Abstract
With the recent introduction of neuraminidase (NA) inhibitors into clinical practice for the treatment of influenza virus infections, considerable attention has been focused on the potential for resistance development and cross-resistance between different agents from this class. A-315675 is a novel influenza virus NA inhibitor that has potent enzyme activity and is highly active in cell culture against a variety of strains of influenza A and B viruses. To further assess the therapeutic potential of this compound, in vitro resistance studies have been conducted and a comparative assessment has been made relative to oseltamivir carboxylate. The development of viral resistance to A-315675 was studied by in vitro serial passage of influenza A/N9 virus strains grown in MDCK cells in the presence of increasing concentrations of A-315675. Parallel passaging experiments were conducted with oseltamivir carboxylate, the active form of a currently marketed oral agent for the treatment of influenza virus infections. Passage experiments with A-315675 identified a variant at passage 8 that was 60-fold less susceptible to the compound. Sequencing of the viral population identified an E119D mutation in the NA gene, but no mutations were observed in the hemagglutinin (HA) gene. However, by passage 10 (2.56 μM A-315675), two mutations (R233K, S339P) in the HA gene appeared in addition to the E119D mutation in the NA gene, resulting in a 310-fold-lower susceptibility to A-315675. Further passaging at higher drug concentrations had no effect on the generation of further NA or HA mutations (20.5 μM A-315675). This P15 virus displayed 355-fold-lower susceptibility to A-315675 and >175-fold-lower susceptibility to zanamivir than did wild-type virus, but it retained a high degree of susceptibility to oseltamivir carboxylate. By comparison, virus variants recovered from passaging against oseltamivir carboxylate (passage 14) harbored an E119V mutation and displayed a 6,000-fold-lower susceptibility to oseltamivir carboxylate and a 175-fold-lower susceptibility to zanamivir than did wild-type virus. Interestingly, this mutant still retained susceptibility to A-315675 (42-fold loss). This suggests that cross-resistance between A-315675- and oseltamivir carboxylate-selected variants in vitro is minimal.
Influenza virus is a negative-stranded RNA virus consisting of two major viral proteins, neuraminidase (NA) and hemagglutinin (HA), that protrude from the surface of the virus and mediate critical binding events involved in the initial infection of host cells as well as the release of newly formed virions (7, 29). Inhibition of viral NA enzymatic activity leads to aggregation of progeny virus at the surface of the initially infected cell, effectively halting the progression of infection (26, 27). Thus, NA plays a critical role in completing the viral replication cycle.
NA makes an attractive target for inhibition because the amino acids that line the active site are highly conserved among all clinically relevant influenza viruses (25). Indeed, the NA inhibitors zanamivir, oseltamivir carboxylate (formerly GS4071), and BCX-1812 have demonstrated broad-spectrum inhibition of influenza virus replication in cell culture as well as in vivo efficacy in animal models of influenza virus infection (1, 2, 10, 19, 22, 24, 30). Importantly, zanamivir and oseltamivir carboxylate have demonstrated efficacy in phase II and III clinical trials for the treatment of naturally acquired influenza virus, which was the basis for the recent registration of these agents for therapeutic use in the treatment of human influenza virus infections (15, 16). The drugs were able to reduce the time for alleviation of major symptoms by 1 to 1.5 days, with greater therapeutic benefit observed in patients who presented immediately after the onset of illness. These data establish that potent inhibitors of influenza virus NA can be effective therapeutic agents for the treatment of influenza virus infections.
In exploring the therapeutic utility of NA inhibitors, significant attention has been focused on the possibility of resistance development. This is due in part to previous studies that have demonstrated the emergence of resistance during treatment with amantadine (14), a drug from an earlier class of compounds that targets the M2 protein of influenza A virus (13). Viral variants resistant to NA inhibitors can be readily isolated in tissue culture, and some comparisons have been made between the in vitro resistance profiles of these agents (3, 4, 10, 12, 21, 22, 29, 32). Although clinical resistance has been observed at low levels in human trials with NA inhibitors (8, 9, 11), only limited data are available to date on the rate of emergence of resistant variants during therapy, the range of viral mutations seen, the extent of cross-resistance and the potential for treatment failures.
A-315675 is a chemically novel and potent inhibitor of NA enzymatic activity that is highly efficacious at inhibiting influenza virus replication in vitro (17). Importantly, A-315765 has relatively uniform activity across the range of medically important influenza virus subtypes (17). In this study, we describe the in vitro selection and characterization of A/N9 influenza virus variants with reduced susceptibilities to A-315675. The A/N9 virus was chosen because X-ray crystallographic structures of A-315675 complexed to the wild-type A/N9 virus NA are available for structure-function correlations and for molecular modeling of amino acid substitutions in the NA active site residues. Specific mutations in NA as well as HA have been identified that confer in vitro resistance to A-315675 and provide a basis for assessing potential cross-resistance with other NA inhibitors.
MATERIALS AND METHODS
Virus and cells.
