Abstract
Virions of human immunodeficiency virus type 1 (HIV-1) and other lentiviruses contain conical cores consisting of a protein shell composed of the viral capsid protein (CA) surrounding an internal viral ribonucleoprotein complex. Although genetic studies have implicated CA in both early and late stages of the virus replication cycle, the mechanism of core disassembly following penetration of target cells remains undefined. Using quantitative assays for analyzing HIV-1 core stability in vitro, we identified point mutations in CA that either reduce or increase the stability of the HIV-1 core without impairing conical core formation in virions. Alterations in core stability resulted in severely attenuated HIV-1 replication and impaired reverse transcription in target cells with only minimal effects on viral DNA synthesis in permeabilized virions in vitro. We conclude that formation of a viral core of optimal stability is a prerequisite for efficient HIV-1 infection and suggest that disassembly of the HIV-1 core is a regulated step in infection that may be an attractive target for pharmacologic intervention.
In order to replicate, viruses must disassemble properly following entry into target cells. This capacity to disassemble is likely an important determinant for cell tropism and viral pathogenicity (20). Although disassembly is an essential step in the replication of viruses, studies of these processes have been underemphasized relative to other steps in the viral life cycle, due in part to a lack of relevant assays for early postentry steps in infection.
Virions of human immunodeficiency virus type 1 (HIV-1) and other lentiviruses contain conical cores (or “cones”) formed by the viral capsid protein (CA). Following assembly of the Gag structural polyprotein at the plasma membrane, HIV-1 virions bud as immature particles. Nascent HIV-1 particles subsequently mature through cleavage of Gag by the viral protease to yield discrete matrix (MA), CA, nucleocapsid (NC), and p6 proteins. In mature virions, MA remains associated with the viral lipid envelope, NC coats the viral RNA genome, and the CA monomers condense around the ribonucleoprotein complex to form the shell of the mature conical core (the capsid).
Structural studies have revealed that CA consists of two distinct, globular domains connected by a flexible linker: an amino-terminal domain, composed of seven α-helices and a β-hairpin, and a carboxyl-terminal domain, composed of a 310 helix and four α-helices (15, 16, 18, 29, 31). Based on X-ray crystallographic data and in vitro assembly models, several intermolecular CA-CA interfaces have been proposed for the assembly of mature HIV-1 cores, including C-terminal domains involved in dimerization of adjacent subunits and N-terminal domains involved in hexamerization of adjacent subunits (15, 28, 31). Mutagenesis studies have shown that the N-terminal domain of CA is essential for capsid formation (11) and the C-terminal domain is essential for particle assembly and capsid formation. Mutations in CA that resulted in abnormal core morphology also severely impaired HIV-1 infectivity (11, 29, 33, 35a), suggesting that proper assembly of the capsid is crucial for the success of early postentry events.
Despite the wealth of structural data for HIV-1 CA, little is known about the functional role of the CA shell during the early phase of viral infection. Infection of target cells by HIV-1 begins with attachment and fusion of viral and cellular membranes, proceeds through reverse transcription of the viral RNA into proviral DNA, and culminates in integration of the proviral DNA into the host genome. Based on electron microscopic analysis of acutely infected cells, it is generally thought that the conical core does not persist for long following fusion of viral and cellular membranes (22). The available biochemical data are also consistent with rapid disassembly of the core, as CA was undetectable in reverse transcription complexes isolated from infected cells (13, 24).
Based on analogies with other viruses (4, 21, 34, 35, 40), we hypothesized that proper disassembly of the HIV-1 core is essential for infectivity. To test this hypothesis, we established assays to quantify the stability of HIV-1 cores in vitro. By using these assays to analyze a series of CA point mutants, we identified mutations that alter the stability of the HIV-1 core. All such mutants were defective for replication in primary T cells. For most of the mutants, the block to infection was localized to a defect in reverse transcription in target cells. We conclude that dissociation of CA from the HIV-1 core is a crucial postentry step in viral replication that controls the efficiency of viral DNA synthesis in target cells.
MATERIALS AND METHODS
Cells and viruses.
293T and HeLa-CD4/LTR-lacZ (P4) cells were cultured in Dulbecco's modified Eagle medium (Cellgro) supplemented with 10% fetal bovine serum, penicillin (50 IU/ml), and streptomycin (50 μg/ml) at 37°C and 5% CO2. Primary CD4+ T cells were isolated from whole blood of HIV-1-seronegative donors and were activated with anti-CD3 and mitomycin C-treated antigen-presenting cells as previously described (36). The activated T cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin (50 IU/ml), streptomycin (50 μg/ml), and interleukin-2 (50 U/ml) at 37°C and 5% CO2. The wild-type HIV-1 proviral DNA construct R9, encoding full-length open reading frames for all HIV-1 structural and accessory genes, was used for these studies. Point mutations in the CA region of R9 were engineered by oligonucleotide-directed single-stranded mutagenesis in the phagemid vector pSL1180-gag and were rebuilt into HIV-1 by transferring BssHII-ApaI restriction fragments into full-length R9 (U. von Schwedler and W. I. Sundquist, unpublished data). Viruses were produced by calcium phosphate transfection of 293T cells (20 μg of plasmid DNA per 2 × 106 cells) as previously described (10). One day after transfection, the culture supernatants were harvested and clarified by being passed through 0.45-μm-pore-size filters, and aliquots were frozen at −80°C. The CA contents of the virus stocks were quantified by p24 enzyme-linked immunosorbent assay (ELISA), as previously described (38). The P4 cell line, a HeLa cell clone engineered to express CD4 and an integrated long terminal repeat (LTR)-lacZ reporter cassette, was used to quantify HIV-1 infectivity as previously described (9) with the following modifications. HIV-1 stocks were serially diluted in culture medium, and samples (0.125 ml) were used to inoculate P4 target cells seeded the day before (20,000 cells per well in 48-well plates). Two hours after inoculation, the cultures were fed with additional medium (0.5 ml) and cultured for another 48 h prior to being stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside to detect infected cells. To determine the number of infected cells per well, individual wells were visualized using a charge-coupled device camera equipped with a macro lens, and blue cells were counted using NIH Image software. Infections were performed in triplicate, and only values within the linear range of the infection assay (up to 1,000 blue cells per well) were used to calculate infectivity.
Isolation of HIV-1 cores.
