Abstract
During the final stages of φX174 morphogenesis, there is an 8.5-Å radial collapse of coat proteins around the packaged genome, which is tethered to the capsid's inner surface by the DNA-binding protein. Two approaches were taken to determine whether protein-DNA interactions affect the properties of the mature virion and thus the final stages of morphogenesis. In the first approach, genome-capsid associations were altered with mutant DNA-binding proteins. The resulting particles differed from the wild-type virion in density, native gel migration, and host cell recognition. Differences in native gel migration were especially pronounced. However, no differences in protein stoichiometries were detected. An extragenic second-site suppressor of the mutant DNA-binding protein restores all assayed properties to near wild-type values. In the second approach, φX174 was packaged with foreign, single-stranded, covalently closed, circular DNA molecules identical in length to the φX174 genome. The resulting particles exhibited native gel migration rates that significantly differed from the wild type. The results of these experiments suggest that the structure of the genome and/or its association with the capsid's inner surface may perform a scaffolding-like function during the procapsid-to- virion transition.
Unlike many double-stranded DNA (dsDNA) viruses which use one scaffolding protein (14, 34), φX174 morphogenesis is dependent on two species (30). Together, these two proteins perform the full spectrum of functions found in one-protein systems. However, after procapsid assembly, the final stages of φX174 morphogenesis differ. In dsDNA systems, procapsids expand during packaging (32, 37, 41), and the genome forms a dense core (18). In contrast, φX174 morphogenesis concludes with the collapse of coat proteins around the single-stranded genome, which is associated with the inner surface of the capsid (15, 16, 31, 38, 39).
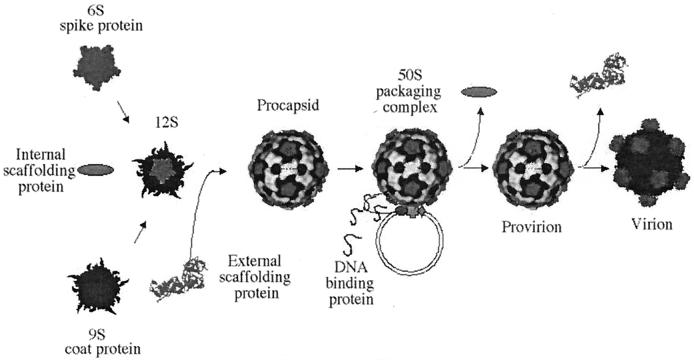
As illustrated in Fig. 1, φX174 genome replication is coupled to DNA packaging. The preinitiation complex, consisting of the host cell rep and viral A and C proteins, associates with the procapsid forming the 50S complex (21, 40). The viral A protein binds the origin of replication in replicative-form DNA. This is both necessary and sufficient for packaging specificity (3, 4, 26). Upon binding, protein A nicks the origin (47) and forms a covalent ester bond with the DNA (19). After one round of rolling circle synthesis, protein A cuts and ligates the newly generated origin, generating a circular single-stranded molecule (7, 20). This packaging mechanism produces precise genomes of identical length (4).
FIG.1.
φX174 morphogenesis.
The highly basic DNA-binding protein (J) enters the procapsid, along with the single-stranded DNA (ssDNA) genome, (27), associating with the genome via charge-charge interactions (13, 33). Once in the procapsid, the C terminus binds to a coat protein cleft (38, 39). This may facilitate further interactions with a small cluster of adjacent basic capsid amino acids. Accordingly, a portion of the packaged DNA (8 to 10%) is icosahedrally ordered, tethered to the inner surface (38, 39). This tether constrains the spatial orientation and secondary structure of the remaining nucleotides (5). Morphogenesis completes with the dissociation of the external scaffolding protein and an 8.5-Å radial collapse of capsid pentamers. We address here the question of whether the tethered genome influences the integrity and/or magnitude of this collapse.
The interaction of icosahedral capsid proteins with single-stranded genomes is a well-documented phenomenon. Many viral RNA capsid proteins have internally localized basic N termini extensions that nonspecifically bind genomes (11, 12, 36, 43, 48, 49). The phenomenon has been documented in single-stranded animal viruses as well (1, 10, 25, 46). To investigate the functions of the φX174 genome tether, both the protein and the nucleic acid components were altered. The protein component was altered by mutations in the DNA-binding protein. The DNA component was altered by packaging foreign, unit-length, single-stranded, circular DNA molecules. In both instances, the altered particles were infectious but exhibited different biophysical properties.
MATERIALS AND METHODS
Phage plating, stock preparation, media, DNA purification, bacterial strains, and plasmids.
The plating protocols, media, and stock preparation used here have been described elsewhere (22). Escherichia coli C strain C122 (sup0) is the wild-type host; BAF5 contains a supE mutation (22). BAF30 is a recA-null derivative of C122 (23). Plasmids pφXB and pφXDJ contain IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible clones of the denoted φX174 genes (8, 9). The slyD host mutation confers resistance to φX174 E-protein-mediated lysis (42). The gro89 host mutation blocks DNA packaging (21).
Phage mutants.
