Abstract
Flavivirus envelope proteins are synthesized as part of large polyproteins that are co- and posttranslationally cleaved into their individual chains. To investigate whether the interaction of neighboring proteins within the precursor protein is required to ensure proper maturation of the individual components, we have analyzed the folding of the flavivirus tick-borne encephalitis (TBE) virus envelope glycoproteins prM and E by using a recombinant plasmid expression system and virus-infected cells. When expressed in their polyprotein context, prM and E achieved their native folded structures with half-times of approximately 4 min for prM and about 15 min for E. They formed heterodimeric complexes within a few minutes after synthesis that were required for the final folding of E but not for that of prM. Heterodimers could also be formed in trans when these proteins were coexpressed from separate constructs. When expressed without prM, E could form disulfide bonds but did not express a specific conformational epitope and remained sensitive to reduction by dithiothreitol. This is consistent with a chaperone-like role for prM in the folding of E. PrM was able to achieve its native folded structure without coexpression of E, but signal sequence cleavage at the N terminus was delayed. Our results show that prM is an especially rapidly folding viral glycoprotein, that polyprotein cleavage and folding of the TBE virus envelope proteins occurs in a coordinated sequence of processing steps, and that proper and efficient maturation of prM and E can only be achieved by cosynthesis of these two proteins.
Flaviviruses belong to those virus genera that encode large polyprotein precursors from which the individual proteins are proteolytically cleaved. Some of the cleavages occur co- or posttranslationally, while others take place later during transit through the secretory pathway. Internal signal sequences and signal peptidase cleavage sites ensure the correct topology of the polyprotein and its cleaved products at the membrane of the endoplasmic reticulum (ER). The use of polyproteins for the generation of structural and nonstructural proteins is dictated (in part) by viral replication strategies that do not allow the generation of multiple mRNAs and by the limited size of the genome. Moreover, in the case of low-pH-activated viral fusion proteins, proteolysis is often needed to render the proteins fusion competent once they have passed the acidic compartments of the late secretory pathway. The individual glycopolypeptide chains are normally found as subunits of multimeric complexes in which they are closely associated with each other.
From a folding point of view, the early cleavages are especially interesting because they usually occur cotranslationally before folding of the individual proteins has been completed. This means that the individual polypeptide chains have the opportunity to undergo folding as part of a larger complex of polypeptide chains derived from the same ribosome. The folding of the two envelope glycoproteins of Semliki Forest virus (p62 and E1), although dependent on different sets of molecular chaperones, seems, for example, to be intimately coordinated (5). Similarly, the folding of hepatitis C virus (HCV) envelope protein E1 appears to be dependent on the second envelope protein, E2 (14, 27). Also, in the case of Uukuniemi virus, a bunyavirus, the two envelope proteins G1 and G2 fold together, although at very different rates (32).
In this study, we have examined the folding of the prM and E envelope proteins of tick-borne encephalitis (TBE) virus, two glycoproteins that follow each other in the polyprotein sequence. TBE virus is a member of the Flavivirus genus of the family Flaviviridae, a family of small enveloped RNA viruses that includes important arthropod-borne human pathogens such as the yellow fever, dengue, West Nile, and Japanese encephalitis viruses. Their positive-stranded RNA genome (ca. 11 kb) contains a single long open reading frame encoding three structural proteins (capsid [C], prM/M, and E) and at least seven nonstructural proteins (reviewed in reference 9). The polyprotein is co- and posttranslationally cleaved into distinct protein products by host- and virus-encoded proteases (24).
The envelope of the mature TBE virus contains the major envelope glycoprotein E (molecular mass, 52 kDa; one N-linked glycan) and the small membrane protein M (molecular mass, 7 to 8 kDa). The E protein mediates virus entry into the cell through receptor binding and low-pH-induced fusion in endosomes after receptor-mediated endocytosis, and it also carries the major antigenic epitopes leading to a protective immune response (reviewed in reference 19). M is synthesized as a precursor protein, prM (molecular mass, 25 kDa; one N-linked glycan), that forms a 1:1 complex with E intracellularly and probably serves as a lock to prevent E from being prematurely activated by low pH during transport through the acidic compartments and vesicles of the trans-Golgi network (15, 17). Shortly before the virus is released from the cell, prM is cleaved, apparently by the cellular protease furin, leading to mature virions (36). Since the TBE virus, like most flaviviruses, probably buds into the ER lumen, the envelope glycoproteins are transported through the Golgi complex to the plasma membrane as part of immature virus particles (24).
The X-ray crystallographic solution of the structure of the E ectodomain purified from mature virions (33) revealed that protein E differs from many other well-characterized viral glycoproteins in that it forms head-to-tail dimers that lie parallel rather than perpendicular to the viral membrane. E is a type I membrane protein with two transmembrane segments at its C terminus. The ectodomain of the TBE virus E protein is composed of three major parts: a central domain (domain I), a dimerization domain (domain II), and a C-terminal immunoglobulin-like domain (domain III). All three domains are rich in β-sheets with two short α-helices in the dimerization domain. There are 12 cysteines forming six intrachain disulfide bonds, and the single glycosylation site is located at the dimerization interface. That this type of overall fold is not unique to flaviviruses was recently demonstrated by the X-ray structure of the E1 protein of Semliki Forest virus, an alphavirus (23). PrM is also a type I membrane protein. It contains six cysteines in the ectodomain, forming three disulfide bonds (30).
A few years ago, a recombinant simian virus 40 (SV40)-based plasmid vector system was developed to express the TBE virus prM and E proteins, as well as C-terminally truncated variants of E (1, 3). This system was used here to analyze the folding of prM and E when expressed individually, in combination with each other, or in cells infected with TBE virus. We found that the maturation of TBE virus envelope proteins prM and E is coordinated and that early interactions between the proteins are needed for complete folding of E but not for that of prM, suggesting that prM serves as a chaperone for the folding of E.
MATERIALS AND METHODS
Plasmids and virus.
The production of recombinant plasmids expressing prM and full-length E or truncated forms of E derived from TBE virus strain Neudoerfl (GenBank accession no. U27495) was described earlier (1, 3). In particular, we used vectors expressing prM and full-length E (SV-PEwt); prM and E truncated after the first transmembrane domain (SV-PE472, i.e., the first 472 amino acids of E); prM and E truncated right after the ectodomain, lacking the stem-anchor elements (SV-PE400); prM alone (SV-prM); or full-length E alone (SV-Ewt). The vector included an SV40 early promoter and an SV40 origin of replication for amplification in COS-1 cells.
TBE virus strain Neudoerfl (25) was grown in primary chicken embryo cells and purified by two cycles of sucrose density gradient centrifugation as described by Heinz and Kunz (18).
Infection of mammalian cells with TBE virus.
COS-1 cells (ATCC CRL 1659) were grown in Dulbecco's modified Eagle medium (DMEM; Life Technologies) supplemented with 10% fetal calf serum (FCS), 4 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml at 37°C in 5% CO2. At 60 to 70% confluency, cells were infected with TBE virus at a multiplicity of infection of ca. 1 in DMEM containing 2% FCS, glutamine, and antibiotics. The virus-containing medium was replaced with fresh medium 1 h after infection. At 24 h postinfection, the cells were used for further experiments.
Expression of TBE virus envelope proteins in mammalian cells.
COS-1 cells were transiently transfected with plasmid DNA complexed to Lipofectamine (Life Technologies) in DMEM containing no serum or antibiotics. At 5 h after transfection, an equal amount of DMEM supplemented with 20% FCS, 8 mM glutamine, 200 U of penicillin per ml, and 200 μg of streptomycin per ml was added to the cells without removing the medium containing the DNA-lipid complexes. At 24 h posttransfection, the cells reached an optimal level of expression of the incorporated DNA and were used for pulse-chase experiments.