The A/N9 reassortant virus containing the HA of A/NWS/33 and the NA of A/tern/Australia/G70c/75 (H1N9) was obtained from W. Graeme Laver, John Curtin School of Medical Research, Australian National University, Australia. Madin-Darby canine kidney (MDCK) cells used for virus propagation were obtained from the American Type Culture Collection (Manassas, Va.). The MDCK cells were routinely passaged in Dulbecco's modified Eagle medium (DMEM) high glucose (GibcoBRL) containing 10% fetal calf serum (JRH Biosciences) and penicillin-streptomycin (GibcoBRL). MDCK cells were infected with virus at a multiplicity of infection (MOI) of 0.05 PFU/cell. After 2 h of adsorption at room temperature, the cells were washed twice with phosphate-buffered saline, placed in medium, and then incubated at 36°C with 5% CO2. Virus was harvested when the cytopathic effect was visually evident, usually 3 to 4 days after inoculation. Virus yields were determined by plaque assay as described below.
Virus plaque assay.
Confluent MDCK cells were infected with A/N9 virus at 40 to 100 PFU in 1.5 ml of media (DMEM with 1% fetal calf serum, 20 mM HEPES, and antibiotics) in a six-well plate in the presence or absence of drug. The plates were rocked for 2 h at room temperature to allow for virus adsorption. After adsorption, 1.5 ml of 2× agarose (final concentration of 0.6%) in overlay media (DMEM with 20 mM HEPES buffer) was added to each well and mixed in the absence of trypsin. The cultures were incubated at 36°C with 5% CO2 for 3 to 4 days. Plaques were fixed with 3.7% formalin in phosphate-buffered saline overnight followed by removal of the agar overlay and staining with 0.1% crystal violet in distilled water. Plaques were counted manually from duplicate wells based on plaque number but not plaque size. The drug concentration required for 50% virus inhibition (50% effective concentration [EC50]) was calculated by using linear regression analysis of a half-log dilution series covering the effective concentrations of the tested drug. Cytotoxicity evaluations of compounds were conducted in parallel with the antiviral assays by using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay (28).
Generation of A-315675- and oseltamivir carboxylate-resistant influenza virus by in vitro passage.
MDCK cells were infected with A/N9 virus at an MOI of 0.05 PFU/cell in 25-cm2 tissue culture flasks in the presence of A-315675 or oseltamivir carboxylate at concentrations of 2 and 5 nM, respectively. Viral replication was monitored by observation for any cytopathic effect present in the culture. Infected cultures were frozen, thawed once, and centrifuged. Aliquots were then stored at −80°C for subsequent analysis. Virus was serially passaged by using 1 aliquot of viral stock from the preceding passage to infect fresh MDCK cells (MOI of 0.05 PFU/cell) in the presence of increasing concentrations of compound, thereby leading to the generation of viral stocks having increased resistance to A-315675 or oseltamivir carboxylate. The drug concentrations used in the selection protocol varied, depending on the level of viral replication present in the preceding passage. The selection was carried out for a total of 15 and 14 passages with A-315675 and oseltamivir carboxylate, respectively. Drug concentrations ranged from 2 or 5 nM to 20.48 μM by passage 15 or 14 for A-315675 and oseltamivir carboxylate, respectively, as detailed in Table 1. Drug sensitivity was determined by the plaque reduction assay with MDCK cells as described above.
TABLE 1.
In vitro selection and phenotypic susceptibility of passaged virus to A-315675 and oseltamivir carboxylate
| Virus | Drug concn used in selection (μM)
|
Total no. of days in culture | EC50 (μM) (fold change)a
|
||
|---|---|---|---|---|---|
| A-315675 | Oseltamivir carboxylate | A-315675 | Oseltamivir carboxylate | ||
| A/N9 (wild type) | 0 | 0 | 4 | 0.002 | 0.005 |
| P1 | 0.002 | 0.005 | 7 | ND | ND |
| P2 | 0.004 | 0.01 | 10 | ND | ND |
| P3 | 0.008 | 0.02 | 13 | ND | ND |
| P4 | 0.016 | 0.04 | 16 | ND | ND |
| P5 | 0.069 | 0.16 | 21 | 0.008 (4) | 0.16 (32) |
| P6 | 0.16 | 0.32 | 26 | ND | ND |
| P7 | 0.32 | 0.64 | 31 | ND | ND |
| P8 | 0.64 | 1.28 | 34 | 0.12 (60) | 0.19 (38) |
| P9 | 1.28 | 2.56 | 37 | ND | ND |
| P10 | 2.56 | 5.12 | 40 | 0.62 (310) | 2.0 (400) |
| P11 | 5.21 | 10.24 | 43 | ND | ND |
| P12 | 10.24 | 20.48 | 46 | ND | ND |
| P13 | 20.48 | 20.48 | 48 | ND | ND |
| P14 | 20.48 | 20.48 | 51 | ND | 30.0 (6,000) |
| P15 | 20.48 | NA | 54 | 0.71 (355) | NA |
Results in parentheses are fold resistances relative to the wild-type influenza A/N9 virus. NA, not applicable; ND, not determined.
Sequence analysis of the NA and HA coding regions from selected passages.