The φX174 J mutants J−3K→LI, J−K→L ALL, and J−3K→LII/su(J)-FS1F have been previously described (33). The su(J)-FS1F mutation was placed into the J−3K→LI background by oligonucleotide-mediated mutagenesis (24). Progeny that had acquired the su(J)-FS1F mutation were identified by selecting against the temperature-sensitive phenotype of the J3K→LI parent. Two rounds of oligonucleotide mutagenesis were performed to place the su(J)-FS1F mutation in the wild-type background. Genotypes of all strains were verified by direct DNA sequence analysis.
Buoyant density gradients.
For small-scale experiments in which two or three different particles were analyzed within the same gradient, 105 to 106 particles were mixed into a 1.4-g/cm3 CsCl solution and spun for 47.5 h at 23,000 rpm in an SW51 rotor (Beckman). Gradients were divided into 80 to 85 fractions. The location of infectious particles was determined by a plaque assay. For large-scale preparations of J−K→L ALL 70S particles, 100 ml of slyD cells (2.0 × 108 cells/ml) were infected (multiplicity of infection [MOI] = 5) at 33°C and incubated for 2 h. To generate wild-type procapsids, gro89 cells were infected with lysis-deficient φX174 am(E)W4. Infected cells were harvested, resuspended in 10 ml of BE buffer, and lysed with T4 lysozyme. Cellular debris was eliminated by centrifugation. The supernatant was layered atop CsCl step gradients and spun as previously described (22). Exogenous virions (1.0 × 106) were added to gradients as a density marker. Procapsid and 70S particle bands were clearly visible after centrifugation. Titers of fractions were determined to ascertain the location of the wild-type marker phage.
Native agarose gel electrophoresis.
To examine particles by native gel electrophoresis, 10-μl samples containing 1.0 × 108 PFU were mixed with 6× gel loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol FF, 30% glycerol). Samples were bidirectionally electrophoresed through 0.8% (wt/vol) agarose-TBE (45 mM Tris-borate, 1.0 mM EDTA) gels for 10 h at 24 V/cm2. After electrophoresis, lanes were cut into 2.0-mm horizontal sections with a modified egg slicer. Particles were eluted from the gel by vortexing in 0.1 ml of HFB buffer. The location of infectious particles was determined by a plaque assay.
Attachment assays.
E. coli C122 cells were grown to a concentration of 1.0 × 108 cells/ml and concentrated fivefold in growth media (1.0% tryptone, 0.5% KCl). MgCl2 and CaCl2 were added to concentrations of 10 and 5.0 mM, respectively. Approximately 1.0 × 108 phage were added to 1.0 ml of cells and incubated at 37°C for specified time intervals (see the figures). At sampling times, attached phage were removed by centrifugation. Supernatants were titered to determine the level of unattached virions.
Construction of Ampr packaging plasmids and transducing particles.
φX174 DNA containing the φX174 origin of replication was amplified. The 5′ PCR primer was designed to introduce an NcoI site into the fragment that was then initially cloned into TOPO 2.1 vector (Invitrogen). The TOPO vector was digested with NcoI and EcoRI (adjacent to cloning site), and the fragment was placed into PSE420 (Invitrogen) digested with the same enzymes (PORI). To bring the size of PORI up to 5,386 bp, additional material was cloned behind the φX174 origin. These DNA sequences were obtained by amplifying pSE420 DNA between bases 3302 and 3776. This PCR fragment was first placed into a TOPO 2.1, and the orientation of the insert was determined. Depending on orientation, plasmids were cut with BamHI and XbaI or with NotI and SacI, and then cloned into PORI that had been prepared with BamHI and NheI or with NotI and SacI. Plasmids were transformed into BAF30 recA cells. Transducing particles were produced by infecting cells harboring the packaging plasmids with wild-type φX174. The presence of the origin of replication in the plasmid is both necessary and sufficient for packaging a single-stranded version of the vector (4, 26). Lysates contain both virion and transducing particles. The titer of transducing particles was determined by infecting 1.0 × 108 slyD cells at an MOI of 0.1 with subsequent plating on ampicillin (100 μg/ml) plates. The low MOI and slyD cells were used to prevent coinfections and a second round of infection by progeny virions, respectively. The ratio of progeny virions to Ampr (ampicillin-resistant) transducing particles was ca. 50:1 in both infections. This represents typical values for these types of experiments (21).
RESULTS
Complete genome encapsidation is not required for the completion of genome biosynthesis.
Microviridae genome biosynthesis and DNA packaging are concurrent processes (27). Previous work with the φX174 J mutants used in these studies demonstrated that mature packaged particle formation was a function of the number of charged residues of the protein (33). For example, mutant J proteins with only nine basic amino acid residues (the wild-type contains 12) could produce packaged particles with wild-type S values (i.e., 114S). However, several other biophysical properties differ (see below). Mutant proteins with only six basic residues produce particles that sediment at 70S (33). Analyzing the nature of the DNA associated with the 70S particles (partial versus complete genomes), its susceptibility to DNase, and the density of the 70S particles would determine if genome biosynthesis terminates with a cessation in packaging or whether these two processes can be partly uncoupled in vivo.