Pulse-chase analysis of TBE virus envelope protein folding.
Subconfluent 60-mm-diameter dishes of COS-1 cells expressing TBE virus envelope proteins were washed once with phosphate-buffered saline (PBS) containing 1 mM Mg2+ and 0.5 mM Ca2+ (PBS++), followed by starvation in cysteine- and methionine-free medium for 30 min. The cells were pulse-labeled at 37°C by adding 200 μCi of 35S-labeled methionine-cysteine (ProMix; Amersham) in cysteine- and methionine-free medium supplemented with 20 mM HEPES, pH 7.4. The pulse was stopped by washing the cells twice with 5 mM unlabeled cysteine and methionine in DMEM containing 10% FCS, glutamine, antibiotics, and 20 mM HEPES (pH 7.4), followed by incubation with this medium at 37°C for chase times varying between 0 and 60 min. For dithiothreitol (DTT) resistance experiments, the regular chase was followed by a 4-min incubation at 37°C in a chase medium supplemented with 5 mM DTT. DTT washout experiments were carried out in a similar way, by using pulse and chase media containing 5 mM DTT, followed by incubation in normal chase medium. After a chase, the cells were flooded with ice-cold PBS++ containing 20 mM N-ethylmaleimide (NEM), an alkylating reagent that prevents postlysis oxidation effects on the newly synthesized proteins (8). After 2 min of incubation, the cells were lysed in 800 μl of ice-cold lysis buffer containing 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid (CHAPS) in HEPES-buffered saline (HBS; pH 7.4) supplemented with 20 mM NEM, 1 mM phenylmethylsulfonyl fluoride, and 10 μg each of chymostatin, leupeptin, antipain, and pepstatin per ml. The nuclei and cellular debris were pelleted by centrifugation, and the postnuclear supernatants were used for immunoprecipitation.
Lysis of cells for sedimentation analysis was carried out with 1% Triton X-100 in 50 mM triethanolamine (pH 8.0) containing 20 mM NEM and protease inhibitors, followed by removal of nuclei and cellular debris as described above.
Immunoprecipitation and SDS-PAGE.
The postnuclear supernatants were precleared on protein A coupled to Sepharose CL-4B beads for 1 to 2 h at 4°C. Volumes of 60 to 120 μl of the precleared lysates were subsequently incubated with protein A-Sepharose CL-4B beads (10-μl bead volume) and 1 μl of rabbit polyclonal antibodies or mouse monoclonal antibodies (MAb) directed against the prM or E protein at 4°C overnight. Immune complexes were pelleted at 8,500 × g for 2 min, followed by washing with agitation three times for 10 min each. The first and second washing step were done with 0.5% CHAPS in HBS (pH 7.4), and the third one was done with HBS (pH 7.4). The washed immune complexes were solubilized by adding 40 μl of sample buffer and incubated at 95°C for 5 min. After pelleting of the protein A-Sepharose beads, the supernatant was divided into two aliquots. For reducing conditions, DTT was added to one of the aliquots to a final concentration of 100 mM. The proteins were analyzed by sodium dodecyl sulfate (SDS)-7.5 and 12% polyacrylamide gel electrophoresis (PAGE) on minigels, followed by autoradiography.
Antibodies.
For the majority of the precipitations, a polyclonal antiserum recognizing both prM and E was used [k-PM(2)] (2). Mouse MAbs specific for E (A5, B1, and C6) (16), prM (8H1) (20), or NS1 (LGT 3-509) (20) were used to detect various epitopes on the proteins and for coprecipitation experiments.
Determination of half-times for folding.
The half-times for folding of prM, E, and NS1 were determined by densitometric analysis of the protein bands from SDS-polyacrylamide gels after immunoprecipitation with conformation-specific antibodies (MAbs 8H1, A5, and LGT 3-509; see above) from at least three independent experiments. The band intensities (arbitrary units) were plotted against the chase time for each protein. The half-time represents the time point at which 50% of the maximal intensity was achieved.
Sedimentation analysis.
To monitor the formation of heterodimers consisting of prM and E, postnuclear supernatants from cell lysates solubilized in 1% Triton X-100 were loaded onto 5 to 20% (wt/wt) sucrose gradients made with 50 mM triethanolamine (pH 8.0), 100 mM NaCl, and 0.5% Triton X-100. After ultracentrifugation in a Beckman SW41 rotor at 38,000 rpm and 15°C for 22 h, fractions of 800 μl were collected by upward displacement and immunoprecipitated with a polyclonal antiserum recognizing both prM and E as described above. The immune complexes were analyzed by reducing SDS-PAGE followed by autoradiography.
Endo Hf digestion.
TBE virus envelope proteins were immunoprecipitated and washed three times as described above. The immune complexes were resuspended in 0.5% SDS-1% β-mercaptoethanol and heated for 10 min at 95°C. After pelleting of the protein A-Sepharose beads, the supernatant was divided into two equal aliquots. A 50 mM concentration of sodium citrate (pH 5.5) was added to both samples, and 20 U of endoglycosidase Hf (Endo Hf; New England Biolabs) was also added to one of them. The samples were incubated at 37°C for 90 min, followed by the addition of reducing sample buffer, separation of the proteins by SDS-12% PAGE, and autoradiography.
RESULTS
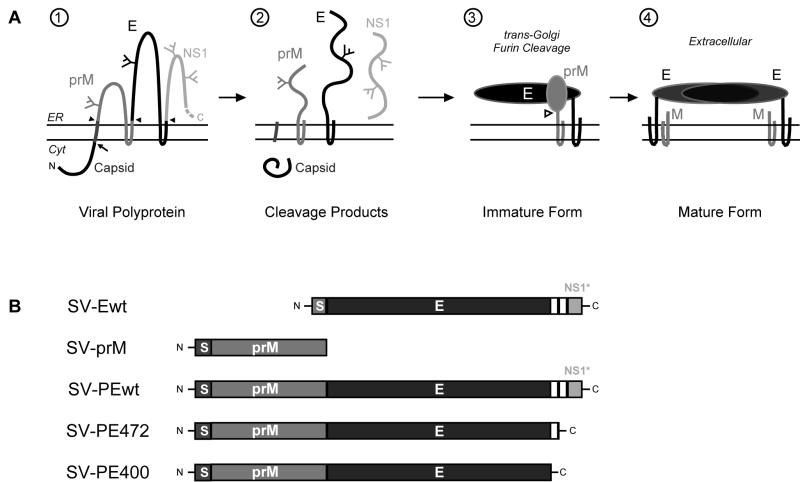
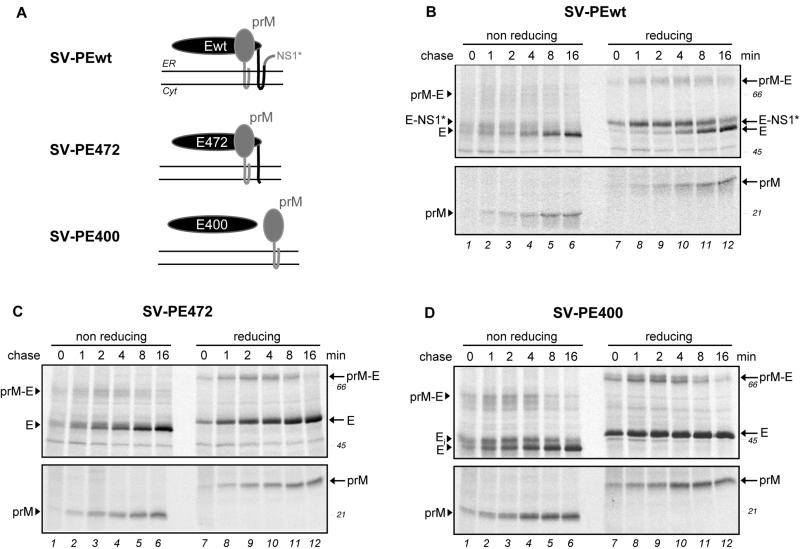
Figure 1A summarizes the biogenesis of the TBE virus envelope proteins, from the polyprotein being cleaved into distinct protein chains (illustrations 1 and 2), heterodimerization of prM and E (illustration 3), and the final processing steps leading to mature virions (illustration 4).