Cores were isolated from concentrated virions as previously described (27). Briefly, supernatants from transfected 293T cells were filtered to remove cellular debris, and HIV-1 particles were concentrated by ultracentrifugation (120,000 × g for 3 h at 4°C) through a cushion of 20% (wt/vol) sucrose in STE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA). Viral pellets were resuspended in 200 μl of STE buffer, and the concentrated virions were subjected to ultracentrifugation (100,000 × g for 16 h at 4°C) through a layer of 1% Triton X-100 into a linear sucrose density gradient (10 ml of STE buffer containing 30 to 70% sucrose). Fractions (1 ml) were collected from the top of the gradient and analyzed for CA content by p24 ELISA. The density of each fraction was determined by measuring the refractive index and converting to density using the formula ρ = (2.6496 × η) − 2.5323, where ρ is the density in grams/milliliter and η is the refractive index. The yield of cores was determined from the CA contents (as determined by p24 ELISA) in the peak fractions of cores as a percentage of the total CA content in the gradient. The peak of isolated cores on the density gradient was initially identified by assaying for exogenous reverse transcriptase (RT) activity as previously described (2). Aliquots (10 μl) of each fraction were mixed with RT reaction cocktail (20 μl). The reaction mixtures were incubated at 37°C for 2 h, spotted on DE81 filters, and washed thrice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and once in 95% ethanol, and 3H was quantified by liquid scintillation counting.
In vitro core disassembly assay.
Samples of purified HIV-1 cores (100 μl) were diluted in STE buffer (1 ml) and incubated at 37°C for various times or under various conditions. Following incubation, samples were centrifuged at 100,000 × g (Beckman TLA-55 rotor at 45,000 rpm) for 20 min at 4°C. The supernatants were removed, the pellets were dissolved in sodium dodecyl sulfate (SDS) sample buffer, and CA in both fractions was quantified by ELISA. The extent of core disassembly was determined as the percentage of CA in the supernatant versus the total quantity of CA in the reaction (supernatant plus pellet).
Protein analyses.
Samples were subjected to electrophoresis on 4 to 20% polyacrylamide denaturing gels, and proteins were transferred electrophoretically to polyvinylidene difluoride membranes. Protein blots were probed with rabbit polyclonal antibodies to individual HIV-1 proteins, including anti-CA (from D. Trono), anti-NC (from L. Henderson), anti-RT (AIDS Repository no. 634), anti-integrase (IN; from D. Grandgenett), and anti-Vpr (reagent no. 3252; National Institutes of Health [NIH] AIDS Research and Reference Reagent Program). Following incubation with peroxidase-conjugated goat antibody specific for rabbit immunoglobulins, protein bands were visualized using chemiluminescence detection (SuperSignal; Pierce Chemical Co.) and exposure to X-ray film.
Endogenous reverse transcription assays.
Assays of endogenous reverse transcription were performed by quantifying incorporation of [3H]TTP, as previously described (41). Concentrated virions (0.5 μg p24) were resuspended in phosphate-buffered saline (PBS; 10 μl) and diluted with 40 μl of endo-RT reaction buffer (5 mM MgCl2; 50 mM NaCl; 10 mM dithiothreitol; 0.5 mM dATP, dCTP, and dGTP; 50 mM Tris [pH 8]; 0.015% Triton X-100; and 10 μM [3H]TTP [62.2 Ci/mmol]). The reaction mixtures were incubated at 37°C and were terminated by the addition of SDS and EDTA to 0.5% and 25 mM, respectively. Samples were spotted on DE81 filters and washed thrice in 2× SSC and once in 95% ethanol, and 3H was quantified by liquid scintillation counting. Activity values were normalized by exogenous RT activity present in the input virions.
Quantitative analysis of HIV-1 reverse transcription in target cells.
For analysis of viral DNA synthesis in target cells, aliquots of viruses containing 100 ng of CA were used to infect 100,000 P4 cells. Viruses were pretreated with 20 μg of DNase I/ml and 10 μg of MgCl2/ml at 37°C for 1 h to remove contaminating plasmid DNA. To harvest the cells, the supernatant was removed and the cells were washed in 1 ml of PBS, followed by trypsinization. The cells were pelleted, washed once with 0.5 ml of PBS, and repelleted. The pellets were dissolved in 100 μl of PCR lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 0.2 mM CaCl2, 0.001% Triton X-100, 0.001% SDS, 1 mg of proteinase K/ml). The lysates were incubated at 58°C for 1 h, followed by heat inactivation at 95°C for 15 min (32).
Viral DNA was quantified by real-time PCR using an ABI 7700 instrument (PE Biosystems) with SYBR Green chemistry. The reaction mixtures (25-μl total volume) contained 2.5 μl of infected lysate, 12.5 μl of 2× SYBR Green PCR Master Mix (PE Biosystems), and 50 nM each primer. A standard curve was prepared from serial dilutions of HIV-1 plasmid DNA using yeast tRNA (30 μg/ml) in water as a diluent. The reactions were amplified as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 2 min and 65°C for 45 s. Plots of threshold cycle number (Ct) values versus log[target] were linear between 2 and 50,000 copies of target DNA. Unknowns were quantified by interpolating target copy numbers from Ct values of unknown samples using the standard curve. The sequences of primers (R and U5) specific for early products were 5′-GGCTAACTAGGGAACCCACTGCTT (forward) and 5′-CTGCTAGAGATTTTCCACACTGAC (reverse). The late-product primer sequences (R and 5NC) were 5′-TGTGTGCCCGTCTGTTGTGT (forward) and 5′-GAGTCCTGCGTCGAGAGAGC (reverse) (6).
Slot blot analysis of viral RNA.
HIV-1 particles were treated with 20 μg of DNase I/ml for 45 min at 37°C to digest plasmid DNA. RNA was isolated from pelleted virions using TriReagent (Molecular Research Center, Inc.). RNA precipitates were dissolved in 500 μl of ice-cold solution containing 10 mM NaOH and 1 mM EDTA and applied to a nitrocellulose membrane by vacuum filtration using a Bio-Dot SF apparatus (Bio-Rad). The membrane was prehybridized overnight at 42°C in hybridization solution (50% formamide, 5× SSC, 1× Denhardt's solution, 50 mM NaHPO4 [pH 6.5], 250 μg of denatured salmon sperm DNA/ml) followed by incubation with HIV-1-specific probe in hybridization solution at 42°C for 16 h. The random-primed 32P-labeled probe was generated by digesting R9 proviral plasmid with XbaI and HpaI, purifying the large fragment corresponding to the 3′ 7 kbp of the proviral genome, and primer extension with [α-32P]dCTP using the Prime-It II kit (Stratagene). For analysis of virion RNA content, samples applied to the filter were normalized by p24 ELISA, with the exception of the K170A mutant, which was normalized by exogenous RT activity due to its poor reactivity in the p24 ELISA. RNA signals were quantified by phosphorimager analysis.
RESULTS
Isolation of HIV-1 cores.