The mutant genotypes and phenotypes used in these studies are summarized in Table 1. DNA associated with J−K→L ALL 70S particles was extracted and analyzed by electrophoresis (data not shown). It was found to be identical to the virion DNA, indicating that ssDNA biosynthesis went to completion. The density of the particles was determined to be 1.36 g/cm3 (Table 2) by buoyant density centrifugation, a value midway between the densities of the 114S virion (1.39 g/cm3) and the 108S procapsid (1.31 g/cm3) used as standards. These data indicate that the mutant J protein had no effect on genome biosynthesis but that the genome was not fully packaged. To test this hypothesis, particle densities were redetermined after DNase treatment (2). Although the densities of the virion and empty procapsid were not affected, the density of the 70S particle was reduced (Table 2).
TABLE 1.
Genotypes and phenotypes of φX174 J mutants in these studies
| Mutant | Genotype | Phenotype |
|---|---|---|
| J−3K→LI | K→L at residues 2, 4, and 5 | Small plaques, ts, csa |
| su(J)-FS1F/J−3K→LI | K→L at residues 2, 4, and 5; extragenic suppressor (S→F) in residue 1 of the capsid protein | Wild type |
| J−K→L ALL | K→L at residues 2, 4, 5, 21, 23, and 25 | Recessive lethal |
ts and cs denote temperature-sensitive and cold-sensitive phenotypes, respectively.
TABLE 2.
Densities of particles generated in J− mutant-infected cells
| Particle | S value | Particle density (g/cm3) with:
|
|
|---|---|---|---|
| No DNase | DNase | ||
| Wild-type virion | 114S | 1.39 | 1.39 |
| Procapsid | 108S | 1.31 | 1.31 |
| J−K→L ALL | 70S | 1.36 | 1.32 |
Further characterization of infectious particles packaged with mutant DNA-binding proteins.
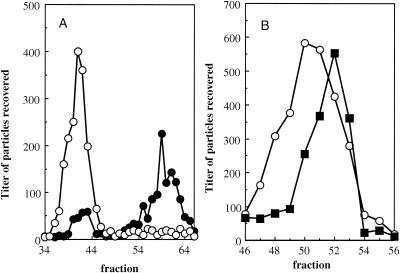
Unlike mutant J proteins with less than six basic residues, proteins with nine basic residues, such as J−3K→LI, produce infectious particles with wild-type S values. However, the mutant has a small plaque phenotype and is both temperature and cold sensitive. Particles packaged with the J−3K→LI and wild-type J protein were characterized by buoyant density centrifugation. The results of these analyses, along with those of stoichiometric studies (see below), could determine whether the genomic DNA is fully encapsidated, more mutant J protein is needed for packaging, or other possibilities. For these experiments particles packaged with wild-type J protein contained an amber mutation in gene B. Since the titers of particles could be independently determined (see the figure legends), it was possible to characterize both types of virion within the same gradient. As seen in Fig. 2A, particles packaged with the mutant J protein are denser than the wild type.
FIG. 2.
(A) Buoyant densities of wild-type and J−3K→LI particles. Both particles were analyzed in the same gradient. An additional genetic marker, amB, was placed in the wild-type background. This allowed the particles to be specifically titered on BAF30 pφXB at 42°C. The J−3K→LI mutant is temperature sensitive. The titer of J−3K→LI was determined on C122 (sup0) at 33°C. (B) Buoyant densities of particles packaged with mutant J proteins with or without the su(J)-FS1F extragenic second-site suppressor. An order of magnitude more of the J−3K→LI particles, which have a temperature-sensitive phenotype, was loaded onto the same gradient with the double mutant. The position of the su(F)S1F/J−3K→LI particles could be ascertained and distinguished from J−3K→LI by determining the titer at 42°C. Particle titers were normalized to the same order of magnitude for the graph. Symbols: •, wild type; ○, J−3K→LI; ▪, su(J)-FS1F/J−3K→LI.
Several hypotheses could explain the observed density difference. In order to compensate for the mutant protein's reduced charge, more J protein may be needed for packaging. If the volume of the mutant particle remains unaltered, additional J protein would replace water, yielding a greater density. Although J protein stoichiometry is dictated by the capsid's icosahedral symmetry in wild-type particles, protein stoichiometry was still assessed via polyacrylamide gel electrophoresis, and the relative amounts of coat (F), spike (G), and J proteins were determined. In a wild-type virion each of these proteins is present in 60 copies. As summarized in Table 3, no significant differences in protein ratios were apparent. The variations seen in the F:J ratio are the same as those observed in the F:G ratio. It is possible that mutant particles may have retained a small amount of internal scaffolding protein that may have been below the limits of detection.
TABLE 3.
Protein composition of wild-type and J−3K→LI particlesa
| Proteinsb | Protein composition of:
|
Variation from wild type | |
|---|---|---|---|
| Wild type | J−3K→LI | ||
| F and G | 1.64 | 1.39 | 0.15 |
| F and J | 3.02 | 2.72 | 0.10 |
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, stained, and digitally photographed. Ratios were calculated by using band intensities derived via one-dimensional image analysis software (Kodak Digital Science).\
F, G, and J denote the major capsid, spike, and DNA-binding proteins, respectively.