FIG. 1.
(A) Schematic diagram of the polyprotein cleavage, folding, and maturation of TBE virus envelope proteins prM and E and the first of the nonstructural proteins, NS1. The most N terminal of the proteins, the C protein, is cytosolic and is cleaved by the viral NS2B/3 protease (arrow). It is followed by a signal sequence cleaved by signal peptidase on the lumenal side (filled arrowhead). The signal peptidase also cleaves prM from the E protein and the E protein from the NS1 protein, leading to the translation products shown in illustration 2. PrM and E fold in the ER and associate with each other, forming heterodimers in immature virions (illustration 3). Cleavage of the N-terminal portion of the prM protein by the cellular protease furin (open arrowhead) after transport to the trans-Golgi network leads to mature virions and formation of E-E dimers on the virus surface (illustration 4). (B) Overview of the plasmids coding for various parts of the TBE virus polyprotein. The two transmembrane segments of the E protein are shown as white boxes. A few amino acids from the carboxy terminus of the C protein and the internal signal sequence (S) for proper translocation into the ER lumen were included at the N terminus of each construct. SV-PE472 and SV-PE400 lack one or both transmembrane regions at the C terminus of E, respectively. Cyt, cytosol; N, N terminus; C, C terminus.
A schematic overview of the plasmid constructs used is shown in Fig. 1B. Both SV-PEwt and SV-Ewt contained the first 30 codons of the NS1 nonstructural protein, which lies downstream of the E sequence in the polyprotein precursor. These nucleotides were included in the constructs to ensure proper translocation and membrane insertion at the C terminus of E.
Folding of TBE virus envelope proteins in infected cells.
To study the folding of TBE virus envelope proteins prM and E when they are expressed as part of the intact viral polyprotein, COS-1 cells infected with the TBE virus were analyzed by a pulse-chase approach. Twenty-four hours after infection, the cells were pulse-labeled for 2 min with 35S-labeled methionine-cysteine and chased for various times (up to 60 min). Before lysis with 2% CHAPS, the cells were treated with 20 mM NEM to alkylate any remaining free sulfhydryl groups, thus preventing further oxidative folding (8). The viral proteins were immunoprecipitated from postnuclear supernatants by using a polyclonal antiserum [k-PM(2)] that recognizes prM, E, and NS1 or by using MAbs specific for these proteins individually. The immunoprecipitates were analyzed by SDS-PAGE under reducing and nonreducing conditions, and the bands were visualized by autoradiography.
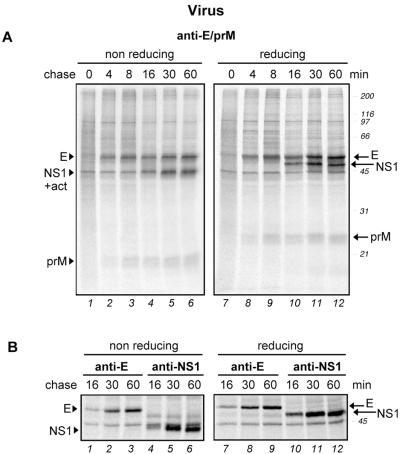
As shown in Fig. 2A, three major virus-derived bands could be detected in the reduced samples, i.e., prM, E, and NS1, all of which migrated with the expected mobilities (25, 52, and 45 kDa, respectively). The identification of the E and NS1 bands was confirmed by immunoprecipitation with specific mouse MAbs (Fig. 2B). In addition, a band at 45 kDa was detected that probably corresponds to labeled actin, which is often seen as a nonspecific contaminant in immunoprecipitations. E and prM were clearly visible after a 4-min chase (lanes 2 and 8), and NS1 was visible after 16 min (lanes 4 and 10). Since no major bands of lower mobility were observed, we concluded that all four of the cleavages needed to generate prM, E, and NS1 occurred rapidly, perhaps even cotranslationally. Furthermore, no cross-linked aggregated forms were detected at the top of the nonreducing lanes.
FIG. 2.
Folding of TBE virus envelope proteins in virus-infected mammalian cells. COS-1 cells were infected with TBE virus strain Neudoerfl at a multiplicity of infection of ca. 1. At 24 h postinfection, the cells were pulse-labeled with 35S-labeled methionine-cysteine for 2 min and chased for 0 to 60 min. This was followed by immunoprecipitation of the postnuclear supernatants with a polyclonal antiserum recognizing both prM and E (A) or MAbs specific for E and NS1, as indicated (B). The immunoprecipitates were analyzed by nonreducing and reducing SDS-PAGE (12% gel for panel A and 7.5% gel for panel B) and exposed for autoradiography. Positions of the individual proteins are marked at the side. In most of the gels, a nonspecific band was detected at ca. 45 kDa comigrating with NS1 under nonreducing conditions (+act) that probably corresponds to labeled actin. Molecular size standards are indicated on the right in kilodaltons.
Analysis of the more weakly labeled bands in the lower part of the gel in Fig. 2A revealed that the cleavages and oxidative folding of prM were rapid. prM underwent a large mobility shift when reduced, suggesting that one or more of the three disulfide bonds in this protein form large loops. Since we were not able to detect any oxidation intermediates of prM even after overexposing the gels for autoradiography, these disulfide bonds must have been formed during the 2-min pulse.
The greater separation of the molecular weight region of E and NS1 obtained with a 7.5% gel (Fig. 2B) revealed further details of their folding. First, it was clear that the nonreduced E band migrated faster than its reduced counterpart (e.g., lanes 3 and 9), consistent with the formation of intrachain disulfide bonds. Mature E contains six disulfide bridges, which are located in each of the three domains (33). That their combined effect on the migration of the protein on SDS-PAGE was relatively minor (but consistently observed) is most likely due to the rather small size of the covalent loops that they form in the polypeptide chain. We also observed that the polyclonal antiserum already reacted well with E after a 4-min chase (Fig. 2A, lane 2), whereas a MAb against domain II of E (A5) yielded a strong band only after 30 min (Fig. 2B, lanes 1 to 3). The intensity of the E band increased significantly between min 16 and 30 of the chase and less strongly between min 30 and 60 of the chase (Fig. 2B, lanes 1 to 3). Taken together, our results suggested that the half-time for the formation of the conformational epitope in domain II of E recognized by the MAb was about 15 to 20 min and that oxidation preceded attainment of the final native structure of the protein.
The NS1 protein was detected by a MAb recognizing a conformational epitope after 16 min (Fig. 2B, lane 4). A slight increase in the electrophoretic mobility of NS1 was observed between min 16 and 60 of the chase in both the nonreducing and reducing gels (lanes 4 to 6 and 10 to 12, respectively). This small shift was probably due to glucose trimming on the carbohydrate side chains of NS1. Recognition of the protein by a MAb started only after min 16 of the chase (lane 4 and data not shown), and folding appeared to be complete after 30 min (lane 5), since no further increase in the amount of precipitated NS1 was observed after 60 min (lane 6).
Our results suggest that the oxidative folding of TBE virus envelope proteins prM and E, as well as that of nonstructural protein NS1, occurs rapidly. However, judging by the appearance of epitopes recognized by the MAb used against E and NS1, conformational alterations continue to occur in these proteins until min 16 to 30 of the chase.