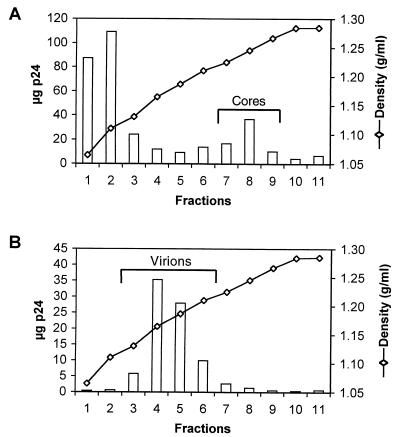
To analyze the role of core stability in HIV-1 infection, we developed methods to quantify the stability of HIV-1 cores in vitro. We have previously described the isolation of HIV-1 cores from mature virions (27). This approach involves layering concentrated virions onto a linear 30 to 70% sucrose gradient containing a layer of 1% Triton X-100 on top of the gradient. Upon centrifugation at 4°C, virions sediment through the layer of detergent, removing the lipid bilayer. This method is effective in removing approximately 97% of phospholipids from virions (J. Zhou and C. Aiken, unpublished results). Furthermore, immunoblot analysis showed that the envelope proteins gp41 and gp120 were efficiently removed by detergent treatment. Intact cores sedimented to a density of 1.24 to 1.26 g/ml (Fig. 1A), as previously reported (27, 39). Electron microscopic analysis of these fractions also revealed the presence of conical cores typical of lentiviruses (27). For comparative purposes, virions also were subjected to equilibrium sedimentation without exposure to detergent. Intact HIV-1 virions were detected in fractions of density 1.16 to 1.18 g/ml, a density typical of native retroviral particles and distinct from that of mature cores (Fig. 1B).
FIG. 1.
Equilibrium density gradient sedimentation of HIV-1 cores and virions. Concentrated virions were layered onto 30 to 70% linear sucrose gradients with (A) or without (B) a layer of Triton X-100. Following ultracentrifugation at 100,000 × g for 16 h at 4°C, fractions (1 ml) were collected from the top of the gradient and were analyzed by p24 ELISA. The density of each fraction was determined by refractometry. Core fractions that were pooled and used for further analysis (Cores) are indicated in panel A.
Mature HIV-1 cores were relatively unstable under these isolation conditions, as reflected in the relatively low yield of CA recovered in the fractions of HIV-1 cores (∼16% of input virions). The majority of the CA protein was present in fractions at the top of the gradient and was therefore not particle associated (Fig. 1A). This relatively low yield of mature cores was reproducible, suggesting that the yield of cores reflects an intrinsic biological property of the virions and might serve as a measure of relative HIV-1 core stability in vitro.
Disassembly of purified HIV-1 cores in vitro via release of CA from the viral ribonucleoprotein complex.
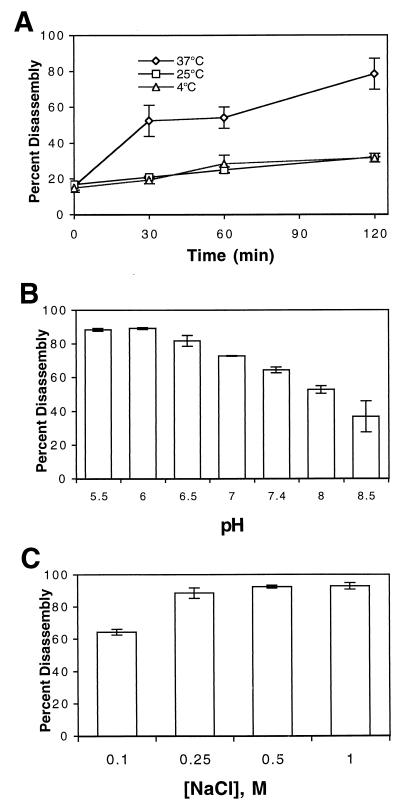
To investigate the dynamics of HIV-1 core disassembly further, we developed a kinetic assay to analyze HIV-1 core disassembly in vitro. Purified HIV-1 cores (pooled fractions 7, 8, and 9 [Fig. 1A]) were diluted in STE buffer and incubated at 37°C. Following incubation, the particles were subjected to ultracentrifugation to separate free CA from intact cores. Both supernatants and pellets were analyzed for CA content by ELISA to determine the percentage of total CA released from the cores during incubation. CA dissociation was found to be temperature dependent, proceeding more rapidly at 37 than at 25 or 4°C (Fig. 2). Furthermore, the reaction was influenced by both pH and ionic strength, as CA dissociation was accelerated at low pH and at high NaCl concentration (Fig. 2).
FIG. 2.
Disassembly of purified HIV-1 cores in vitro. Cores were incubated in STE buffer at the indicated temperatures for the times shown (A) or at 37°C for 1 h in STE buffer prepared at the indicated pH values (B) or NaCl concentrations (C). Following incubation, the samples were subjected to ultracentrifugation at 100,000 × g for 15 min at 4°C. The extent of disassembly was determined as the percentage of the total CA protein in the reaction present in the supernatant. (A) Mean values of triplicate determinations, with error bars representing 1 standard deviation. (B and C) Mean values of duplicate determinations, with error bars representing the range of values.
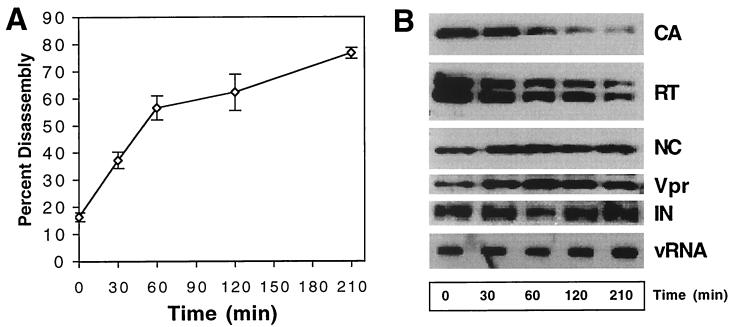
To determine whether CA dissociation was accompanied by dissolution of the internal viral ribonucleoprotein complex, we analyzed the pellets from disassembled cores for other HIV-1 structural components. Immunoblot analysis of the pellets from this reaction confirmed the dissociation of CA protein from the cores (Fig. 3). Loss of particle-associated CA was not due to nonspecific degradation of the protein, as the supernatants showed a corresponding and quantitative increase in CA when analyzed by both immunoblotting and quantitation by ELISA, with all of the CA protein being accounted for (data not shown). Further analysis of the pellets by immunoblotting for core-associated proteins revealed that RT was released at a rate similar to that of CA, but the quantities of NC, Vpr, and IN were unchanged in the pellets despite nearly complete solubilization of CA (Fig. 3B). To determine whether genomic RNA also remained particle associated, RNA was extracted from parallel samples and immobilized by vacuum filtration followed by probing with a 32P-labeled HIV-1-specific probe (Fig. 3B). Quantitative phosphorimager analysis of the RNA slot blot confirmed that similar quantities of RNA were present in pellet samples across the time course (data not shown). These results suggest that HIV-1 cores disassembled in vitro through dissociation of CA from the internal viral ribonucleoprotein complex, which remained intact for at least 3 h at 37°C.