Viral RNA was isolated from cell supernatant from selected passages by using the QIAamp Viral RNA Mini kit (Qiagen Inc.) according to the manufacturer's protocol. RNA was also isolated from the cell supernatant of plaque-purified viruses. Reverse transcription-PCR was carried out with the SuperScript One-Step RT-PCR system (GibcoBRL). cDNA was reversed transcribed from the RNA at 50°C for 30 min and then predenatured at 94°C for 2 min. In the PCR, a fragment containing the entire NA (1,429 nucleotides) coding region was amplified with primers 1 and 2 (5′-ATCAAAAGCAGGGTCAAGATG-3′ and 5′-GTCGACCAGTAGATACAAGGG-3′, respectively). Similarly, a fragment containing the entire HA (1,696 nucleotides) coding region was amplified with primers 3 and 4 (5′-ATGAAGGCAAAACTACTGGTC-3′ and 5′-CTTATATTTCTGAAATCCTAATCTCA-3′, respectively). All PCRs were performed for 35 cycles of melting at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 68°C for 1.5 min followed by a single cycle of final extension at 72°C for 7 min. The amplified products were purified and ligated into a pCR-TOPO cloning vector (Invitrogen). The ligation mixture was then used to transform TOP10F′ cells. Individual bacterial colonies were picked, and miniprep DNA was purified and then sequenced by the dideoxy-chain termination method in an automated DNA sequencer. The PCR-amplified product was also directly sequenced for population sequencing. The HA H1 numbering system of Winter et al. was used (35).
NA enzyme assay.
Purified influenza virus preparations were the sources of NA for the enzyme assay. The substrate used was methylumbelliferyl sialic acid, which provides a rapid, sensitive fluorometric assay of enzyme activity (18). Enzymatic reactions were run in white, 96-well plates with a Fluoroskan II plate reader (Titertek) equipped with 355- and 460-nm excitation and emission filters controlled by DeltaSoft software (Biometallics) according to the procedure of Kati et al. (18). In all cases, Ki values were measured with high precision by using 8 to 16 concentrations of inhibitor.
NA crystal structure.
The crystal structure of A-315675 bound to N9 virus NA was determined according to the literature procedure (6). A model of the E119D mutant was created by replacing E119 with Asp with the residue replacement option of the Insight Molecular Modeling software package (Accelrys, San Diego, Calif.).
RESULTS
Antiviral activity of A-315675 and oseltamivir carboxylate against the A/N9 strain of influenza virus.
The anti-influenza virus activities of the NA inhibitors against the wild-type A/N9 strain of influenza virus were determined with a standard cell culture plaque reduction assay. The EC50s for A-315675 and oseltamivir carboxylate were 2 and 5 nM, respectively (Table 1). These data demonstrate that both compounds are highly potent inhibitors of influenza virus replication in tissue culture and suggest appropriate inhibitor concentrations for passaging experiments. The EC50 obtained for oseltamivir carboxylate is within the range of potencies observed previously for this compound against type A strains of influenza virus (22, 32).
Selection of A-315675 and oseltamivir carboxylate-resistant influenza virus (A/N9) by in vitro passage.
The A/N9 virus was serially passaged in the presence of increasing concentrations of A-315675 or oseltamivir carboxylate to select for drug-resistant virus. As a control, wild-type virus was passaged in the absence of drug. Virus was initially cultured in the presence of A-315675 or oseltamivir carboxylate at levels corresponding to their EC50s (2 and 5 nM, respectively). During the course of the selection procedure, the drug concentrations were increased to 20.48 μM for both drugs (passage 13 [P13] for A-315675 and P12 for oseltamivir carboxylate) (Table 1). Following the selection, the titers of viral stocks from selected passages and the susceptibilities to both drugs were determined with a plaque reduction assay (Table 1). The A-315675-selected virus from P5 displayed a fourfold decrease in susceptibility to A-315675. Viruses selected at P8 and P10 exhibited 60- and 310-fold-higher resistance, respectively, to A-315675 than the wild-type virus. The final passaged (P15) virus displayed 355-fold-higher resistance to A-315675. Similarly, virus passaged in the presence of oseltamivir carboxylate exhibited 32-fold-higher resistance to oseltamivir carboxylate at P5. However, by P10, a highly resistant population emerged that differed from the A/N9 wild-type precursor virus in its susceptibility to oseltamivir carboxylate by 400-fold. Continued passage resulted in a population of virus that was 6,000-fold more resistant to oseltamivir carboxylate (P14).
Identification of mutations in the NA and HA genes responsible for phenotypic changes.
Mutations in both the NA and HA genes have been reported following passage with NA inhibitors (10, 22, 31, 32). The NA sequences from A/N9 wild-type and passaged viruses were compared to that of A/tern/Australia/G70C/75 (GenBank accession no. M11445). The HA sequences from wild-type and passaged viruses were compared to that of NWS/G70C (GenBank accession no. U37727). No mutations in NA or HA were observed when A/N9 virus was passaged in the absence of drug. PCR amplification and sequencing of the NA gene indicated that the E119D mutation was present in the A-315675-treated virus populations by P8 but could have been present in earlier passages (Table 2). To determine the relative abundance of this E119D mutation within each passaged virus population, 10 viruses from selected passages were plaque purified and sequenced. At P10 and P15, 10 of 10 viruses contained the E119D mutation in the NA gene. No additional mutations were found in the NA gene at P10 or P15. However, the HA gene, which was wild type at P8, had acquired a double mutation at positions R233K and S339P by P10 which was maintained through P15. These results suggest that the 60-fold-greater resistance displayed by the P8 virus might result from the E119D mutation in the NA gene and that the further 6-fold increase in resistance found in P10 and P15 may be due to one or both of the mutations in the HA gene. However, because the P8 virus was not plaque purified, we cannot rule out the possibility that a small amount of wild-type virus might be present in the viral population. In this case, the observed 60-fold-greater resistance at P8 might be a lower value than expected relative to a pure viral population containing only the E119D NA mutation.
TABLE 2.