Folding of recombinant prM and E expressed individually.
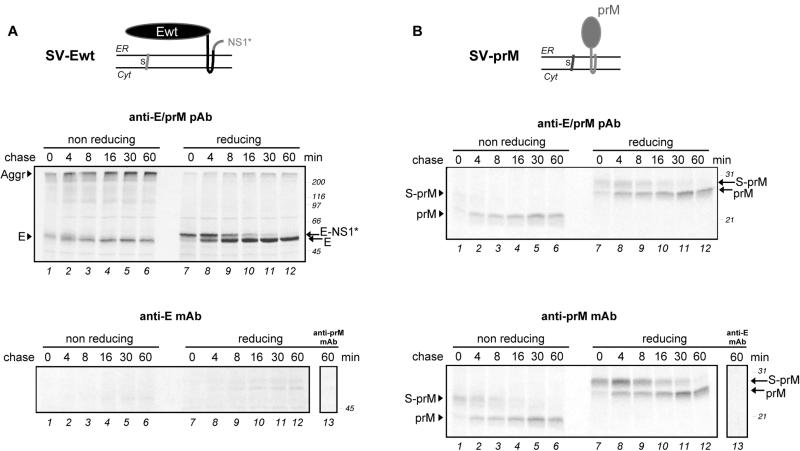
To examine the folding and maturation of prM or E alone, we used recombinant plasmids expressing various parts of the TBE virus polyprotein as schematically depicted in Fig. 1B. They all lacked the C protein at the N terminus, but two of them (SV-PEwt and SV-Ewt) contained a short 30-amino-acid stretch of the NS1 protein (called NS1*). This short sequence of NS1 was included to ensure the correct topology of the C-terminal end of the full-length E protein.
The constructs were expressed in COS-1 cells with a transient expression system. A pulse-chase protocol similar to that described above was used. When prM and E were expressed individually, relatively simple patterns were obtained (Fig. 3A and B, top). In the reduced gel, the E protein gave a single band (56 kDa) immediately after the pulse, which is labeled E-NS1* in Fig. 3A. The E-NS1* band disappeared with a half-time of 4 to 6 min and was replaced by a faster-migrating band (52 kDa) corresponding to the mobility of the mature wild-type E protein. Since the shift was not seen with the constructs SV-PE472 and SV-PE400, which produced C-terminally truncated forms of E and therefore also lacked the additional NS1 sequence (see below), we concluded that this shift was due to removal of the C-terminal NS1* stretch. Evidently, the signal peptidase-mediated cleavage was delayed compared to the corresponding cleavage in the full-length polyprotein (Fig. 2A). As described below, this delay was similar whether prM was expressed together with E or not.
FIG. 3.
Folding of recombinant TBE virus envelope proteins expressed individually. COS-1 cells were transfected with plasmids encoding either E (A) or prM (B). A schematic diagram of the topology of the constructs at the ER membrane is shown at the top of each panel. At 24 h posttransfection, the cells were pulse-labeled with 35S-labeled methionine-cysteine for 2 min and chased for 0 to 16 min. The postnuclear supernatants were immunoprecipitated either with a polyclonal antiserum recognizing both prM and E (upper panels) or with a MAb against prM or E (lower panels). Immunoprecipitation of E with an anti-prM MAb and vice versa (lower panels, lanes 13) confirmed that the antibodies were specific. The proteins were separated by nonreducing and reducing SDS-PAGE and visualized by autoradiography. Positions of the individual proteins are marked at the left, and molecular size standards are indicated on the right in kilodaltons.
Under nonreducing conditions, only a single band was observed in the SDS-PAGE gel. We therefore believe that this band represents the E protein and that most of the E-NS1* protein was present in the large disulfide-linked aggregates that remained at the top of the gel (Fig. 3A, lanes 1 to 3). If this is indeed the case, the gradual increase in electrophoretic mobility of the putative E band that occurs in the first 8 min of the chase can be attributed to oxidative folding.
A difference was also observed for prM expressed separately, compared to the full-length polyprotein in infected cells. As shown in Fig. 3B, a shift from a 29-kDa form to a 26-kDa form occurred under reducing conditions, with the latter corresponding to the authentic molecular mass of wild-type prM. Given the construct, this shift was clearly due to delayed cleavage at the N terminus of prM, which consists of a few C-terminal residues of the C protein and the internal signal sequence of prM. The half-time of this cleavage was about 4 to 8 min. Interestingly, the delayed cleavage was not seen when the prM sequence was followed by E (see Fig. 4), indicating that the timing of this cleavage depended on the downstream sequences.
FIG. 4.
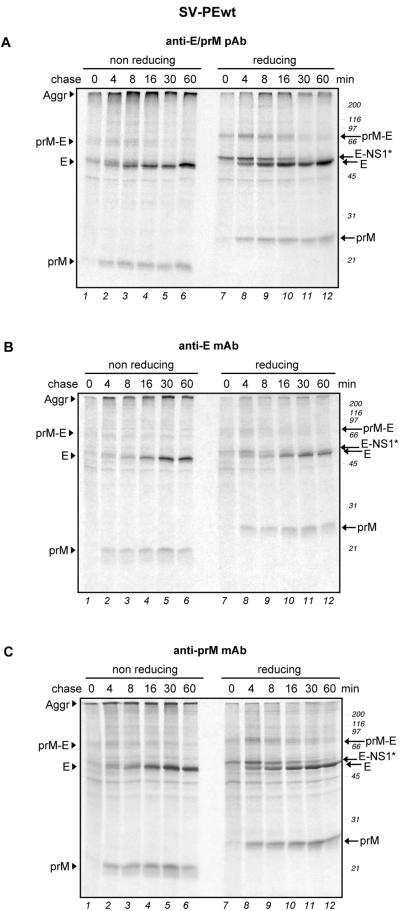
Folding of TBE virus envelope proteins expressed as a polyprotein precursor. COS-1 cells were transfected with a recombinant plasmid encoding prM and E. At 24 h posttransfection, the folding of prM and E was assayed by a pulse-chase approach, followed by immunoprecipitation of the postnuclear supernatants with a polyclonal antiserum recognizing both prM and E (A) or MAb specific for prM and E, as indicated (B and C). The immunoprecipitates were analyzed by nonreducing and reducing SDS-PAGE. Positions of the proteins are marked at the left, and molecular size standards are indicated on the right in kilodaltons.
We have also analyzed the prM and E proteins expressed individually by immunoprecipitation with anti-prM or anti-E mouse MAbs. Although recognized by MAbs against domains I and III (B1 and C6, respectively; data not shown), E synthesized alone was hardly precipitated by the MAb against domain II (A5). Only faint bands were visible after a 30- to 60-min chase (Fig. 3A, bottom). This was a clear difference from E expressed as part of the full-length polyprotein (Fig. 2B), suggesting that E is not able to attain full maturity when synthesized alone. On the other hand, prM was precipitated with an anti-prM MAb, indicating that, at least by this criterion, prM was able to fold independently of other viral components even though the signal sequence cleavage at its N terminus was delayed.
Thus, the two TBE virus envelope proteins matured to different extents when produced outside of their usual context within the polyprotein. The E protein expressed alone could not attain its final conformation, while prM synthesized individually appeared to be completely folded, but proteolytic cleavage at its N terminus was delayed.
Folding of prM and E expressed from the same polyprotein precursor.
To test the cleavage processes and folding of prM and E when they were expressed together but without other viral proteins, we performed pulse-chase experiments with the SV-PEwt plasmid (Fig. 1B), which encodes a polyprotein consisting of a few amino acid residues from the C terminus of the C protein, followed by the internal prM signal sequence, prM, E, and the NS1* 30-amino-acid segment. Figure 4A shows the autoradiography of an immunoprecipitation with an antiserum recognizing both prM and E. Both proteins oxidized rapidly and with the same kinetics as when they were expressed individually. Under reducing conditions, the band shift from E-NS1* to E was observable again and aggregates were again observed at the top of the lanes under nonreducing conditions. This, together with the experiment shown in Fig. 3A, indicated that the cleavage of NS1* from the C terminus of E occurred independently of prM.