The greater density could have been caused by an increase of Cs+ ions within the mutant capsid. In this model, the positively charged counter ion would supplant the missing basic amino acids in the mutant protein. To address this hypothesis, the densities of J−3K→LI and su(J)S1F/J−3K→LI particles were compared. The su(J)S1F mutation is a nonallele-specific, extragenic, second-site suppressor of charge-altered J proteins, which has been previously characterized and described elsewhere (33). su(J)S1F suppresses both the small plaque and the temperature-sensitive phenotypes, raising plating efficiencies from 10−3 to 0.5 at the restrictive temperatures. An order of magnitude more J−3K→LI particles was loaded on the same gradient with the double mutant. The position of the su(F)S1F/J−3K→LI could be ascertained and distinguished from J−3K→LI particles by determining the titers at 42°C. As seen in Fig. 2B, the particles were readily separated by their densities, demonstrating that the suppressor is affecting the density of the capsid, restoring it to a more wild-type value. Since both particles are packaged with the same mutant J protein, the observed density differences are probably not caused by Cs+ ions or an excess of J protein in the J−3K→LI particles.
The extragenic suppressor is a Ser→Pro substitution located at amino acid 1 of the coat protein (33). In wild-type virion, this serine participates, via the γ O, in a three-way polar interaction (coat-coat-coat) directly atop the threefold axis of symmetry (38, 39). The suppressing substitution most likely alters this interaction. Buoyant density gradients were performed to determine the effect, if any, of this substitution alone in a wild-type background (Fig. 3). Wild-type, su(J)-F S1F/wild type J, and J−3K→LI particles were analyzed in the same gradient. The su(J)-F S1F/wild type J is slightly more dense than the wild type.
FIG. 3.
Buoyant densities of wild-type, su(J)-F S1F/wild type J, and J−3K→LI particles. All three types of particles were analyzed within the same gradient. An additional genetic marker, amB, was placed in the wild-type background. This allowed the titers of the particles to be specifically determined on BAF30 pφXB at 42°C, where the J−3K→LImutant is temperature sensitive. An order of magnitude more of the J−3K→LI and wild-type (amB) particles was loaded onto the gradient than the su(J)-F S1F/wild type J particles. The position of the su(F)SIF/wild type J particles could be distinguished from the other particles by determining their titers at 42°C on a sup0 host. Particle titers were normalized to the same order of magnitude for the graph. Symbols: •, wild type; ○, ▪, J−3K→LI; ○, su(J)-FS1F packaged with wild-type J protein.
Mutant and wild-type particles migrate differently in native gels.
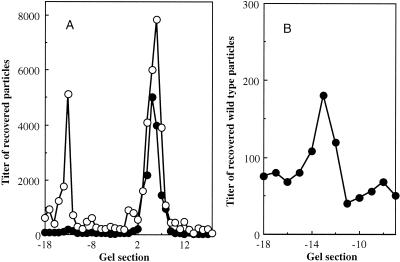
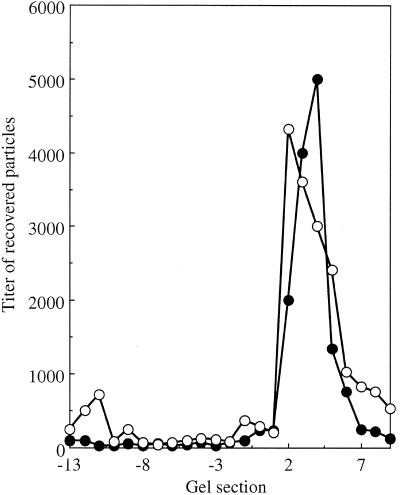
The altered densities could be caused by general distortions affecting the entire capsid. If this were the case, alterations in genome-capsid interactions might be expressed on the capsid's external surface. The migrations of wild-type and mutant virions were analyzed side-by-side in bidirectional, native agarose gels, which assay for differences in size and net surface charge (45). After electrophoresis, each lane was cut into 2-mm sections with a modified egg slicer, particles were eluted, and titers were determined. For both wild-type and mutant particles, a major population peak was detected migrating toward the positively charged anode, referred to as the negative or major population (Fig. 4). However, migration rates differed. With longer run times, the negatively charged particle peaks can be more clearly separated (data not shown). Mutant virions were not more sensitive than the wild-type virions to DNase treatment. Therefore, protruding DNA is probably not responsible for the faster migration rate. There were also positively charged populations in each sample. While these positively charged particles represent a minor portion of the total wild-type population (1:1,000 ratio), they are quite prominent in the mutant sample (1:3 ratio). As seen in Fig. 5, the extragenic second-site suppressor restores mutant population ratios toward wild-type levels.
FIG. 4.
(A) Native gel migration of wild-type and J−3K→LI particles. (B) Enlargement of the wild-type minor population of positively charged particles. Negative gel section numbers indicate movement toward the negative electrode; positive gel section numbers indicate movement toward the positive electrode. Symbols: •, wild type; ○, J−3K→LI.
FIG. 5.
Native gel migration of wild-type and su(J)-FS1F/J−3K→LI particles. Negative gel section numbers indicate movement toward the negative electrode; positive gel section numbers indicate movement toward the positive electrode. Symbols: •, wild type; ○, su(J)-FS1F/J3K→LI.
Attachment assays as a probe for external capsid alterations.