FIG. 3.
Kinetic and biochemical analyses of HIV-1 core disassembly in vitro. Purified HIV-1 cores were incubated at 37°C for the indicated times, followed by separation of free and core-associated CA by ultracentrifugation. (A) Dissociation of CA from pelletable cores during incubation at 37°C. Supernatants and pellets were analyzed by p24 ELISA. The extent of disassembly was determined as the percentage of the total CA protein in the reaction detected in the supernatant. (B) Biochemical analysis of particles remaining following disassembly of HIV-1 cores. Pellets from the disassembly reactions shown in panel A were subjected to Western blotting and RNA slot blot analysis. The Western blot membrane was probed sequentially with rabbit anti-CA, anti-RT, anti-NC, and anti-Vpr antibodies. RNA was extracted from the pellets, denatured, and immobilized by vacuum filtration. Relative HIV-1 RNA levels were determined by hybridization with a 32P-labeled HIV-1 probe, followed by detection by autoradiography. In panel A, the values represent means of triplicates, with error bars representing 1 standard deviation. Shown are representative data from one of three independent experiments.
Point mutations in CA alter the yield of HIV-1 cores.
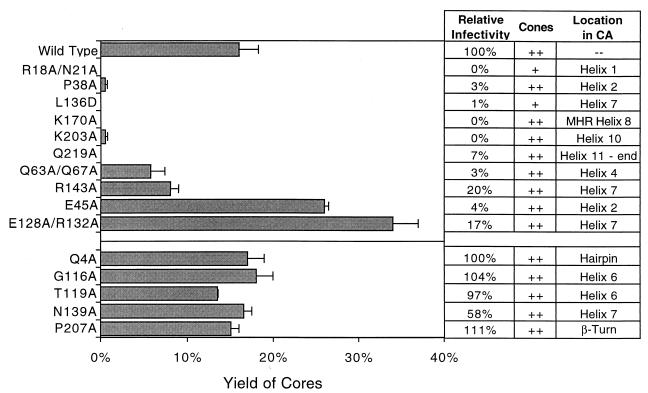
We hypothesized that proper dissociation of the CA shell from the HIV-1 core is required for efficient infection. To test this hypothesis, we employed a panel of viruses containing point mutations that map throughout the CA coding region of gag and collectively scan different surfaces of CA. We sought mutants that were defective for replication due to specific postentry defects, reasoning that these mutants might exhibit alterations in core stability. Accordingly, all mutants used in this study were previously found to be competent for particle assembly, to exhibit normal processing of Gag, and to incorporate envelope protein (Env) at levels similar to those for wild-type HIV-1 (von Schwedler and Sundquist, unpublished). These residues are all highly conserved in HIV-1 isolates from HIV-infected individuals. Furthermore, ultrastructural analysis (by transmission electron microscopy of ultrathin sections of virions) revealed that most of the mutations did not impair cone formation in virions. Although cone formation was not rigorously quantified in these mutants, cones were observed at a frequency similar to that in wild-type virions (von Schwedler and Sundquist, unpublished). Core assembly differed significantly for only two of the mutants: R18A/N21A and L136D. The R18A/N21A mutant formed a greater percentage of aberrant cores, such as rods, while the L136D mutant appeared to have few detectable cones. Despite the lack of apparent defects in assembly or morphology for most of the mutants, the infectivity of each of the HIV-1 mutants was markedly reduced relative to wild-type HIV-1 (Fig. 4) (von Schwedler and Sundquist, unpublished), suggesting that they might exhibit defects in core disassembly in target cells.
FIG. 4.
Identification of CA residues that influence the stability of HIV-1 cores. Concentrated virions were subjected to ultracentrifugation through a detergent layer into a sucrose density gradient. Yield of Cores, percentage of total CA detected in the peak fractions of cores; Relative infectivity, infectivity relative to wild-type HIV-1 as determined by a single-cycle infection assay; Cones, presence of similar quantities (compared to the wild type) of conical cores in virions (as determined by transmission electron microscopy). For R18A/N21A (+), few cones were apparent, and they were frequently aberrantly shaped; for L136D (+), cones were rarely detected. Location refers to the location of the mutation in the structure of HIV-1 CA. Shown are the mean values of at least two determinations, with error bars representing 1 standard deviation of the mean.
To determine whether the reduced infectivity of the mutants might arise from alterations in core stability, we attempted to isolate cores from each of the mutant viruses. Core yields were calculated by determining the amount of CA in the dense core fractions as a percentage of the total CA content of the gradient. As observed previously (Fig. 1), this yield was approximately 16% for wild-type HIV-1. In contrast, the yields from CA mutant viruses deviated markedly from those of wild-type virus (Fig. 4). Cores from mutants R18A/N21A, P38A, L136D, K170A, K203A, and Q219A could not be recovered, presumably due to their reduced stability. Two other mutants (Q63A/Q67A and R143A) yielded quantities of cores that were approximately half that of the wild type. Two mutants (E45A and E128A/R132A) were more stable than the wild type, yielding cores in greater quantities (∼30% of input virions). Although yields of mutant cores exhibited significant deviations from that of the wild type, the CA mutations did not detectably alter the gradient profiles of equilibrium density of the cores (data not shown). All of the mutants exhibiting altered yields of cores were markedly impaired in single-cycle HIV-1 infectivity assays (Fig. 4). As an additional control to determine whether HIV-1 infection requires formation of a core of wild-type stability, we evaluated five CA mutants that exhibited normal core morphology by electron microscopy and minimal impairments in infectivity. In every case, these mutants (Q4A, G116A, T119A, N139A, and P207A) also yielded cores in quantities that were very similar to that of wild-type HIV-1 (Fig. 4). These results demonstrate a strong correlation between HIV-1 core stability and viral infectivity.
Mutations in CA affecting core yield result in altered disassembly kinetics.