PrM folded properly within 2 min after synthesis and displayed a clear shift between nonreducing and reducing conditions, as shown for prM expressed alone. However, there was no double band visible for prM, as was the case for prM expressed alone. This suggests that signal sequence cleavage at the N terminus of prM is accelerated when the downstream E protein is present on the polyprotein. The transiently expressed band around 70 kDa detected under both nonreducing and reducing conditions between min 0 and 16 of the chase probably corresponded to the noncleaved prM-E precursor because it was precipitated with antibodies to both prM and E (Fig. 4B and C, respectively). Small amounts of this oxidized precursor were obtained with all of the constructs expressing prM and E together.
Moreover, in contrast to the individually expressed E protein (Fig. 3A, lower panel), E from the SV-PEwt construct was efficiently precipitated by MAb A5 when cosynthesized with prM (Fig. 4B). This indicated that E was able to reach a conformation detectable by this antibody only when prM was present.
The half-times of prM and E folding were determined by densitometric analysis of the bands shown in Fig. 4C and B, respectively. We obtained half-times of 3 to 4 min for prM and ca. 15 min for E.
The interaction between prM and E involved formation of complexes, since the two proteins could be coprecipitated after a 4-min chase with either anti-E or anti-prM MAb (Fig. 4B and C, respectively). A small fraction of E and E-NS1* was precipitated with the anti-prM antibody after a 0-min chase (Fig. 4C, lanes 1 and 7).
Our results indicated that, when synthesized together, prM and E begin to form stable complexes within a few minutes after synthesis and that this association is needed for E to achieve its final conformation. The presence of E also results in faster signal sequence cleavage at the N terminus of prM.
Folding of C-terminally truncated variants of E.
Earlier studies have revealed several functional elements in the C-terminal stem-anchor region of the E protein (3), including a small helix involved in trimerization of E under acidic conditions, a conserved region of unknown function, and a second helix required for interaction with prM. To test whether the transmembrane domain at the C terminus of E has an effect on its folding, we analyzed the folding of prM and C-terminally truncated variants of E lacking the second transmembrane segment (SV-PE472, i.e., prM and the first 472 amino acids of protein E) or both transmembrane segments and the stem region (SV-PE400) of E in a pulse-chase experiment.
Figure 5A depicts the predicted ER membrane topology of these proteins, and Fig. 5B, C, and D show the SDS-PAGE of immunoprecipitates with an anti-prM/E polyclonal antiserum. To maximize the resolution for each of the proteins, SDS-7.5% acrylamide gels were used for E and SDS-12% acrylamide gels were used for prM. The shift from the 56-kDa band to the 52-kDa band under reducing conditions was present only for the SV-PEwt construct, confirming, as discussed above, that this shift was indeed due to C-terminal cleavage of NS1* from E. Moreover, an additional faint band, migrating slightly slower than E and probably corresponding to E-NS1*, was detected for SV-PEwt (Fig. 5B) under nonreducing conditions. E472 showed a folding pattern similar to that of Ewt (Fig. 5C), but it achieved its fully oxidized state a little earlier than Ewt. E400 oxidized more rapidly, with some completely oxidized protein form being visible after a 0- to 1-min chase (Fig. 5D), and also showed a higher degree of aggregation at the top of the nonreducing gel (data not shown). An additional transient band with slightly less mobility than the fully oxidized form of E400 was visible under nonreducing conditions (Fig. 5D, lanes 1 to 5). This band probably corresponded to incompletely oxidized folding intermediates, which were further oxidized by a 16-min chase. For all three constructs, some prM-E precursor was detected that became cleaved after an 8- to 16-min chase. PrM displayed identical folding kinetics with all three constructs and was fully oxidized after a 2-min pulse and a 0- to 4-min chase. In a separate experiment, both E472 and E400 could be precipitated by the A5 MAb, indicating that E can fold without the stem-anchor region (data not shown). Taken together, the data suggest that truncations at the C-terminal end of E had a minor effect on its folding kinetics and no influence on the folding of prM.
FIG. 5.
Folding of C-terminally truncated forms of TBE virus E proteins. COS-1 cells were transfected with recombinant plasmids encoding prM and full-length E (SV-PEwt), prM and E lacking the second transmembrane domain (SV-PE472), or prM and stem- and membrane anchor-free E (SV-PE400). A schematic view of the topology of the proteins at the ER membrane is depicted in panel A. The degree of interaction of the E protein with prM is diminished when both transmembrane domains are deleted. Folding of the E variants and prM was assessed by a pulse-chase experiment, followed by anti-prM/E immunoprecipitation as described in the legend to Fig. 4. For each construct, the proteins were resolved by SDS-7.5 and 12% PAGE (top and bottom, respectively), followed by autoradiography (B, C, and D). A folding intermediate for E400 (Ei) can be seen under nonreducing conditions (D). Positions of the proteins are marked at the left, and molecular size standards are indicated on the right in kilodaltons.
Folding of prM and E expressed in trans.
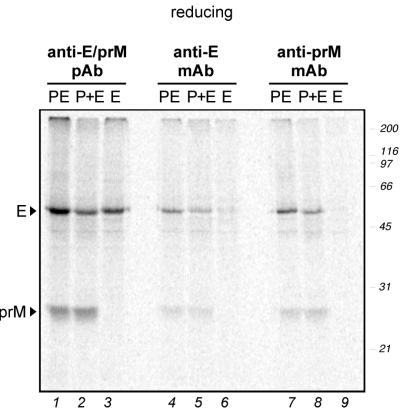
The results shown so far suggested that the E protein needs prM to achieve its final native conformation. To test whether this effect depends on expression of prM and E on the same polyprotein, a pulse-chase experiment was carried out with COS-1 cells transfected with the SV-PEwt plasmid (expression of prM and E in cis), with SV-prM and SV-Ewt (expression in trans), or with SV-Ewt alone. The cells were pulsed for 2 min and chased for 60 min. Aliquots of the cellular extracts were immunoprecipitated with three different antibodies: a rabbit anti-prM/E polyclonal antiserum, a mouse anti-E MAb, and a mouse anti-prM MAb.
The autoradiogram of a reducing SDS-PAGE with the immunoprecipitates is shown in Fig. 6. PrM and E were coprecipitated with both of the anti-E MAb when coexpressed either in cis (lanes 4 and 7) or in trans (lanes 5 and 8). This indicates that prM and E can form complexes whether synthesized on the same polyprotein or not.
FIG. 6.
Folding of TBE virus envelope proteins expressed in trans. COS-1 cells transfected with a plasmid coding for prM and E (SV-PEwt) as a polyprotein (expression in cis; PE), with plasmids encoding prM (SV-prM) and E (SV-Ewt) separately (expression in trans; P+E), or with a plasmid coding for E alone (SV-Ewt; E) were pulsed for 2 min and chased for 60 min. Total cellular extracts were immunoprecipitated with either a rabbit anti-prM/E polyclonal antiserum, an anti-E MAb, or a mouse anti-prM MAb as indicated above the lanes, and this was followed by analysis of the proteins by reducing SDS-PAGE. Positions of the proteins are marked at the left, and molecular size standards are indicated on the right in kilodaltons.
DTT sensitivity of the E protein.