Genotypic changes resulting from passage of influenza virus in the presence of A-315675 or oseltamivir carboxylatef
| Passage no. | A-315675
|
Oseltamivir carboxylate
|
||
|---|---|---|---|---|
| NA mutationd | HA mutation(s)e | NA mutation(s)d | HA mutation(s)e | |
| 8 | E119Dc | Wild typec | ND | ND |
| 10 | E119Da | R233K + S339Pc | ND | S158N + G430Ec |
| 14 | ND | ND | E119V + R305Qb | H154Qc |
| E119V + R292Kb | ||||
| 15 | E119Da | R233K + S339Pc | N/A | N/A |
Ten A-315675-resistant variants from P10 and P15 were plaque purified and sequenced. Sequence analysis of the NA gene confirmed the presence of the E119D mutation in 100% of the purified influenza virus variants selected with A-315675.\
Ten oseltamivir carboxylate-resistant variants from P14 were plaque purified and sequenced. Sequence analysis of the NA gene confirmed the presence of the E119V-R305Q double mutation in 90% and the E119V-R292K double mutation in 10% of the purified influenza virus variants.\
Data are from population sequencing.\
A/Tokyo/3/67 (H3N2) numbering system.\
NWS/G70C numbering system.\
ND, not determined; N/A, not applicable.
The sequencing of oseltamivir carboxylate-selected P10 and P14 revealed a different set of mutations in the NA and HA genes (Table 2). All 10 plaque-purified viruses from P14 contained the E119V mutation in the NA gene, with 9 of the 10 also containing the R305Q mutation. Only 1 of the 10 viruses contained the R292K mutation in our study, although this was the major NA mutation found in a previous study after passage of a different subtype of influenza virus in the presence of zanamivir or oseltamivir carboxylate (23, 32). The R305Q residue lies outside of the active site near the surface of the protein, so it is not clear if this mutation was selected by virtue of drug treatment or simply arose as a spontaneous, neutral mutation. Sequence analysis of the HA gene revealed the S158N-G430E double mutation and the H154Q single mutation at P10 and P14, respectively, from viruses selected by oseltamivir carboxylate.
Susceptibility of passaged viruses to A-315675 and oseltamivir carboxylate in enzyme inhibition assay and in cell culture.
The phenotypic susceptibility of the resistant virus populations to NA inhibitors was evaluated in the standard cell culture plaque reduction assay. The viruses used in this assay were from plaque-purified virus of P15 (A-315675-resistant variants) and the population of virus of P14 (oseltamivir carboxylate-resistant variants). The results shown in Table 3 indicate that the oseltamivir carboxylate-resistant variants were much more susceptible to A-315675 and zanamivir than to oseltamivir carboxylate. Likewise, the A-315675-selected variant was more susceptible to oseltamivir carboxylate than to A-315675 and zanamivir. However, the comparison of EC50s between viruses can be misleading because of different mutations in the HA gene (see above). To dissect the effects of the NA mutations alone, we tested the potency of these compounds against the NA activity associated with the resistant viruses in a biochemical enzyme inhibition assay.
TABLE 3.
Sensitivity of plaque-purified influenza virus variants to A-315675 and oseltamivir carboxylate in both cell culture and in vitro enzyme inhibition assaya
| Passage no. | Virus | NA mutation(s) | EC50 (nM) (fold change)
|
Ki (nM) (fold change)
|
||||
|---|---|---|---|---|---|---|---|---|
| A-315675 | Oseltamivir carboxylate | Zanamivir | A-315675 | Oseltamivir carboxylate | Zanamivir | |||
| 0 | A/N9 | Wild type | 2 | 5 | 57 | 0.082 | 0.19 | 1.1 |
| 15 | A-315675 resistant | E119D | 710 (355) | 28 (6) | >10,000 (>175) | 13.3 (162) | 0.13 (0.7) | 150 (136) |
| 14 | Oseltamivir carboxylate resistant | E119V + R305Q E119V + R292K | 83 (42) | 30,000 (6,000) | 10,000 (175) | 1.15 (14) | 210 (1,100) | 26 (24) |
Viruses resistant to the indicated drugs were generated through in vitro passage. Results in parentheses are fold resistances relative to the wild-type A/N9 virus enzyme.
Table 3 is a comparative assessment of the in vitro susceptibilities and NA Ki values for the late-passage variants against A-315675, oseltamivir carboxylate, and zanamivir. The data indicate that the E119D NA mutation selected by A-315675 results in 162- and 136-fold losses in sensitivity toward A-315675 and zanamivir, respectively. However, oseltamivir carboxylate retains wild-type activity towards this mutant in an in vitro enzyme inhibition assay. Similarly, the Kis of A-315675 and zanamivir toward the NA enzyme from the oseltamivir carboxylate-resistant virus were only 14- and 24-fold, respectively, above the Ki for wild-type NA, whereas the Ki of oseltamivir carboxylate was about 1,100-fold higher than that of the wild-type enzyme. This trend carries over to the observed in vitro EC50s for the late-passage oseltamivir carboxylate-resistant virus where 42-, 175-, and 6,000-fold losses of in vitro potency were observed for A-315675, zanamivir, and oseltamivir carboxylate, respectively. Collectively, these biochemical and cell culture data suggest minimal in vitro cross-resistance between A-315675 and oseltamivir carboxylate.