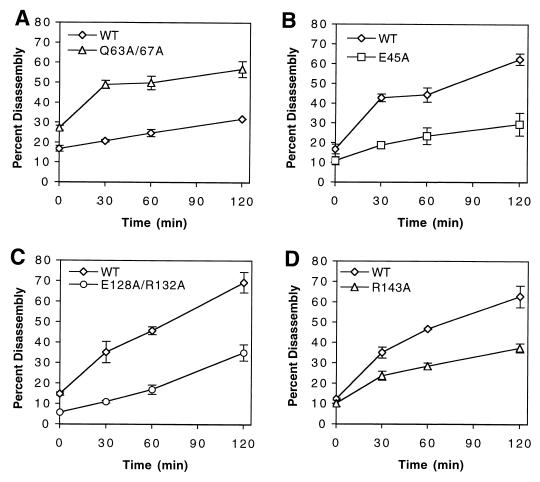
To evaluate the relative stability of mutant HIV-1 cores further, we performed kinetic assays of core disassembly using the CA mutants from which cores could be isolated. As shown in Fig. 5, any mutant that showed aberrant core yields also displayed disassembly kinetics which deviated significantly from those of wild-type HIV-1. Cores isolated from the Q63A/Q67A mutant disassembled more rapidly than the wild type. Disassembly of this mutant was quantified at 25°C owing to its instability (Fig. 5A); all other assays were performed at 37°C (Fig. 5B, C, and D). Cores isolated from CA mutants E45A and E128A/R132A (Fig. 5B and C) disassembled more slowly than wild-type cores, consistent with the greater yields of core observed with these mutants. Surprisingly, however, the behavior of the R143A mutant was paradoxical. R143A cores were isolated in lower yields than wild-type cores (Fig. 4); however, the R143A cores that were recovered disassembled more slowly than the wild type in the kinetic assay (Fig. 5D). The increased core stability exhibited for these three viruses was not due to elevated levels of unprocessed Gag, as core preparations from E45A, E128A/R132A, R143A, and wild-type virions contained similar levels of Pr55Gag (data not shown). Importantly, cores from the five control mutants (Q4A, G116A, T119A, N139A, and P207A), all of which exhibited wild-type infectivity, disassembled at rates that were similar to that of wild-type cores (data not shown). Collectively, these results demonstrate that the cores from the CA mutant viruses exhibiting deviations from wild-type core yields also disassembled in vitro at rates that differed from that of wild-type HIV-1 cores and further support the hypothesis that HIV-1 infectivity is dependent on formation of a core of optimal stability.
FIG. 5.
Kinetics of disassembly of wild-type (WT) and CA mutant cores in vitro. Isolated cores were diluted in STE buffer and incubated at 25 (A) or 37°C (B, C, and D). Shown are the mean values of triplicate determinations, with error bars representing 1 standard deviation.
Alterations in HIV-1 core stability block HIV-1 replication in primary T cells.
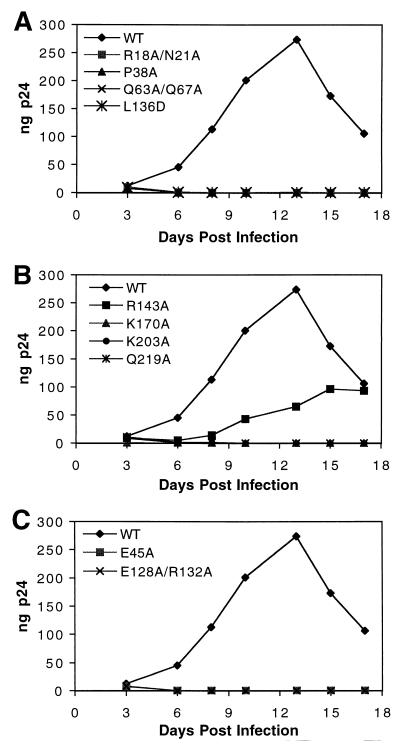
To evaluate the effects of alterations in core stability on the efficiency of HIV-1 replication, viral spread in primary T cells was studied. CD4+ primary T cells were purified from peripheral blood mononuclear cells from uninfected donors and were inoculated with equivalent quantities of each of the viruses (1-ng p24 input). HIV-1 replication was monitored by quantifying p24 production in culture supernatants. The CA mutants exhibiting unstable cores were also impaired for replication in T cells (Fig. 6A and B). Among this class of mutant viruses, only R143A replicated to any significant extent in these cells. Interestingly, this mutant showed only a twofold reduction in the yield of cores (Fig. 4) and was therefore the least impaired of all the mutants containing unstable cores. However, the possibility that the observed replication of R143A is due to reversion or pseudoreversion has not been tested. Mutants containing hyperstable cores (E45A and E128A/R132A) also failed to replicate in T cells (Fig. 6C). These results demonstrate that mutations in CA that alter the stability of the core also result in severe defects in replication in primary T cells, as well as impairments in single-cycle infectivity.
FIG. 6.
Replication kinetics of CA mutants in primary T cells. Cultures of activated primary T cells were inoculated with equal quantities (1 ng) of the indicated viruses. Samples were collected on the days indicated and analyzed for p24 content by ELISA. (A and B) Viruses with mutations that lead to decreased core stability in vitro; (C) viruses with mutations that lead to increased core stability in vitro. Shown is a representative growth curve for each virus from duplicate analyses. WT, wild type.
Alterations in core stability do not impair HIV-1 reverse transcription in vitro.
Because the core stability mutant viruses were impaired in both single-cycle and replication assays, we sought to determine whether these CA mutants exhibited defects in the reverse transcription machinery of the virions. Virion-associated RT activity was measured using an enzymatic assay for RT. Following normalization for the amount of p24 in the samples, no significant defects in exogenous RT activity were observed for the CA mutants (Table 1). To determine whether the mutations affected virion RNA packaging efficiency, virion-associated viral RNA was analyzed by slot blot detection of isolated RNA, followed by phosphorimager quantification. As shown in Table 1, no significant reductions in viral RNA packaging were observed. We conclude that the quantities of active RT enzyme and viral RNA incorporated in the mature particles are similar to those in wild-type HIV-1.
TABLE 1.
RT activity and RNA packaging efficiency of CA mutant particles
| Virus | Mean % wild-type HIV-1 activity (±SD)
|
||
|---|---|---|---|
| Exogenous RT activity/unit of p24 | Endogenous RT activity | Viral RNA content | |
| R18A/N21A | 133 ± 28 | 63 ± 8 | 85 ± 33 |
| P38A | 106 ± 15 | 184 ± 24 | 86 ± 21 |
| E45A | 96 ± 25 | 115 ± 6 | 102 ± 23 |
| Q63A/Q67A | 101 ± 25 | 60 ± 2 | 121 ± 22 |
| E128A/R132A | 110 ± 9 | 111 ± 12 | 92 ± 33 |
| L136D | 68 ± 19 | 65 ± 16 | 93 ± 58 |
| R143A | 89 ± 17 | 108 ± 7 | 80 ± 36 |
| K170A | NDa | 113 ± 2 | 102 ± 18b |
| K203A | 88 ± 9 | 103 ± 12 | 103 ± 30 |
| Q219A | 97 ± 30 | 155 ± 16 | 130 ± 29 |
ND, not determined due to inefficient detection by p24 ELISA.\
K170A was normalized by exogenous RT due to poor p24 ELISA reactivity.