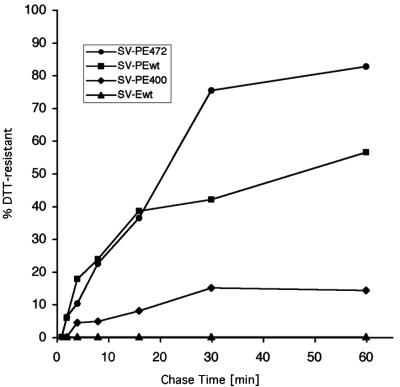
One parameter that has been used to assess the folding state of a protein is its resistance to reducing agents such as DTT added to the medium (21, 38). When proteins are incompletely folded, their disulfide bonds are more exposed and therefore much more sensitive to such treatment than when the proteins have achieved their native conformation. Earlier studies on influenza virus hemagglutinin (HA) have shown that the subunits become resistant soon after reaching the fully oxidized state but before trimer formation (38, 39). We measured the DTT sensitivity of E expressed from SV-PEwt, SV-PE472, SV-PE400, or SV-Ewt after different chase times. The fraction of each variant resistant to 5 mM DTT is plotted in Fig. 7. When E was expressed from the SV-PE472 construct, 80% became resistant after a 60-min chase. By that time, 50% of the full-length E protein (SV-PEwt) acquired DTT resistance. In contrast, only 15% of the membrane anchor-free form of E (SV-PE400) was resistant to reduction 60 min after synthesis. Interestingly, E expressed without prM (SV-Ewt) did not become resistant to DTT treatment at all. We conclude that prM is required for E to reach the DTT-resistant conformation. This result is consistent with the observation described above that the final stages of E folding depend on association with prM.
FIG. 7.
DTT resistance of C-terminally truncated TBE virus E proteins. Cells were transfected with recombinant plasmids as indicated and subjected to a pulse-chase experiment with 2-min pulses and chases of 0 to 60 min. After a chase in normal nonreducing medium, the cells were chased in DTT-containing medium for an additional 4 min. The ratio of DTT-resistant E protein to total E protein was plotted for the Ewt, E472, and E400 forms.
We also tested the effect of DTT on the folding of prM and E by a DTT washout pulse-chase experiment (data not shown). When DTT is washed out of cells, the oxidative environment in the ER is rapidly restored and reduced proteins can resume their oxidative folding program (7). When DTT was washed out after a 2-min pulse and a 4-min chase, both prM and E resumed the correct oxidation but aggregation was strongly increased, as shown by analysis of the proteins under nonreducing SDS-PAGE. Reuptake of the folding process combined with increased aggregation was also observed in DTT washout experiments with the SV-PE472 and SV-PE400 constructs (data not shown).
These results suggested that the proteins were able to fold posttranslationally upon removal of the reducing milieu, but this led to a decrease in folding efficiency, as well as to a significantly higher degree of aggregation, probably due to the formation of intermolecular disulfide bonds.
Formation of heterodimers between TBE virus envelope proteins.
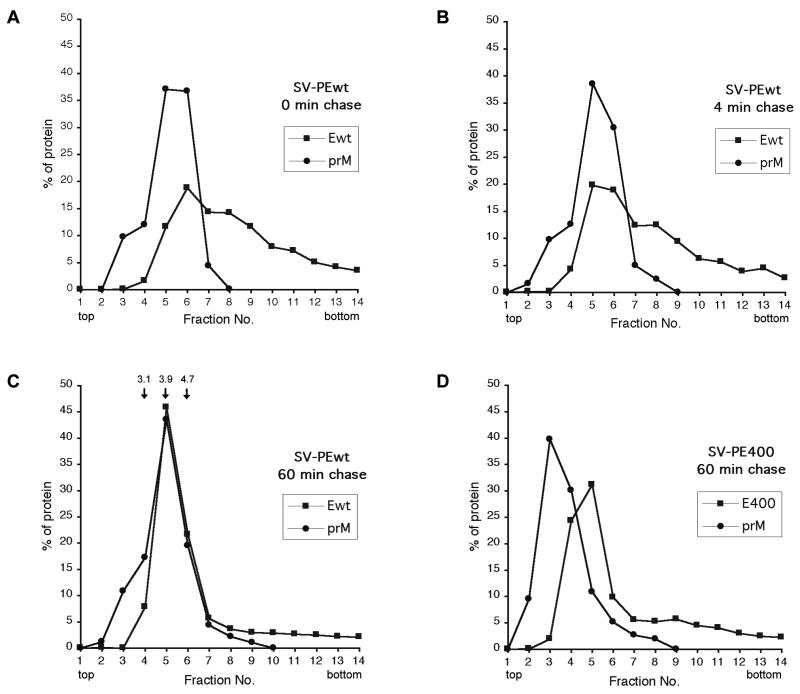
Our results from coimmunoprecipitations of prM and E suggested that stable complexes of the two envelope proteins formed early during folding. To further characterize the prM-E heterodimer formation, we subjected the postnuclear supernatants from a pulse-chase experiment to sucrose velocity gradient centrifugation in gradients containing 0.5% Triton X-100. As shown in Fig. 8A, the peaks of prM and E overlapped partially at 0-min chase. The prM peak was seen in fractions 5 and 6. The E protein peaked in fraction 6, but it was also detected in faster-sedimenting fractions. This indicated that weak interactions between some prM and E molecules occurred, while a larger portion of E formed higher-molecular-weight clusters or aggregates. Consistent with the results obtained by coimmunoprecipitations (cf. Fig. 4B and C), interaction between prM and E was further increased after a 4-min chase. (Fig. 8B). After a 60-min chase, prM and E cosedimented almost completely in fractions 5 and 6 with an average S20 sedimentation coefficient of 4.0, indicating that most of the newly synthesized prM and E molecules had dimerized (Fig. 8C).
FIG. 8.
Cosedimentation analysis of prM and E at various chase times. Cells transfected with recombinant plasmids encoding prM and Ewt (A to C) or prM and stem- and membrane anchor-free E400 (D) were pulsed for 2 min and chased for the indicated times. The postnuclear supernatants were subjected to velocity sedimentation on 5 to 20% (wt/wt) sucrose gradients. Subsequently, the proteins were immunoprecipitated from the gradient fractions with a polyclonal antiserum recognizing both prM and E, analyzed by reducing SDS-PAGE, and visualized by fluorography. The relative amount of prM and E protein from each fraction (out of a total of 100% over all 14 fractions) was quantitated. The arrows above fractions 4 to 6 in panel C indicate the S20 sedimentation coefficient standards, as determined by quantification of a velocity sedimentation gradient run with a protein molecular weight marker.
Earlier studies have shown that the membrane anchor-free E400 protein does not form a stable complex with the prM protein (2). We confirmed this result by analyzing the sedimentation behavior of E400 and prM (Fig. 8D). In this case, the proteins peaked at different fractions and E400 and prM showed average sedimentation coefficients of 3.6 and 2.8, respectively. Thus, prM and E form heterodimers at very early stages of folding but stable association does not occur without the stem region.
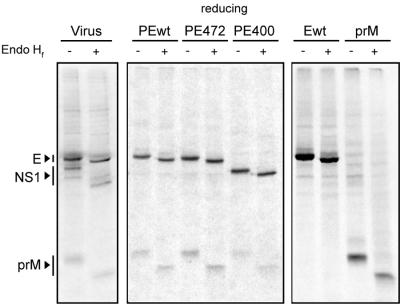
Endo Hf digestion of TBE virus envelope proteins.
Finally, to check the further processing of the newly synthesized TBE virus envelope proteins, the glycosylation state of prM and E was examined by an Endo Hf digestion assay of the immunoprecipitated TBE virus envelope proteins expressed from different DNA plasmids and virus-infected cells. Both prM and E remained fully sensitive to Endo Hf treatment after a 60-min chase with all of the constructs tested in this study, as well as in virus-infected cells (Fig. 9). The proteins did not acquire resistance to digestion with Endo Hf after a 24-h chase (data not shown). This indicated that both prM and E were retained in a pre-Golgi compartment.