DISCUSSION
In this study, we have shown that in vitro selection of the A/N9 influenza virus with increasing concentrations of the NA inhibitor A-315675 leads to the accumulation of the E119D mutation in the NA gene. We have also shown in this study that the in vitro selection of A/N9 virus with increasing concentrations of oseltamivir carboxylate leads to the accumulation of the E119V mutation in NA. Highly A-315675-resistant viruses retained nearly complete sensitivity to oseltamivir carboxylate. Likewise, highly oseltamivir carboxylate-resistant viruses were much less resistant to A-315675. Previous reports have shown that several different mutations can be selected at residue 119 after passaging strains of influenza A and B viruses in vitro in the presence of increasing concentrations of zanamivir (3, 4, 10, 12, 31). Whereas the E119D mutation confers equivalent resistance to A-315675 and zanamivir, A/N2 mutants carrying a substitution of E119G or E119A are resistant to zanamivir but sensitive to A-315675 in NA enzyme inhibition assays (L. V. Gubareva, personal communication).
Previous structural studies have shown that the loss in affinity of the mutant enzyme for zanamivir derives in part from the loss of stabilizing interactions between the guanidine moiety in zanamivir and the carboxylate residue at position 119 of the NA and in part from alterations to the solvent structure at the active site (4, 34). The present results confirm the selection of the E119V mutation by oseltamivir carboxylate found previously (12, 23) as well as the E119V mutation-containing virus isolated from oseltamivir-treated patients (8, 9). Taken together, these results implicate E119 as a primary site for generation of resistance to NA inhibitors. However, the R292K catalytic site mutation selected by zanamivir or oseltamivir carboxylate in vitro (12, 32) has also been observed in virus isolated from patients during oseltamivir treatment (9). This mutation was also found together with the E119V mutation during in vitro selection with oseltamivir carboxylate in the present study (Table 2). The oseltamivir carboxylate-resistant virus population consisting of 90% E119V-R305Q and 10% E119V-R292K was highly resistant to oseltamivir carboxylate but displayed only a low level of resistance to A-315675 (Table 3). The R292K mutation has been selected previously from in vitro passage studies in an N2 background and exhibits a high level of resistance to oseltamivir carboxylate (32). Because the R292K mutation is present in only 10% of our oseltamivir carboxylate-resistant virus population, the effect of the R292K mutation against A-315675 could be masked. Preliminary data from an NA inhibition assay with an N2 enzyme containing the R292K mutation results in only a low level of resistance to A-315675 and zanamivir and a high level of resistance to oseltamivir carboxylate (L. V. Gubareva, personal communication). The substitutions at the catalytic residues of the NA active site are more likely to lead to cross-resistance than the substitutions at the framework residues. Both types of substitutions, at framework and catalytic residues, result in impairment of the NA function (19).
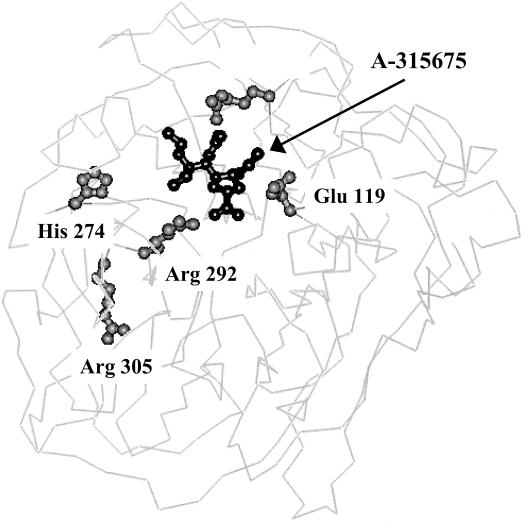
The availability of the X-ray structures of A-315675 bound to the A/N9 virus NA provides a qualitative understanding of the structural basis for mutations leading to resistance. Figure 1 shows A-315675 bound to NA, highlighting the locations of the four residues listed in Table 2 except H274. The enzyme subsite associated with E119 mutations accommodates dramatically different interactions from each of the respective inhibitors. The propenyl group of A-315675 makes exclusively hydrophobic contacts within the enzyme subsite in contrast to oseltamivir carboxylate and zanamivir, which both make predominantly hydrophilic hydrogen bonding and electrostatic interactions in this subsite via amine and guanidine substituents (1, 19, 34). However, with respect to E119, the guanidine substituent of zanamivir does not make a direct hydrogen bond to E119 like the amine of oseltamivir carboxylate.
FIG. 1.
Three-dimensional protein structure of NA showing the active site-bound orientation of A-315675 and the location of mutations.
The propenyl group of A-315675 and the guanidinyl group of zanamivir project into this subsite within the same plane of space along a vector that interacts with the hydrophobic portion of the E119 side chain as well as the hydrophobic face of the carboxylate functional group. This specific orientation of the E119 side chain is stabilized by intramolecular hydrogen bonds to R156 and it allows both substituents to expel water from the pocket without the requirement for compensating hydrogen bonding interactions.