A putative function of the HIV-1 capsid is to organize the ribonucleoprotein complex during virion maturation (17). Therefore, to test the hypothesis that the CA mutations impaired the formation of the viral ribonucleoprotein complex, we utilized the endogenous reverse transcription assay (41). This approach involves permeabilization of the viral membrane with detergent and incubation of the permeabilized virions with deoxynucleoside triphosphates and Mg2+ to promote reverse transcription. In this assay, viral DNA synthesis depends on the endogenous tRNA primer and template (viral genomic RNA) and is therefore thought to simulate reverse transcription more accurately than measuring RT activity from fully solubilized virions. The endogenous reaction has been used in studies of retroviral minus- and plus-strand DNA synthesis and to identify defects in Vif-defective virions (19) and an MA point mutant (25, 26). Furthermore, this assay has been used to reveal defects in the reverse transcription machinery of Rous sarcoma virus (RSV) CA mutants (7).
Wild-type and CA mutant virions were concentrated by ultracentrifugation, and equal quantities (normalized by exogenous RT activity) were tested in endogenous RT assays. Reverse transcription was quantified by measuring incorporation of [3H]TTP into product DNA. All of the mutants synthesized quantities of DNA similar to that of wild-type HIV-1; however, several of the mutants (R18A/N21A, Q63A/Q67A, and L136D) exhibited reductions of approximately 35% (Table 1). In addition, two mutants (P38A and Q219A) were enhanced by up to twofold relative to wild-type virions. Endogenous reverse transcription was effectively blocked by the nonnucleoside RT inhibitor nevirapine, demonstrating that the signal was specific for HIV-1 RT enzyme (data not shown). Collectively, these results suggest that the CA mutations do not markedly impair RNA packaging or formation of a functional ribonucleoprotein complex within the virion.
Alterations in core stability inhibit HIV-1 reverse transcription in vivo
Our data indicated that the impaired viral replication of the core stability mutants could not be explained by intrinsic defects of the reverse transcription machinery of the viruses. Therefore, we sought to identify the step in the viral life cycle affected by the mutations. All of the mutant virions were shown to incorporate HIV-1 envelope proteins as efficiently as wild-type HIV-1 particles (von Schwedler and Sundquist, unpublished). Furthermore, using a reporter assay of virus-cell fusion, we observed that all of the CA mutants used in this study were competent for entry into T cells (M. Miller and C. Aiken, unpublished observations). Therefore, we hypothesized that the mutants would be impaired for a postentry step in infection occurring prior to or during reverse transcription.
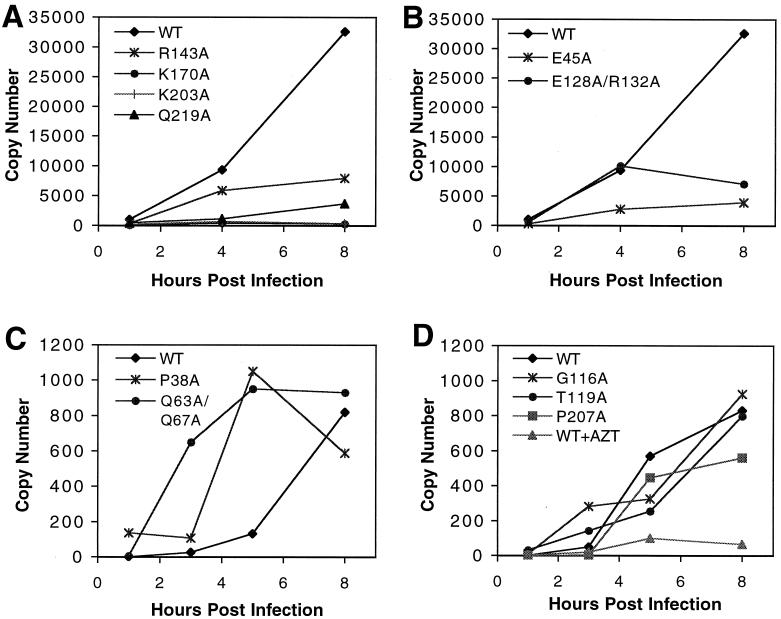
To probe the abilities of these CA mutant viruses to undergo reverse transcription in target cells, viral DNA synthesis was analyzed following infection of HeLa-CD4 (P4) target cells. Because retroviral reverse transcription occurs in distinct temporal stages, we employed quantitative real-time PCR using primers designed to specifically amplify early or late products of reverse transcription. Primers in the 5′ LTR (R and U5) were used in PCR to detect minus-strand strong-stop DNA, an early step in reverse transcription. Primers in U5 and the noncoding region upstream of gag were used to detect DNA synthesis occurring after the second-strand transfer event, at a relatively late stage of reverse transcription. Most of the mutant viruses exhibiting alterations in core stability also exhibited severe defects at both early and late stages of reverse transcription compared to wild-type HIV-1 (Fig. 7A and B and data not shown). Mutants with aberrant core morphologies (R18A/N21A and L136D) were also severely impaired for viral DNA synthesis (data not shown). Surprisingly, two of the mutants, P38A and Q63A/Q67A, were competent for efficient reverse transcription. However, both of these mutants exhibited unusually rapid kinetics (Fig. 7), with DNA synthesis peaking several hours earlier than for wild-type HIV-1. Both mutants were less infectious than the wild type, so the accelerated DNA synthesis observed must occur through a pathway that is not productive for infection. As expected, CA mutants exhibiting wild-type infectivity (G116A, T119A, and P207A) also displayed reverse transcription kinetics similar to those of the wild type (Fig. 7D). As a control for contaminating viral DNA and PCR specificity, infections by wild-type HIV-1 were also performed in the presence of the reverse transcription inhibitor azidothymidine, which effectively blocked viral DNA synthesis in target cells (Fig. 7D). Based on the results of the endogenous reverse transcription assays, we conclude that the defects in reverse transcription exhibited by the CA mutants in cells are not likely to be due to major defects in the ribonucleoprotein complex present in the mutant particles. Rather, the impaired reverse transcription probably reflects a specific difference in viral core stability.
FIG. 7.
Kinetics of viral DNA synthesis in vivo. Cultures of CD4-expressing HeLa cells were inoculated with equivalent quantities (100 ng of p24) of the indicated viruses. Individual cultures were harvested at the indicated times, and DNA was isolated for PCR. HIV-1 DNA was quantified by real-time PCR using the SYBR Green protocol. (A and B) Results from analysis using early-stage primers (R and U5); (C and D) DNA products corresponding to a later stage of reverse transcription (U5 and gag). The results are representative of three independent experiments. WT, wild type.
DISCUSSION
In this study, we have employed a series of biochemical approaches to dissect the mechanism of HIV-1 core disassembly. Using purified HIV-1 particles, we established novel assays for quantifying HIV-1 core stability in vitro and applied these assays to a collection of HIV-1 CA point mutants that exhibited reduced infectivity but still supported conical-core formation in virions. Our data revealed specific CA residues that influence the stability of the HIV-1 core. Without exception, mutants that deviated from wild-type core stability, either positively or negatively, were impaired in single-round infectivity as well as in replication in primary T cells. In most cases, CA mutants with altered core stability were competent for fusion with target cells but were blocked at a stage of infection prior to or during reverse transcription. Our data indicate that formation of a core of optimal stability is a strict requirement for efficient HIV-1 infection. These findings imply that, following delivery into the cytoplasm, the HIV-1 core must remain intact for some period of time in order for productive reverse transcription to occur.