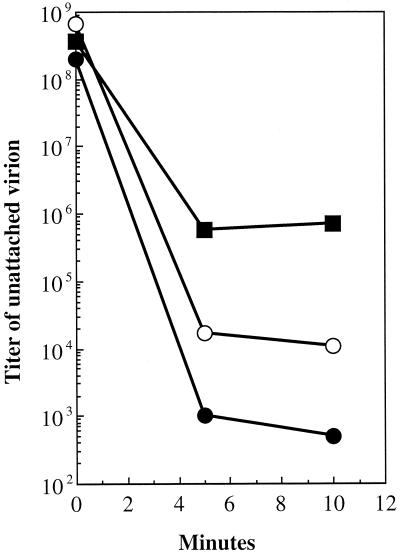
To further explore possible alterations on the outer surface, attachment assays were performed. For these assays, particles were incubated with exponentially growing cells at 37°C as described in Materials and Methods. At 5 and 10 min postinfection, aliquots were removed and separated into pellet and supernatant fractions. The level of unattached virion was determined by determining the titer of the supernatant. The attachment of wild-type particles is 3 orders of magnitude greater than that of the mutant particles (Fig. 6). The extragenic suppressor appears to correct for defects in host cell recognition, restoring attachment to intermediary efficiency. The level of unattached phage does not reflect the population ratios observed in the native gel migration experiments. Similar experiments were conducted with separated negatively and positively charged mutant and wild-type particles (data not shown). The results of these experiments did not differ significantly from the data presented in Fig. 7, wherein whole populations were assayed. In each instance, wild-type particles exhibited greater attachment efficiencies than mutant particles. While the positively charged particles of both the mutant and wild-type strains attached more efficiently than their negatively charged counterparts, it is not known whether this is a consequence of nonspecific binding to the negatively charged lipopolysaccharide. These simple assays do not determine whether the mutant particles are defective in DNA ejection, nor can they be used to interpret the small plaque phenotypes of the mutants, which may be a consequence of both extracellular and intracellular factors.
FIG. 6.
Attachment assays. Symbols: •, wild type; ▪, J−3K→LI; ○, su(J)-FS1F/J−3K→LI.
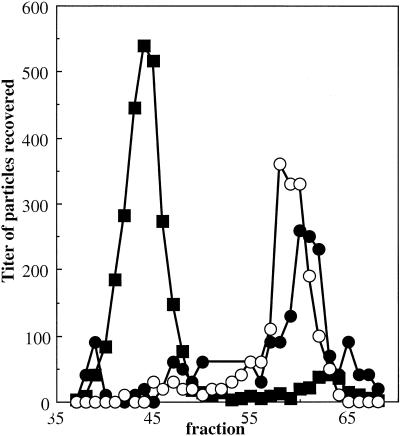
FIG. 7.
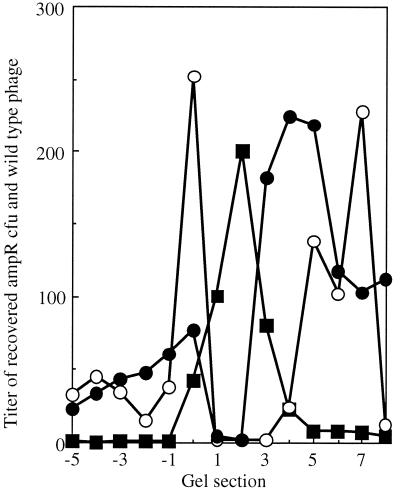
Native gel migration of wild-type φX174 and Ampr transducing particles. The titer of wild-type phage has been normalized to the same order of magnitude as the transducing particles. Negative gel section numbers indicate movement toward the negative electrode; positive gel section numbers indicate movement toward the positive electrode. Symbols: ▪, wild-type phage; ○ and •, transducing particles.
Packaging foreign DNA also alters the biophysical properties of capsids.
The DNA-binding protein connects the genome to the inner surface of the capsid at each asymmetric unit. These 60 interactions allow the remainder of the ssDNA genome to form limited secondary structure by base pairing upon itself within the virion. Hence, the base-pairing properties of the genome may influence the properties of the mature virion. To investigate this, φX174 was packaged with foreign DNA. Two packaging plasmids identical in length to the φX174 genome were constructed. These plasmids contain the φX174 origin of replication, which is both necessary and sufficient for packaging a single-stranded version of the plasmid (4, 26), and a gene encoding ampicillin resistance. The two plasmids differed in one segment. This segment is repeated, in either a parallel-parallel or a parallel-antiparallel manner. The parallel-antiparallel configuration may introduce a large hairpin loop. Particles were packaged in vivo by infecting cells harboring the plasmids with wild-type φX174. Particles were analyzed by native gel electrophoresis, and the gels were processed as described above. The titer of each fraction was determined for phage and Ampr transducing particles (Fig. 7). Two population peaks, positively and negatively charged, were detected for both sets of transducing particles (Fig. 7). However, population ratios differ between the two constructs. In addition, the negatively charged populations migrate considerably faster than the internal φX174 control. Therefore, to avoid running the transducing particles off the gel, run times were shorter than in the experiments described above. These data suggest that the nucleotide arrangement of the genome can change the biophysical properties of the capsid, perhaps by promoting or inhibiting genome secondary structure.
DISCUSSION
Interactions between the capsid inner surface and the packaged genome.