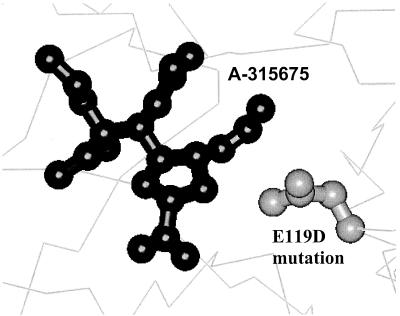
Figure 2 displays a model of the E119D mutation to demonstrate the detrimental impact of this active site change on the binding of inhibitor A-315675. The hydrophobic interactions between the cis propenyl group of A-315675 and the E119 side chain are abolished in the E119D mutant. Perhaps most importantly, the D119 side chain carboxylate is not able to interact intramolecularly with R156. The uncompensated negative charge on the D119 carboxylate results in a mismatch in polarity with the propenyl group of A-315675 (Fig. 2). Although the guanidine group of zanamivir is polarity matched to provide some electrostatic stabilization of the D119 carboxyl, the distance and angle requirements for efficient hydrogen bond formation are not contributing to stabilizing the D119 carboxyl, thereby resulting in the loss of inhibitory potency. The oseltamivir carboxylate P14 virus retains sensitivity to A-315675, which implies that the E119V hydrophobic amino acid substitution in this virus has a minimal effect on the ability of the propenyl group of A-315675 to make hydrophobic interactions within the subsite.
FIG. 2.
Close-up view of the active site of NA complexed with A-315675 and the location of the E119D mutation. The propenyl group of A-315675 makes hydrophobic contact with the methylene of wild-type Glu 119, which is replaced by the carboxylate of Asp with a mismatched polarity contact.
For oseltamivir carboxylate, the amino group interacts with the enzyme via hydrogen bonds to D151 and E119 (20) and tightly bound water molecules within the subsite (20). According to the model we have proposed for the E119D mutant, oseltamivir carboxylate retains the potential for hydrogen bonding to D119 either directly or via an intervening water molecule. What remains to be explained is which amino acid substitution is primarily responsible for the oseltamivir carboxylate P14 virus resistance. Gubareva et al. (12) characterized the mutants with E119A and E119G substitutions isolated from virus passaged in the presence of zanamivir. The NA inhibition activity of zanamivir was approximately 20- to 200-fold less active, respectively, than amino-Neu5Ac2en, whose amino group interaction with this subsite is more analogous to oseltamivir carboxylate. Therefore, one might believe that the primary source of this passaged virus's resistance is the R292K mutation.
The R292 side chain interacts with the carboxylate functional groups of all the NA inhibitors. Although R292 normally makes a hydrogen bonding interaction with E277, the R292K mutant, as a consequence of a different conformation, now causes an electrostatic interaction between the K292 side chain and E276 instead. The R292K mutation stabilizes the E276 side chain to prevent it from moving to a conformation that can accommodate hydrophobic groups such as the 3-pentyl side chain of oseltamivir carboxylate (34). The affinity of a compound such as oseltamivir carboxylate, which depends on this movement for optimal binding (19), is expected to be significantly affected by this mutation in N9. Structure-activity relationship data for zanamivir suggest that E276 is not required to move for high potency in N1, N2, N9, and B enzymes, so the fold resistance of zanamivir is minimal for K292 mutants (12, 21, 23, 32, 33).
The lack of cross-resistance of A-315675 for the oseltamivir carboxylate P14 virus is a consequence of the structure-based design strategy for the hydrophobic side chain of A-315675 that does not require an alternate conformation for E276 in N9 or any other strains for high affinity binding. A viral variant selected in previous in vitro studies for reduced susceptibility to oseltamivir carboxylate contains a mutation, H274Y, that is located in the vicinity of E276 (Z. M. Wang, C. Y. Tai, and D. B. Mendel, Prog. Abstr. 13th Int. Conf. Antivir. Res., vol. 46, abstr. 81, p. A60, 2000). This H274 residue is close to the active site but does not contact any NA inhibitor directly. However, it has been shown that side chains larger than His at position 274 create a steric clash with the E276 side chain whenever E276 tries to alter its conformation to accommodate large hydrophobic groups of inhibitors (Wang et al., Prog. Abstr. 13th Int. Conf. Antivir. Res.). Thus, it is not surprising that the binding of compounds that induce an alternative E276 conformation, such as oseltamivir carboxylate, would be affected by this mutation (Wang et al., Prog. Abstr. 13th Int. Conf. Antivir. Res.). However, compounds that do not depend on the E276 conformational change, such as A-315675, may show a different resistance profile to this mutation.
Previous studies have shown that mutants selected during in vitro passage by NA inhibitors have mutations not only in the NA gene but also in the HA gene (3, 5, 10, 12, 22, 23, 25, 32). The present study extends these observations to a fourth NA inhibitor and suggests that, in response to the selective pressure exhibited on the virus by this class of compounds, mutation in HA may facilitate the growth of virus containing impaired NA function. Either NA or HA mutations can occur first during in vitro passage. In vivo mutations in HA preceded the mutation in NA during zanamivir therapy (11). The results of the present study provide further evidence for the potential role of genetic changes in HA in producing viral resistance during therapy with NA inhibitors.
Importantly, the mutations selected by A-315675 and oseltamivir carboxylate in vitro were distinct. The passaged virus produced by selection with A-315675 displayed little cross-resistance to oseltamivir carboxylate. Similarly, the virus passaged in the presence of oseltamivir carboxylate remained largely susceptible to A-315675, particularly in the enzyme inhibition assay, which directly measures the effect of mutations in the NA gene. The lack of cross-resistance between these two NA inhibitors may prove important if widespread clinical resistance to the NA inhibitor class emerges.