FIG. 9.
Endo Hf sensitivity of TBE virus envelope proteins. Two-minute pulse and 60-min chase samples were immunoprecipitated with a polyclonal antiserum recognizing both prM and E, followed by digestion of the immune complexes with Endo Hf. Subsequently, the proteins were analyzed by reducing SDS-PAGE and visualized by autoradiography. All recombinant constructs plus a sample from virus-infected cells were tested, as indicated at top of the lanes.
DISCUSSION
Generation of proteins in the form of a polyprotein in the ER involves a variety of complexities that are not encountered when proteins are synthesized individually. The cleavage reactions must be orchestrated correctly on both sides of the membrane, and folding must be organized in such a way that the protein components support rather than interfere with each other during maturation. Our results indicate that the TBE virus envelope proteins must associate with each other shortly after synthesis for efficient cleavage of prM from the polyprotein precursor and for proper folding of E. However, expression of prM and E without other viral components is sufficient for the attainment of their fully mature form.
The three signal peptidase cleavages involved in the synthesis of the two TBE virus envelope proteins were found to occur immediately after synthesis in infected cells, and judging by our assays, the folding process was efficiently completed within 30 min after chain termination. At this time, the proteins were already heterodimers, a prerequisite for virus production and secretion. Although complete folding of prM appeared to be possible in the absence of E, interaction with prM and the formation of a heterooligomeric complex were required for E to attain its final conformation. prM and E had to form heterooligomeric complexes before they could reach their final conformations, but it was not necessary for them to be synthesized from the same polyprotein precursor.
It can be estimated that translation takes about 35 s for prM and about 110 s for E (8). As a rule, both prM and E were already visible as cleaved products after a 2-min pulse and a 0- to 4-min chase in recombinant constructs and virus-infected cells. This suggests that the amino-terminal and carboxy-terminal cleavages occurred as soon as the synthesis of each segment was completed.
Other studies have shown that cleavage at the C-prM junction of flaviviruses involves two coordinated cleavages (4, 37). Cleavage of the N-terminal signal sequence of prM by a host cellular peptidase requires prior removal of the C protein from this signal sequence by the viral NS2B/3 protease. Our experiments with virus-infected cells suggest that the cleavages in the C-prM region occurred fast and efficiently, because (i) no uncleaved precursor forms were detected, (ii) the ratio of prM to E did not increase with longer chase times, and (iii) the amounts of prM and E found in virus-infected cells were similar to those obtained with recombinant constructs in which the C protein was not present.
Based on our data, we further suggest that processing of the C-prM junction by NS2B/3 occurs in trans rather than in cis. The NS2B/3 protease sequence ranges from amino acid 1359 to amino acid 2111 in the polyprotein (26). Thus, approximately 7 min is required for complete translation of NS2B/3 after the C and prM proteins have been synthesized. However, in our experiments, the C protein has already been cleaved from prM by that time, essentially ruling out in-cis cleavage.
Elimination of the E sequence following the prM protein in one of the recombinant constructs resulted in slower cleavage at the N terminus of prM, suggesting that the timing of this cleavage is dependent on the presence of downstream E sequences. This implies that processing of the prM signal sequence occurs only after at least part of the E protein has been synthesized.
In all of the constructs that contained both prM and full-length E or truncated forms of E, a distinct uncleaved prM-E side product was observed as a band that disappeared with a half-time of 30 min, suggesting that if the cleavage between prM and E did not occur cotranslationally immediately after passing the translocon, it was delayed. This phenomenon may be explained by loss of easy access to the signal peptidase after the newly synthesized protein has left the translocon complexes.
When cleaved, the prM protein was already in a form that migrated fast on nonreducing SDS-PAGE. This means that of the three potential disulfide bonds, at least one that forms a large loop must have been in place. It is possible that the protein was, in fact, completely oxidized by the time it was released from the translocon because no further changes in mobility were seen. That disulfide bond formation can take place cotranslationally is well known from previous studies with influenza virus HA and Semliki Forest virus glycoproteins (10, 29). To be oxidized, the prM protein did not have to separate from its signal peptide or be associated with E. The prM protein thus emerged as a rapid and robust folder.
Oxidative folding of E was much slower than that of prM, extending over a time period of 8 to 16 min after chain termination. Although the structure revealed by X-ray crystallography shows the presence of six disulfide bonds in the mature ectodomain, the shifts in mobility of the protein remained modest. The reason was most likely the small relative size of the loops formed. Although oxidation did take place in the absence of prM, it was apparent that full maturation of E and expression of a conformational epitope could only take place once a stable heterodimer with prM had been formed. The data thus indicate that the folding of E takes place in two phases, one before the formation of a stable heterodimer and one that proceeds slowly after formation of the dimer. The need for cosynthesis for folding and secretion has been previously demonstrated in other flaviviruses (22, 31). Based on those reports and the results presented in this study, prM may be considered a chaperone for the folding and dimerization of flavivirus envelope proteins.
By measuring the appearance of a conformational epitope on each of the individual proteins, we obtained folding half-times of 3.5 to 4 min for prM, 15 to 20 min for E, and ca. 18 min for NS1. The folding of another member of the Flavivirus genus has recently been analyzed (11). In the case of dengue virus, the half-times for folding were 15 to 20 min for prM and 60 min for E. Thus, the dengue glycoproteins fold considerably more slowly than their TBE counterparts. The reason for the differences in the observed folding rates is not clear, but since the methods used in the two studies were similar, it is likely that different flaviviruses exhibit different glycoprotein folding efficiencies, a phenomenon that has also been observed with HA from different influenza virus strains (8).
Compared with envelope proteins from other virus families that are synthesized as polyproteins, TBE virus E folds at comparable rates, while prM is an unusually rapid and highly efficient folder. Semliki Forest virus envelope proteins E1 and p62 are folded after 10 and 20 min, respectively (28). Envelope protein E1 of HCV is a slow folder, getting oxidized only after 60 min, whereas HCV E2 can form its disulfide bonds within 10 min of synthesis (14). In the case of Uukuniemi virus of the Bunyamwera family, G1 folds rapidly, within 10 min, while G2 only reaches its folded form after 60 min (32).
Heterodimer formation of TBE virus envelope proteins starts within a few minutes after translation, probably as soon as prM is fully folded. In the case of dengue virus, the two envelope proteins also dimerize within 15 min after synthesis but the precise dimerization half-time was not determined (40). Our results showed that interaction between prM and E can also occur when the two proteins are expressed from different messengers; i.e., they can interact productively without being synthesized as part of a common polyprotein. Whether such dimerization in trans occurs in infected cells, where the proteins emerge from the same polyprotein, is not clear from our data. However, we believe that, in a natural infection, in-cis dimerization is more likely because the sucrose gradient data show the prM protein already associating with E within 4 min of synthesis. Moreover, it does so with the simultaneously labeled E protein. If dimerization occurred in trans, one would expect dimers to form between the newly synthesized prM and previously synthesized, unlabeled E proteins that are already further matured. This has been reported for Uukuniemi virus G1 and G2 (32). Furthermore, dimerization in cis is probable because the ratio of labeled prM to E protein remained the same whether E was coprecipitated with an anti-prM antibody or vice versa.
A recent study has shown striking structural similarities between the TBE virus E protein and the E1 protein of the alphavirus Semliki Forest virus (23). Although alphavirus envelope proteins make use of distinct chaperones, they also interact with each other a few minutes after synthesis by forming heterodimers in cis (6). For HCV, another member of the Flaviviridae family, interactions between E1 and E2 occur early but these proteins seem to be aggregated and it takes much longer for them to form native, stable heterodimeric complexes (12, 13). In the case of Uukuniemi virus, a bunyavirus, the envelope glycoproteins G1 and G2 form heterodimers rapidly after synthesis although dimerization occurs in trans rather than in cis (32).