Single-stranded φX174 DNA does not exist as a dense core in the capsid, as is observed in dsDNA bacteriophages (18). Instead, it is tethered to the capsid's inner surface by the highly basic DNA-binding protein (J) and a group of basic capsid amino acid residues (38, 39). There are 60 copies of protein J per virion: one associated with each coat protein. In the atomic model, the protein forms an S-shaped polypeptide chain devoid of secondary structure. The C terminus of the protein is tightly associated with a cleft, located near the center of the coat protein. Moving toward the N terminus, the protein traces a path toward the fivefold axis of symmetry, crosses over to the adjacent capsid protein, and veers toward the C terminus of the adjacent J protein. This motif suggests that the DNA-binding protein guides the incoming genome into a somewhat ordered conformation. Accordingly, between 8 and 10% of the genome is ordered in the X-ray structure (38, 39). Here and in other systems where higher percentages of the genome are ordered—such as Flock house virus (FHV) (25); Pariacoto virus (46), and Satellite tobacco mosaic virus (36)—the genome is icosahedrally ordered via interactions with structural proteins. This phenomenon is thought to contribute to capsid assembly and stability.
Altering the protein components of the tether.
To investigate whether genome-capsid interactions affect the final stages of virion morphogenesis and/or the structure of the mature virion, particles packaged with mutant DNA-binding proteins were characterized. The mutant particles were significantly more dense than wild type, but the protein composition of the two particles is probably identical. Therefore, the altered density was probably not caused by a gross excess of protein contained in a volumetrically fixed capsid. The effects of possible Cs+ permeability were more difficult to discern. However, extragenic second-site suppressor procapsids packaged with the mutant DNA-binding protein restored particle densities to near wild-type values. Therefore, a simple model, in which counter ions compensate for the loss of basic amino acids, is not supported by the data. This leaves the intriguing possibility that the dimensions or shape of the mutant particles may be altered.
When suppressor capsids are packaged with wild-type J protein, the density values are slightly greater than with the wild-type virion. Since the mutant particles are still more dense than wild-type particles, the suppressor does not act via a compensatory mechanism. This suggests that there is an alternative maturation pathway that minimizes the effect of the genomic tether on the magnitude of capsid collapse.
To investigate whether differences were expressed on the capsid exterior, host cell attachment assays were performed. The particles packaged with the mutant DNA-binding protein exhibited dramatically lower attachment efficiencies. As seen in the density experiments, the presence of the extragenic second-site suppressor restored values toward that of the wild type. Differences in native gel migration, which is a function of size and net surface charge (45), were also observed. While small migration differences were seen between the wild-type and mutant particles migrating toward the anode, the most dramatic differences involved the presence of a minor population migrating in the opposite direction. In wild-type populations these particles represent a small subpopulation (1:1,000). However, the ratio was ca. 1:3 in mutant samples. As seen in the other assays, the extragenic suppressor restored the ratio to nearly wild-type values.
Altering the nucleic acid component of the tether.
Naked φX174 DNA is substantially richer in secondary structure than packaged DNA (5). Benevides et al. (5) hypothesized that the DNA-binding protein inhibited the formation of secondary structure. Therefore, it is likely that an interplay between base pairing and DNA-capsid association occurs. To investigate this hypothesis, two species of φX174 ampicillin-transducing particles were generated by packaging single-stranded versions of unit length plasmids. The plasmids differed in the orientation of one cloned section, which was designed to introduce a large hairpin loop. Differences in migration rates and subpopulation ratios were observed between the transducing particles and wild-type virions.
While this may be one of the first reports of genome-capsid interactions affecting ssDNA viral structure and morphogenesis, the phenomenon has been well documented in ssRNA viruses. FHV RNA stabilizes contact regions (25) via nonspecific interactions with coat protein grooves at twofold axes of symmetry. Deletions of the RNA-interacting residues results in the production of polymorphic structures (17). Packaging FHV with foreign RNA also leads to altered particles (6). Thus, genome-capsid interactions can effect the fidelity of virion morphogenesis, a function commonly associated with viral scaffolding proteins. A dramatic example of a scaffolding-like function for nucleic acids has been observed in Southern cowpea mosiac virus. Deleting the highly basic, RNA-interacting N terminus of the coat protein results in the production of T=1, as opposed to T=3, capsids (44). Similarly, the nature of the RNA packaged in Brome mosaic virus can determine whether T=3 or 120-subunit capsids are formed (35).
The role of DNA-capsid interactions in φX174 is obviously not as dramatic. Procapsid morphogenesis does not require the genome (8, 28, 29); however, two scaffolding proteins mediate this stage of assembly. In the procapsid there are no discernible pentamer-pentamer interactions. The integrity of the capsid appears to be maintained by the scaffolding proteins. After packaging, the internal scaffolding protein is extruded from the structure and replaced by the DNA-binding protein and the tethered genome. This may supplant scaffolding function in the provirion. The provirion-to-virion transition is marked by the release of the external scaffolding protein and the completion of the 8.5-Å radial collapse of coat protein pentamers (15, 16, 31, 38, 39). Genome-capsid interactions may be mediating the magnitude and preserving integrity during the collapse.
Acknowledgments
We thank Bryan L. Jennings for technical assistance.
This work was supported by a grant from the National Science Foundation (B.A.F.).