Several lines of evidence support the biological relevance of the core stability phenotypes elucidated in the present study. Examination of purified wild-type cores by electron microscopy revealed a heterogeneous population of cones with a size distribution and morphologies similar to those of cores within intact virions. Biochemical analyses of the cores further demonstrated the presence of the predicted core-associated proteins in quantities similar to those in virions (1, 27, 39). Preliminary studies in our laboratory indicate that purified HIV-1 cores undergo endogenous reverse transcription as efficiently as permeabilized virions (B. M. Forshey and C. Aiken, unpublished observations), confirming their biochemical activity in vitro. Most importantly, CA mutants that exhibited wild-type infectivity yielded normal quantities of cores upon isolation; however, mutations that affected core stability invariably had deleterious effects on HIV-1 replication. Thus, the stability of HIV-1 cores measured in vitro is strongly correlated with a biological function of the viral core during HIV-1 infection.
Similar studies have recently been performed on the CA protein of RSV (3, 7). Several mutations in the major homology region of RSV resulted in efficient assembly of virions that were blocked for reverse transcription in target cells and exhibited core stability defects (7). Subsequent analysis of CA mutant pseudorevertants revealed restoration of wild-type replication kinetics (3). In the case of one pseudorevertant, replication competence was not accompanied by an increase in core stability. These results suggest that, at least for RSV, core stability as assayed by detergent resistance may not be an absolute requirement for infection. However, the relevance of these findings to HIV remains unclear; there are significant differences in the architectures of HIV and RSV cores, suggesting that these capsids may mediate different functions during infection or may interact differently with host cell factors following delivery into the cytosol. In a recent study by Fitzon et al. (14), mutagenesis of proline residues in HIV-1 CA led to defects in core stability, which correlated with reduced infectivity, similar to the effects observed in our study.
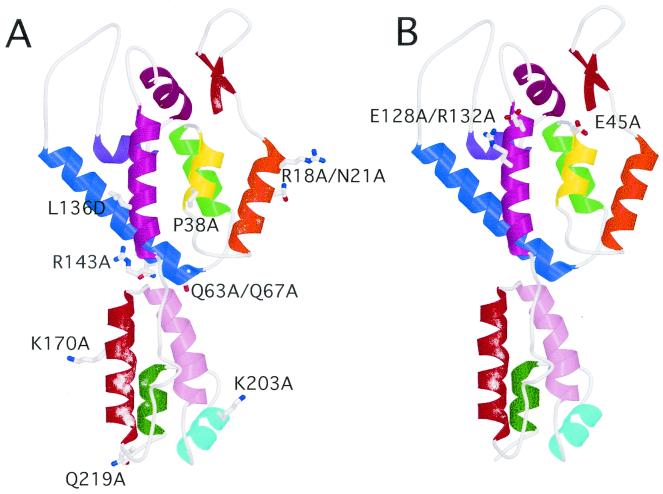
The mutations observed to affect HIV-1 core stability in the present study were found to map to various surfaces of the CA protein (Fig. 8). Based on cryoelectron microscopy and image analysis of cylinders assembled in vitro from recombinant HIV-1 CA, we have recently proposed a structural model of the HIV-1 core (28). In this model, intersubunit contacts in both amino-terminal and carboxy-terminal CA domains are involved in determining the three-dimensional arrangement of CA hexamers within the capsid. Consistent with this model, we observed mutations in both domains that affected core stability. Nevertheless, mutations not predicted to be localized to CA-CA interfaces also had effects on core stability, suggesting that core stability is very sensitive to subtle structural alterations in CA.
FIG. 8.
Location of core stability mutations in proposed complete structure of CA. The structure of the intact CA molecule was based on the work of Gamble et al. (15, 16). (A) Mutations that destabilize the HIV-1 core; (B) mutations that stabilize the core.
Two of the CA mutants characterized in this study, E45A and E128A/R132A, exhibited cores that were more stable than wild-type HIV-1 cores. E45A is found within helix 2 of the amino-terminal domain (Fig. 8). Based on the structural model for the arrangement of CA in the HIV-1 core, helix 2 is predicted to mediate contacts with hexameric CA rings. In contrast, the E128A/R132A mutations lie within helix 7 of the N-terminal domain (Fig. 8). The close proximity of E45 and E128 to one another on the CA surface (Fig. 8) is reminiscent of the role of carboxylate groups in the disassembly of the tobacco mosaic virus (TMV) coat protein (35). TMV is a helical virus that undergoes cotranslational disassembly, a process that is proposed to be driven by changes in calcium ion concentration and pH encountered by the virus following entry into target cells. Electrostatic repulsion of carboxylate side chains in adjacent subunits drives the disassembly of the TMV coat protein. Further biochemical and structural analyses will be required to precisely define the mechanism by which E45 and E128 facilitate HIV-1 core disassembly.
In this study, we observed defects at early stages of reverse transcription for HIV-1 mutants exhibiting deviations from wild-type core stability. Both classes of mutants—those exhibiting either increased or decreased core stability—displayed defects in synthesis of minus-strand strong-stop DNA, a very early product of reverse transcription. The defects observed for DNA synthesis in cultured cells are not likely due to defects in the reverse transcription machinery of the mutant viruses, because all of the mutant virions were able to undergo reverse transcription in vitro to an extent similar to that of wild-type virus. Furthermore, the viral RNA content and ratio of RT to CA were similar for wild-type and mutant viruses, further arguing against defects in RNA incorporation or in processing and function of the RT enzyme in the mutants. Therefore, a more likely explanation for the defect in reverse transcription exhibited by the mutants in target cells is that proper disassembly of the viral core is critical for efficient proviral DNA synthesis in vivo.
In contrast to the reverse transcription defects exhibited by most of the CA mutants, two mutants, P38A and Q63A/Q67A, were capable of efficient reverse transcription in cells. However, the kinetics of DNA synthesis deviated significantly from that of wild-type virus, as these mutants actually synthesized DNA at an accelerated rate. Since P38A and Q63A/Q67A virions were much less infectious than the wild type, the rapid DNA synthesis observed likely represents a nonproductive pathway for reverse transcription. Conceivably, these mutants could be blocked at a later stage of reverse transcription, such as improper formation of LTR ends due to incorrect initiation of plus-strand synthesis. Alternatively, the mutants may be defective for nuclear transport or integration of the provirus. Consistent with the latter hypothesis, other groups have reported CA mutations that are competent for second-strand transfer yet show defects in two-LTR circle formation, a marker of HIV-1 nuclear import (8, 14). Further analysis of the unusual phenotype of the P38A and Q63A/Q67A mutants may reveal additional roles for CA during HIV-1 infection.
