan, D. P. Bolognesi, R. V. Gilden, and P. J. Fischinger. 1986. Prospect for prevention of human immunodeficiency virus infection: purified 120-kDa envelope glycoprotein induces neutralizing antibody. Proc. Natl. Acad. Sci. USA 83:7023-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roebuck, K. A., and M. Saifuddin. 1999. Regulation of HIV-1 transcription. Gene Expr. 8:67-84. [PMC free article] [PubMed] [Google Scholar]
- 42.Romano, G., M. Kasten, G. De Falco, P. Micheli, K. Khalili, and A. Giordano. 1999. Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression. J. Cell. Biochem. 75:357-368. [PubMed] [Google Scholar]
- 43.Sattentau, Q. J. 1996. Neutralization of HIV-1 by antibody. Curr. Opin. Immunol. 8:540-545. [DOI] [PubMed] [Google Scholar]
- 44.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 45.Scheffold, A., M. Lohning, A. Richter, M. Assenmacher, R. Manz, F. Austrup, and A. Hamann. 1998. Analysis and sorting of T cells according to cytokine expression. Eur. Cytokine Netw. 9:5-11. [PubMed] [Google Scholar]
- 46.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 47.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204-210. [DOI] [PubMed] [Google Scholar]
- 49.Steimer, K. S., C. J. Scandella, P. V. Skiles, and N. L. Haigwood. 1991. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to Gp120. Science 254:105-108. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z.-Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 51.VanCott, T. C., F. R. Bethke, D. S. Burke, R. R. Redfield, and D. L. Birx. 1995. Lack of induction of antibodies specific for conserved, discontinuous epitopes of HIV-1 envelope glycoprotein by candidate AIDS vaccines. J. Immunol. 155:4100-4110. [PubMed] [Google Scholar]
- 52.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 53.Weissenhorn, W., S. A. Wharton, L. J. Calder, P. L. Earl, B. Moss, E. Aliprandis, J. J. Skehel, and D. C. Wiley. 1996. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 15:1507-1514. [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, L., A. Sanchez, Z. Yang, S. R. Zaki, E. G. Nabel, S. T. Nichol, and G. J. Nabel. 1998. Immunization for Ebola virus infection. Nat. Med. 4:37-42. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, Z., R. Delgado, L. Xu, R. F. Todd, E. G. Nabel, A. Sanchez, and G. J. Nabel. 1998. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 279:1034-1037. [DOI] [PubMed] [Google Scholar]
- 57.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]