REFERENCES
- 1.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossman. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 2.Aoyama, A., R. K. Hamatake, and M. Hayashi. 1981. Morphogenesis of φX174: in vitro synthesis of infectious phage from purified viral components. Proc. Natl. Acad. Sci. USA 78:7285-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoyama, A., R. K. Hamatake, and M. Hayashi. 1983. In vitro synthesis of bacteriophage φX174 by purified components. Proc. Natl. Acad. Sci. USA 80:4195-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoyama, A., and M. Hayashi. 1985. In vitro packaging of plasmid DNAs into φX174 bacteriophage capsid. Nature 297:704-707. [DOI] [PubMed] [Google Scholar]
- 5.Benevides, J. M., P. L. Stow, L. L. Ilag, N. L. Incardona, and G. J. Thomas, Jr. 1991. Differences in secondary structure between packaged and unpackaged single-stranded DNA of bacteriophage φX174 determined by Raman spectroscopy: a model for φX174 DNA packaging. Biochemistry 30:4855-4862. [DOI] [PubMed] [Google Scholar]
- 6.Bothner, B., A. Schneemann, D. Marshall, V. Reddy, J. E. Johnson, and G. Siuzdak. 1999. Crystallographically identical virus capsids display different properties in solution. Nat. Struct. Biol. 6:114-116. [DOI] [PubMed] [Google Scholar]
- 7.Brown, D. R., M. J. Roth, D. Reinberg, and J. Hurwitz. 1984. Analysis of bacteriophage φX174 gene A protein mediated termination and reinitiation of φX174 DNA synthesis. I. Characterization of the termination and reinitiation reactions. J. Biol. Chem. 259:10545-10555. [PubMed] [Google Scholar]
- 8.Burch, A. D., J. Ta, and B. A. Fane. 1999. Cross-functional analysis of the Microviridae internal scaffolding protein. J. Mol. Biol. 286:95-104. [DOI] [PubMed] [Google Scholar]
- 9.Burch, A. D., and B. A. Fane. 2000. Foreign and chimeric external scaffolding proteins as inhibitors of Microviridae morphogenesis. J. Virol. 74:9347-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman, M. S., and M. G. Rossmann. 1995. Single-stranded DNA-protein interactions in canine parvovirus. Structure 3:151-162. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z. G., C. Stauffacher, Y. Li, T. Schmidt, W. Bomu, G. Kamer, M. Shanks, G. Lomonossoff, and J. E. Johnson. 1989. Protein-RNA interactions in an icosahedral virus at 3.0 Å resolution. Science 245:154-159. [DOI] [PubMed] [Google Scholar]
- 12.Choi, Y. G., and A. L. N. Rao. 2000. Packaging of tobacco mosaic virus subgenomic RNAs by brome mosaic virus coat protein exhibits RNA controlled polymorphism. Virology 275:249-257. [DOI] [PubMed] [Google Scholar]
- 13.Dalphin, M. E. 1989. Bacteriophage φX174: cross-linking studies of the virion and prohead and biophysical characterization of the gene J protein. Ph.D. thesis. University of California, San Diego.
- 14.Desai, P., S. C. Watkins, and S. Person. 1994. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J. Virol. 68:5365-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dokland, T., R. A. Bernal, A. Burch, S. Pletnev, B. A. Fane, and M. G. Rossmann. 1999. The role of scaffolding proteins in the assembly of the small single-stranded DNA virus φX174. J. Mol. Biol. 288:595-608. [DOI] [PubMed] [Google Scholar]
- 16.Dokland, T., R. McKenna, L. L. Ilag, B. R. Bowen, N. L. Incardona, B. A. Fane, and M. G. Rossmann. 1997. Structure of a viral assembly intermediate with molecular scaffolding. Nature 389:308-313. [DOI] [PubMed] [Google Scholar]
- 17.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasi-equivalent icosahedral virus. J. Virol. 72:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earnshaw, W. C., and S. R. Casjens. 1980. DNA packaging by the double-stranded DNA bacteriophages. Cell 21:319-331. [DOI] [PubMed] [Google Scholar]
- 19.Eisenberg, S., and A. Kornberg. 1979. Purification and characterization of φX174 gene A protein: a multifunctional enzyme of duplex DNA replication. J. Biol. Chem. 254:5328-5332. [PubMed] [Google Scholar]
- 20.Eisenberg, S., J. Griffith, and A. Kornberg. 1977. φX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc. Natl. Acad. Sci. USA 74:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekechukwu, M. C., D. J. Oberste, and B. A. Fane. 1995. Host and φX174 mutations affecting the morphogenesis or stabilization of the 50S complex, a single-stranded DNA synthesizing intermediate. Genetics 140:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fane, B. A., and M. Hayashi. 1991. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage φX174. Genetics 128:663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fane, B. A., S. Head, and M. Hayashi. 1992. The functional relationship between the J proteins of bacteriophages φX174 and G4 during phage morphogenesis. J. Bacteriol. 174:2717-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fane, B. A., S. Shien, and M. Hayashi. 1993. Second-site suppressors of a cold sensitive external scaffolding protein of bacteriophage φX174. Genetics 134:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176-179. [DOI] [PubMed] [Google Scholar]