Although it is not apparent whether CA dissociation from HIV-1 cores occurs spontaneously upon fusion with target cells, the thermodynamics of CA assembly are consistent with this hypothesis. Within the constraints of an HIV-1 particle, the concentration of CA is quite high, favoring CA multimerization. Results from our study suggest relatively weak CA-CA interactions, as seen for CA assemblies in vitro (16, 17, 23, 37), since detergent treatment of virions induced the solubilization of the majority of the virion-associated CA. Similarly, fusion of the virus with a target cell would release the volume constraints on the particle. Capsid dissociation may therefore be driven by dilution effects, although the participation of cellular factors in core disassembly in vivo remains an intriguing possibility.
The available data suggest that CA dissociates from the core soon after penetration. Studies of acutely infected cells by electron microscopy imply that the CA shell does not persist during infection (22). Biochemical studies are consistent with this interpretation, as reverse transcription complexes isolated from cells as early as 1 h after infection exhibited minimal quantities of CA (13, 24, 30). Despite this apparently transient association of the CA protein with the viral genomic complex after entry into the cells, our data suggest that the CA shell must remain intact for some period of time following penetration in order to facilitate downstream postentry events. The specific function of CA during this phase remains to be determined. The impaired reverse transcription observed for mutants containing hyperstable cores suggests that the CA shell must dissociate in order for the virus to undergo early stages of reverse transcription. This interpretation is consistent with the work of Zhang and coworkers, which suggests that reverse transcription may not take place within the intact viral core (42). However, in our studies, cores that disassembled too rapidly were also defective for reverse transcription in cells. A model that may reconcile this apparent paradox is that upon delivery into the cytoplasm, the CA shell targets the reverse transcription complex to a specific intracellular compartment that is conducive to successful infection. In this model, HIV-1 cores that disassemble too rapidly are not transported. However, following delivery to this compartment, the CA shell must also dissociate in order for the viral ribonucleoprotein complex to initiate reverse transcription. Consistent with a putative requirement for transport in reverse transcription, emerging evidence suggests that the target cell cytoskeleton facilitates early steps of HIV-1 infection. In a study by Bukrinskaya and coworkers, HIV-1 reverse transcription complexes were found to localize to actin microfilament components, and disruption of the actin cytoskeleton inhibited HIV-1 reverse transcription in target cells (5). Other investigators have suggested the incoming HIV-1 particles associate with microtubules, an interaction that may facilitate intracellular transport of the intact core or ribonucleoprotein complex (T. Hope, personal communication).
In summary, our data strongly suggest that a core of optimal stability is necessary for HIV-1 infection. The quantitative assays described in this report also provide a potentially valuable experimental system for identifying cellular factors that regulate HIV-1 core disassembly and for evaluating the effects of antiviral drugs on HIV-1 core stability.
Acknowledgments
We thank Paul Spearman, Terry Dermody, and members of the Aiken laboratory for helpful suggestions; Michael Miller for performing assays of virus-cell fusion; and Derya Unutmaz for purified CD4+ primary human T cells. We also thank Lou Henderson for rabbit antiserum to HIV-1 NC. The following reagents were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: antisera to HIV-1 RT (catalog no. 634), HIV-1 Vpr (catalog no. 3252) from Velpandi Ayyavoo, and HIV-1 IN (catalog no. 758) from Duane P. Grandgenett.
This work was supported by grants to C.A. and W.I.S. from NIH and by a Discovery Grant to C.A. from Vanderbilt University School of Medicine. B.M.F. was supported by an NIH institutional training grant.
REFERENCES
- 1.Accola, M. A., Å hagen, and H. G. Göttlinger. 2000. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6gag. J. Virol. 74:6198-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., J. W. Wills, and R. C. Craven. 2001. Second-site suppressors of Rous sarcoma virus CA mutations: evidence for interdomain interactions. J. Virol. 75:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui, M., G. Whittaker, and A. Helenius. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70:8391-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinskaya, A., B. Brichacek, A. Mann, and M. Stevenson. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 188:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, T. M., and R. C. Craven. 2001. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J. Virol. 75:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartier, C., P. Sivard, C. Tranchat, D. Decimo, C. Desgranges, and V. Boyer. 1999. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J. Biol. Chem. 274:19434-19440. [DOI] [PubMed] [Google Scholar]
- 9.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C., and H. Okayama. 1987. High-efficiency tranformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorfman, T., and H. G. Gottlinger. 1996. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J. Virol. 70:5751-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 15.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 16.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 17.Ganser, B. K., S. Li, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 18.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 19.Goncalves, J., Y. Korin, J. Zack, and D. Gabuzda. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 70:8701-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greber, U. F., I. Singh, and A. Helenius. 1994. Mechanisms of virus uncoating. Trends Microbiol. 2:52-56. [DOI] [PubMed] [Google Scholar]
- 21.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 22.Grewe, C., A. Beck, and H. R. Gelderblom. 1990. HIV: early virus-cell interactions. J. Acquir. Immune Defic. Syndr. 67:965-974. [PubMed] [Google Scholar]
- 23.Gross, I., H. Hohenberg, and H.-G. Krausslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 24.Karageorgos, L., P. Li, and C. Burrell. 1993. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res. Hum. Retrovir. 9:817-823. [DOI] [PubMed] [Google Scholar]
- 25.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiernan, R. E., A. Ono, and E. O. Freed. 1999. Reversion of a human immunodeficiency virus type 1 matrix mutation affecting Gag membrane binding, endogenous reverse transcriptase activity, and virus infectivity. J. Virol. 73:4728-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotov, A., J. Zhou, P. Flicker, and C. Aiken. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J. Virol. 73:8824-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 29.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 32.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh, I., and A. Helenius. 1992. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J. Virol. 66:7049-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubbs, G. 1999. Tobacco mosaic virus particle structure and the initiation of disassembly. Phil. Trans. R. Soc. Lond. B 354:551-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Schwedler, U. K., T. L. Stemmler, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wehrly, K., and B. Chesebro. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12:288-293. [DOI] [PubMed] [Google Scholar]
- 39.Welker, R., H. Hohenberg, U. Tessmer, C. Huckhagel, and H. G. Krausslich. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 74:1168-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wengler, G., C. Gros, and G. Wengler. 1996. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology 222:123-132. [DOI] [PubMed] [Google Scholar]
- 41.Yong, W. H., S. Wyman, and J. A. Levy. 1990. Optimal conditions for synthesizing complementary DNA in the HIV-1 endogenous reverse transcriptase reaction. AIDS 4:199-206. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, H., G. Dornadula, J. Orenstein, and R. J. Pomerantz. 2000. Morphologic changes in human immunodeficiency virus type 1 virions secondary to intravirion reverse transcription. J. Hum. Virol. 3:165-172. [PubMed] [Google Scholar]