Our results confirmed that the ectodomain of E without a membrane anchor cannot stably associate with prM (3). Although the protein appeared to be folded, as judged by MAb reactivity (data not shown), most of it remained DTT sensitive. Since E expressed without prM did not acquire DTT resistance, we concluded that interaction of prM with E was required for the latter to reach its final folded state.
We also observed that deletion of the stem and membrane anchor of E caused oxidation of the protein to proceed more rapidly. This generated a partially oxidized intermediate that was prominent during folding of the E400 form but could not be detected for the membrane-anchored forms of E.
The single N-linked glycans on prM and E expressed individually or together remained fully sensitive to digestion with Endo Hf several hours after synthesis. The secreted forms of E, however, acquire resistance to treatment with Endo Hf (34). Influenza virus HA, which has folding kinetics similar to those of E, becomes resistant to Endo Hf digestion with a half-time of 15 min, indicating that it has reached the Golgi by that time (35). Therefore, it is likely that prM and E are retained in the ER unless they are incorporated into virus particles, which is consistent with the observation that flavivirus assembly occurs at the ER membrane (reviewed in reference 24).
In summary, we have provided evidence for coordinated maturation of the glycoproteins of the TBE virus. The results indicate that E requires prM to reach the native conformation efficiently, suggesting a chaperone-like role for prM. On the other hand, prM needs E for rapid signal sequence cleavage at its N terminus. PrM can exert its assisting function on E when coexpressed either in cis or in trans. Dimerization of the two proteins occurs rapidly and is required for the final folding steps. However, the proteins flanking prM and E in the TBE virus polyprotein do not have a major influence on the folding process.
Acknowledgments
We thank Connie Schmaljohn (USAMRIID, Fort Detrick, Md.) for providing anti-prM and anti-NS1 antibodies; Karin Mench, Silvia Röhnke, and Walter Holzer for excellent technical assistance; and Karin Stiasny, Lars Ellgaard, and Srinivas Venkitraman for critical reading of the manuscript.
This work was supported by a grant from the Swiss Federal Institute of Technology to A.H. I.C.L. was a recipient of a short-term fellowship from the European Molecular Biology Organization.
REFERENCES
- 1.Allison, S. L., C. W. Mandl, C. Kunz, and F. X. Heinz. 1994. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes 8:187-198. [DOI] [PubMed] [Google Scholar]
- 2.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amberg, S. M., and C. M. Rice. 1999. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J. Virol. 73:8083-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson, H., B. U. Barth, M. Ekstrom, and H. Garoff. 1997. Oligomerization-dependent folding of the membrane fusion protein of Semliki Forest virus. J. Virol. 71:9654-9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barth, B. U., J. M. Wahlberg, and H. Garoff. 1995. The oligomerization reaction of the Semliki Forest virus membrane protein subunits. J. Cell Biol. 128:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braakman, I., J. Helenius, and A. Helenius. 1992. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 11:1717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braakman, I., H. Hoover-Litty, K. R. Wagner, and A. Helenius. 1991. Folding of influenza hemagglutinin in the endoplasmic reticulum. J. Cell Biol. 114:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 10.Chen, W., J. Helenius, I. Braakman, and A. Helenius. 1995. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc. Natl. Acad. Sci. USA 92:6229-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courageot, M. P., M. P. Frenkiel, C. D. Dos Santos, V. Deubel, and P. Despres. 2000. Alpha-glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J. Virol. 74:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guirakhoo, F., R. A. Bolin, and J. T. Roehrig. 1992. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology 191:921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guirakhoo, F., F. X. Heinz, and C. Kunz. 1989. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology 169:90-99. [DOI] [PubMed] [Google Scholar]
- 17.Guirakhoo, F., F. X. Heinz, C. W. Mandl, H. Holzmann, and C. Kunz. 1991. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol. 72:1323-1329. [DOI] [PubMed] [Google Scholar]
- 18.Heinz, F. X., and C. Kunz. 1981. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. J. Gen. Virol. 57:263-274. [DOI] [PubMed] [Google Scholar]
- 19.Heinz, F. X., and C. W. Mandl. 1993. The molecular biology of tick-borne encephalitis virus. APMIS 101:735-745. [DOI] [PubMed] [Google Scholar]
- 20.Iacono-Connors, L. C., J. F. Smith, T. G. Ksiazek, C. L. Kelley, and C. S. Schmaljohn. 1996. Characterization of Langat virus antigenic determinants defined by monoclonal antibodies to E, NS1 and preM and identification of a protective, non-neutralizing preM-specific monoclonal antibody. Virus Res. 43:125-136. [DOI] [PubMed] [Google Scholar]
- 21.Kaji, E. H., and H. F. Lodish. 1993. In vitro unfolding of retinol-binding protein by dithiothreitol. Endoplasmic reticulum-associated factors. J. Biol. Chem. 268:22195-22202. [PubMed] [Google Scholar]
- 22.Konishi, E., and P. W. Mason. 1993. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J. Virol. 67:1672-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 24.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 25.Mandl, C. W., F. X. Heinz, and C. Kunz. 1988. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology 166:197-205. [DOI] [PubMed] [Google Scholar]
- 26.Mandl, C. W., F. X. Heinz, E. Stockl, and C. Kunz. 1989. Genome sequence of tick-borne encephalitis virus (Western subtype) and comparative analysis of nonstructural proteins with other flaviviruses. Virology 173:291-301. [DOI] [PubMed] [Google Scholar]
- 27.Michalak, J. P., C. Wychowski, A. Choukhi, J. C. Meunier, S. Ung, C. M. Rice, and J. Dubuisson. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299-2306. [DOI] [PubMed] [Google Scholar]
- 28.Molinari, M., and A. Helenius. 2000. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science 288:331-333. [DOI] [PubMed] [Google Scholar]
- 29.Molinari, M., and A. Helenius. 1999. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature 402:90-93. [DOI] [PubMed] [Google Scholar]
- 30.Nowak, T., and G. Wengler. 1987. Analysis of disulfides present in the membrane proteins of the West Nile flavivirus. Virology 156:127-137. [DOI] [PubMed] [Google Scholar]
- 31.Ocazionez Jimenez, R., and B. A. Lopes da Fonseca. 2000. Recombinant plasmid expressing a truncated dengue-2 virus E protein without co-expression of prM protein induces partial protection in mice. Vaccine 19:648-654. [DOI] [PubMed] [Google Scholar]
- 32.Persson, R., and R. F. Pettersson. 1991. Formation and intracellular transport of a heterodimeric viral spike protein complex. J. Cell Biol. 112:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 34.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, I., R. W. Doms, K. R. Wagner, and A. Helenius. 1990. Intracellular transport of soluble and membrane-bound glycoproteins: folding, assembly and secretion of anchor-free influenza hemagglutinin. EMBO J. 9:631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stocks, C. E., and M. Lobigs. 1995. Posttranslational signal peptidase cleavage at the flavivirus C-prM junction in vitro. J. Virol. 69:8123-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatu, U., I. Braakman, and A. Helenius. 1993. Membrane glycoprotein folding, oligomerization and intracellular transport: effects of dithiothreitol in living cells. EMBO J. 12:2151-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatu, U., C. Hammond, and A. Helenius. 1995. Folding and oligomerization of influenza hemagglutinin in the ER and the intermediate compartment. EMBO J. 14:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, S., R. He, and R. Anderson. 1999. PrM- and cell-binding domains of the dengue virus E protein. J. Virol. 73:2547-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]