- 26.Fluit, A. C., P. D. Baas, and H. S. Jansz. 1985. The complete 30-base-pair origin region of bacteriophage φX174 in a plasmid is both required and sufficient for in vivo rolling circle DNA replication. Eur. J. Biochem. 149:579-584. [DOI] [PubMed] [Google Scholar]
- 27.Fujisawa, H., and M. Hayashi. 1976. Viral DNA-synthesizing intermediate complex isolated during assembly of bacteriophage φX174. J. Virol. 19:409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamatake, R. K., A. Aoyama, and M. Hayashi. 1985. The J gene of φX174: in vitro analysis of J protein function. J. Virol. 54:345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamatake, R. K., K. J. Buckley, and M. Hayashi. 1988. The J gene of φX174: isolation and characterization of a J gene mutant. Mol. Gen. Genet. 211:72-77. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi, M., A. Aoyama, D. L. Richardson, and M. N. Hayashi. 1988. Biology of the bacteriophage φX174, p. 1-71. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Publishing, New York, N.Y. [Google Scholar]
- 31.Ilag, L. L., N. H. Olson, T. Dokland, C. L. Music, R. H. Cheng, Z. Brown, R. McKenna, M. G. Rossman, T. S. Baker, and N. L. Incardona. 1995. Bacteriophage φX174 procapsid: purification and structure at 25 Å resolution. Structure 3:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jardine, P. J., and D. H. Combs. 1998. Capsid expansion follows the initiation of DNA packaging in bacteriophage T4. J. Mol. Biol. 284:661-672. [DOI] [PubMed] [Google Scholar]
- 33.Jennings, B., and B. A. Fane. 1997. Genetic analysis of the φX174 DNA binding protein. Virology 227:370-377. [DOI] [PubMed] [Google Scholar]
- 34.King, J., and S. Casjens. 1974. Catalytic head assembling protein in virus morphogenesis. Nature 251:112-119. [DOI] [PubMed] [Google Scholar]
- 35.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96:13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larson, S. B., J. Day, A. Greenwood, and A. McPherson. 1998. Refined structure of satellite tobacco mosaic virus at 1.8 Å resolution. J. Mol. Biol. 277:37-59. [DOI] [PubMed] [Google Scholar]
- 37.Lata, R., J. F. Conway, N. Cheng, R. L. Duda, R. W. Hendrix, W. R. Wikoff, J. E. Johnson, H. Tsuruta, and A. C. Steven. 2000. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell 100:253-263. [DOI] [PubMed] [Google Scholar]
- 38.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, H. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, J. B. Wolfinbarger, and M. Agbandje-McKenna. 1992. Atomic structure of single-stranded DNA bacteriophage φX174 and its functional implications. Nature 355:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna, R., L. L. Ilag, and M. G. Rossman. 1994. Analysis of the single-stranded DNA bacteriophage φX174 at a resolution of 3.0 Å. J. Mol. Biol. 237:517-543. [DOI] [PubMed] [Google Scholar]
- 40.Mukai, R., R. K. Hamatake, and M. Hayashi. 1979. Isolation of the bacteriophage φX174 prohead. Proc. Natl. Acad. Sci. USA 76:4877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad, B. V., P. E. Prevelige, E. Marietta, R. O. Chen, D. Thomas, J. King, and W. Chiu. 1993. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 231:65-74. [DOI] [PubMed] [Google Scholar]
- 42.Roof, W. D., S. M. Horne, K. D. Young, and R. Young. 1994. slyD, a host gene required for φX174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J. Biol. Chem. 269:2902-2910. [PubMed] [Google Scholar]
- 43.Rossmann, M. G., and J. E. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533-573. [DOI] [PubMed] [Google Scholar]
- 44.Savithri, H. S., and J. W. Erickson. 1983. The self-assembly of the cowpea strain of southern bean mosaic virus: formation of T = 1 and T = 3 nucleoprotein particles. Virology 126:328-335. [DOI] [PubMed] [Google Scholar]
- 45.Serwer, P., and M. E. Pichler. 1978. Electrophoresis of bacteriophage T7 and T7 capsids in agarose gels. J. Virol. 28:917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, L., K. N. Johnson, L. A. Ball, T. Lin, M. Yeager, and J. E. Johnson. 2001. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 8:77-83. [DOI] [PubMed] [Google Scholar]
- 47.Van Mansfeld, A. D., P. D. Baas, and H. S. Jansz. 1984. Gene A protein of bacteriophage φX174 is a highly specific single-stranded DNA nuclease and binds via a tyrosyl residue to DNA after cleavage. Adv. Exp. Med. Biol. 197:221-230. [DOI] [PubMed] [Google Scholar]
- 48.Vriend, G., B. J. Verduin, and M. A. Hemminga. 1986. Role of the N-terminal part of the coat protein in the assembly of cowpea chlorotic mottle virus: a 500 MHz proton nuclear magnetic resonance study and structural calculations. J. Mol. Biol. 191:453-460. [DOI] [PubMed] [Google Scholar]
- 49.Wikoff, W. R., C. J. Tsai, G. Wang, T. S. Baker, and J. E. Johnson. 1997. The structure of cucumber mosaic virus: cryoelectron microscopy, X-ray crystallography, and sequence analysis. Virology 232:91-97. [DOI] [PubMed] [Google Scholar]