Acknowledgments
We thank W. Graeme Laver for providing the influenza A/N9 virus strain. We are grateful to Larisa Gubareva (Department of Internal Medicine, University of Virginia Health Sciences Center, Charlottesville, Va.) for providing unpublished data and for valuable comments and suggestions.
REFERENCES
- 1.Babu, Y. S., P. Chand, S. Bantia, P. Kotian, A. Dehghani, Y. El-Kattan, T-H. Lin, T. L. Hutchison, A. J. Elliott, C. D. Parker, S. L. Ananth, L. L. Horn, G. W. Laver, and J. A. Montgomery. 2000. BCX-1812: discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 43:3482-3486. [DOI] [PubMed] [Google Scholar]
- 2.Bantia, S., C. D. Parker, S. L. Ananth, L. L. Horn, K. Andries, P. Chand, P. L. Kotian, A. Dehghani, Y. El-Kattan, T. Lin, T. L. Hutchison, J. A. Montgomery, D. L. Kellog, and Y. S. Babu. 2001. Comparison of the anti-influenza virus activity of RWJ-270201 with those of oseltamivir and zanamivir. Antimicrob. Agents Chemother. 45:1162-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, J. M., A. Cadman, F. M. Burrell, S. H. Madar, A. P. Lewis, M. Tisdale, and R. Bethell. 1999. In vitro selection and characterization of influenza B/Beijing/1/87 isolates with altered susceptibility to zanamivir. Virology 265:286-295. [DOI] [PubMed] [Google Scholar]
- 4.Blick, T. J., T. Tiong, A. Sahasrabudhe, J. N. Varghese, P. M. Colman, G. J. Hart, R. C. Bethell, and J. L. McKimm-Breschkin. 1995. Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity for the neuraminidase-specific inhibitor 4-guanidino-neu5Ac2en. Virology 214:475-484. [DOI] [PubMed] [Google Scholar]
- 5.Blick, T. J., A. Sahasrabudhe, M. McDonald, I. J. Owens, P. J. Morley, R. J. Fenton, and J. L. McKimm-Breschkin. 1998. The interaction of neuraminidase and hemagglutinin mutations in influenza virus in resistance to 4-guanidino-Neu5Ac2en. Virology 246:95-103. [DOI] [PubMed] [Google Scholar]
- 6.Bossart-Whitaker, P., M. Carson, Y. S. Babu, C. D. Smith, W. G. Laver, and M. Gillian. 1993. Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid. J. Mol. Biol. 232:1069-1083. [DOI] [PubMed] [Google Scholar]
- 7.Burnet, F. M. 1948. Mucin and mucoids in relation to influenza virus actin. IV. Inhibition by purified mucoid of infectin and haemagglutinin with the virus strain WSE. Aust. J. Exp. Biol. Med. Sci. 26:381-387. [DOI] [PubMed] [Google Scholar]
- 8.Carr, J., J. Ives, N. A. Roberts, C. Y. Tai, D. B. Mendel, L. Kelly, R. Lambkin, and J. Oxford. 1999. An oseltamivir treatment-selected influenza A/Wuhan/359/95 virus with an E119V mutation in the neuraminidase gene has reduced infectivity in vivo, p. 23. In Second International Symposium on Influenza and other Respiratory Viruses, Cayman Island, December 10-12. The Macrae Group, New York, N.Y.
- 9.Covington, E., D. B. Mendel, P. Escarpe, C. Y. Tai, K. Soderbarg, and N. A. Roberts. 1999. Phenotypic and genotypic assay of influenza neuraminidase indicate a low incidence of viral drug resistance during treatment with oseltamivir, p. 21. Second International Symposium on Influenza and other Respiratory Viruses, Cayman Island, December 10-12. The Macrae Group, New York, N.Y.
- 10.Gubareva, L. V., R. C. Bathell, G. J. Hart, K. G. Murti, C. R. Penn, and R. G. Webster. 1996. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J. Virol. 70:1818-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gubareva, L. V., M. N. Matrosovich, M. K. Brenner, R. C. Bethell, and R. G. Webster. 1998. Evidence of zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178:1257-1262. [DOI] [PubMed] [Google Scholar]
- 12.Gubareva, L. V., M. J. Robinson, R. C. Bethell, and R. G. Webster. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J. Virol. 71:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay, A. J. 1992. The action of adamantamines against influenza A viruses: inhibition of the m2 ion channel protein. Semin. Virol. 3:21-30. [Google Scholar]
- 14.Hayden, F. G., and A. J. Hay. 1992. Emergence and transmission of influenza A viruses resistant to amantadine and rimantidine. Curr. Top. Microbiol. Immunol. 176:119-130. [DOI] [PubMed] [Google Scholar]
- 15.Hayden, F. G., A. D. M. E. Osterhaus, J. Treanor, D. M. Fleming, G. Y. Aoki, K. G. Nicholson, A. M. Bohnen, H. M. Hirst, O. Keene, and K. Wightman. 1997. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenza virus infections. N. Engl. J. Med. 337:874-880. [DOI] [PubMed] [Google Scholar]
- 16.Hayden, F. G., J. J. Treanor, R. S. Fritz, M. Lobo, R. F. Betts, M. Miller, N. Kinnersley, R. G. Mills, P. Ward, and S. E. Straus. 1999. Use of the oral neuraminidase inhibitor oseltamivir in experimental human influenza. JAMA 282:1240-1246. [DOI] [PubMed] [Google Scholar]
- 17.Kati, W. M., D. Montgomery, R. Carrick, L. Gubareva, C. Maring, K. McDaniel, K. Steffy, W. G. Laver, A. Molla, F. Hayden, D. Kemp, and W. Kohlbrenner. 2002. In vitro characterization of A-315675, a highly potent inhibitor of A and B strain influenza virus neuraminidases and influenza virus replication. Antimicrob. Agents Chemother. 46:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kati, W. M., A. S. Saldivar, F. Mohamadi, H. L. Sham, W. G. Laver, and W. E. Kohlbrenner. 1998. GS4071 is a slow-binding inhibitor of influenza neuraminidase from both A and B strains. Biochem. Biophys. Res. Commun. 244:408-413. [DOI] [PubMed] [Google Scholar]
- 19.Kim, C. U., X. Chen, and D. B. Mendel. 1999. Neuraminidase inhibitors as anti-influenza virus agents. Antivir. Chem. Chemother. 10:141-154. [DOI] [PubMed] [Google Scholar]
- 20.Kim, C. U., W. Lew, M. A. Williams, H. Liu, L. Zhang, S. Swaminathan, N. Bischofberger, M. S. Chen, D. B. Mendel, C. Y. Tai, W. G. Laver, and R. C. Stevens. 1997. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 119:681-690. [DOI] [PubMed] [Google Scholar]
- 21.McKimm-Breschkin, J. L. 2000. Resistance of influenza viruses to neuraminidase inhibitors--a review. Antivir. Res. 47:1-17. [DOI] [PubMed] [Google Scholar]
- 22.McKimm-Breschkin, J. L., T. J. Blick, A. Sahasrabudhe, T. Tiong, D. Marshall, G. J. Hart, R. C. Bethell, and C. R. Penn. 1996. Generation and characterization of variants of NWS/G70c influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu-5Ac2en. Antimicrob. Agents Chemother. 40:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKimm-Breschkin, J. L., A. Sahasrabudhe, T. J. Blick, M. McDonald, P. M. Colman, G. J. Hart, R. C. Bethell, and J. N. Varghese. 1998. Mutations in a conserved residue in the influenza virus neuraminidase active site decrease sensitivity to Neu5Ac2en-derived inhibitors. J. Virol. 72:2456-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendel, D. B., C. Y. Tai, P. A. Escarpe, W. Li, R. W. Sidwell, J. H. Huffman, C. Sweet, K. J. Jakeman, J. Merson, S. A. Lacy, W. Lew, M. A. Williams, L. Zhang, M. S. Chen, N. Bischofberger, and C. U. Kim. 1998. Oral administration of a prodrug of the influenza virus neuraminidase inhibitor GS4071 protects mice and ferrets against influenza infection. Antimicrob. Agents Chemother. 42:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobusawa, E., T. Aoyama, H. Kato, Y. Suzuki, Y. Tateno, and K. Nakajima. 1991. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182:475-485. [DOI] [PubMed] [Google Scholar]
- 26.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397-410. [DOI] [PubMed] [Google Scholar]
- 27.Palese, P., and R. W. Compans. 1976. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J. Gen. Virol. 33:159-163. [DOI] [PubMed] [Google Scholar]
- 28.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. De Clercq. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309-321. [DOI] [PubMed] [Google Scholar]
- 29.Penn, C. R., J. M. Barnett, R. C. Bethell, R. Fenton, K. L. Gearing, N. Healy, and A. J. Jowett. 1996. Selection of influenza virus with reduced sensitivity in vitro to the neuraminidase inhibitor GG167 (4-guanidino-Neu5A2en): changes in haemagglutinin may compensate for loss of neuraminidase activity, p. 735-740. In L. E. Brown, A. W. Hampson, and R. G. Webster (ed.), Options for control of influenza III. Elsevier, Amsterdam, The Netherlands.
- 30.Sidwell, R. W., J. H. Huffman, D. L. Barnard, K. V. Bailey, M. Wong, A. Morrison, T. Syndergaard, and C. U. Kim. 1998. Inhibition of influenza virus infections in mice by GS4104, an orally effective influenza virus neuraminidase inhibitor. Antivir. Res. 37:107-120. [DOI] [PubMed] [Google Scholar]
- 31.Staschke, K. A., J. M. Colacine, A. J. Baxter, G. M. Air, A. Bansal, W. J. Hornback, J. E. Munroe, and W. G. Laver. 1995. Molecular basis of resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology 214:642-646. [DOI] [PubMed] [Google Scholar]
- 32.Tai, C. Y., P. A. Escarpe, R. W. Sidwell, M. A. Williams, W. Lew, H. Wu, C. U. Kim, and D. B. Mendel. 1998. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS4071. Antimicrob. Agents Chemother. 42:3234-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, N. R., A. Cleasby, O. Singh, T. Skarzynski, A. J. Wonacott, P. W. Smith, S. L. Sollis, P. D. Howes, P. C. Cherry, R. Bethell, P. Colman, and J. Varghese. 1998. Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus type A and B. J. Med. Chem. 41:798-807. [DOI] [PubMed] [Google Scholar]
- 34.Varghese, J. N., V. C. Epa, and P. M. Colman. 1995. Three dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase. Protein Sci. 4:1081-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter, G., S. Fields, and G. G. Brownlee. 1981. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature 292:72-75. [DOI] [PubMed] [Google Scholar]