Abstract
Human parvovirus B19 frequently causes acute and chronic arthritis in adults. The molecular mechanism of B19 arthritis, however, remains poorly understood. We previously showed that the transmission of B19 from rheumatoid synoviocytes to monocytic cells is associated with enhanced secretion of tumor necrosis factor alpha (TNF-α), which triggers inflammation, and interleukin-6. To determine the role of B19 in the production of TNF-α, we focused on the function of its nonstructural protein, NS1, and established monocytic U937 lines transduced with the NS1 gene under the control of an inducible promoter. Production of TNF-α mRNA and protein was elevated in a manner associated with NS1 expression. Reporter assays revealed that AP-1 and AP-2 motifs on the TNF-α promoter were responsible for NS1-mediated up-regulation. Electrophoretic mobility shift assay showed specific binding of nuclear proteins from NS1 gene-transduced cells with the AP-1 or AP-2 probe. Antibodies against transcription factors AP-1 and AP-2 and anti-NS1 antibody inhibited the binding of nuclear proteins to the corresponding probes. These data indicate that NS1 up-regulates TNF-α transcription via activation of AP-1 and AP-2 in monocytic cells. The molecular mechanisms of NS1-mediated TNF-α expression would explain the pathogenesis of B19-associated inflammation.
Human parvovirus B19 belongs to the family Parvoviridae and the genus Erythrovirus. The B19 genome includes three major open reading frames coding for the nonstructural protein NS1 in the left half and structural proteins VP1 and VP2 in the right half (44). B19 is the only parvovirus that has been clearly linked with disease in humans. Common manifestations caused by B19 infection include transient aplastic crisis in patients with histories of chronic hemolytic anemia (37), erythema infectiosum (7), nonimmune hydrops fetalis (9), chronic pure red cell aplasia in patients with immunosuppression (24), and arthralgia or arthropathy (59). Acute onset of polyarthritis is common in adults (mostly women) (60). Joint symptoms last for 1 to 3 weeks, although they may persist for months or years. B19 arthritis often meets clinical diagnostic criteria for rheumatoid arthritis and can be erosive (12, 19, 35, 53, 59). Because it has been observed that joint symptoms caused by B19 infection coincide with the appearance of specific immunoglobulin G (IgG) and the disappearance of viremia (2) and joint symptoms occur in chronically infected subjects after treatment with immunoglobulin (17), immune complex is thought to cause acute polyarthropathy. However, molecular mechanisms for the involvement of immune complexes in joint inflammation are unclear.
Tumor necrosis factor alpha (TNF-α), a representative proinflammatory cytokine, is an important mediator in the inflammatory process associated with many types of infections and autoimmune diseases through induction of a variety of cytokines, chemokines, and proteases in autocrine and paracrine pathways (16, 55). Although various cell types are capable of producing TNF-α, monocytes and macrophages are the main sources (29, 55). The regulatory mechanisms controlling TNF-α expression seem to depend on both the cell type and differentiation state of the cells. They also depend on activation signals induced by stimuli, including infection by viruses or other agents, interaction with adhesion molecules, or treatment with phorbol esters, endotoxins, or cytokines such as interleukin-1 or even TNF-α itself (55, 58, 62).
A recent study has demonstrated elevated levels of TNF-α in circulation during the acute and convalescent phases of B19 infection (23). We have detected B19 VP antigen in macrophages, follicular dendritic cells, and lymphocytes of the synovial tissue from patients with rheumatoid arthritis after several years of B19 infection (33). Expression of late viral proteins suggests persistent infection. In addition, it has been shown that B19 in the rheumatoid synoviocytes can be transmitted to monocytic U937 and THP-1 cells in a membrane-separated coculture system and that transmission is inhibited by anti-B19 VP (50). Higher levels of TNF-α and interleukin-6 were secreted in this system, suggesting that B19 protein produced in newly infected cells mediated enhanced production of proinflammatory cytokines. NS1 is the most plausible activator of TNF-α expression because of its ability to activate several promoters including the B19 p6 promoter (15, 18, 31, 46).
In the present study, we addressed the question of whether TNF-α transcription is regulated by B19 NS1, and if so, which transcription factors are responsible for it. A histiocytic (promonocytic) cell line, U937 (49), was used as a host cell line. As isolation and maintenance of stable NS1-producing cell lines are tightly limited because of the cytotoxicity of NS1 (36, 48), we adopted an inducible expression system with a lactose operator and repressor.
MATERIALS AND METHODS
Cells.
The promonocytic cell line U937 (49) was provided by the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University. The U937 cell line was cultured in RPMI 1640 medium (Gibco BRL, Rockville, Md.) supplemented with 10% heat-inactivated fetal calf serum and 50 μg (each) of streptomycin and penicillin (Gibco BRL) per ml in a 5% CO2 atmosphere at 37°C.
Plasmid construction.
The NS1 expression plasmid pOPRSV1-N8NS was constructed with a vector plasmid of the B19-cloned plasmid N8 (54), pOPRSV1CAT (Stratagene, La Jolla, Calif.), and the NS1 coding region. The sequence of the NS1 coding region was amplified by PCR and inserted into the NotI site of pOPRSV1CAT by the method of Moffatt et al. (31). The reporter plasmid with the full-length human TNF-α promoter (pTNF1051; from −1051 bases to +126 bases relative to the transcription initiation site) was obtained by amplifying the region of peripheral leukocyte DNA from a healthy donor who had given informed consent by the method described elsewhere (51). The amplified DNA fragment was cloned into the NotI site of pGEM-T Easy (Promega, Madison, Wis.). The MluI-NcoI fragment of the resulting plasmid was then subcloned into a luciferase reporter plasmid, pGL3-basic (Promega). A deletion series of the TNF-α promoter region (pTNF593, pTNF128, pTNF106, and pTNF58) was constructed from pTNF1051 by removing restriction fragments. A substitution mutation series (pTNF593/Egr-1m, pTNF593/Etsm, pTNF593/CREm, pTNF593/NF-κBm, pTNF593/AP-1m, pTNF593/SP-1m, and pTNF593/AP-2m) was constructed by inducing site-directed mutations in pTNF593 according to the protocol for the GeneEditor in vitro site-directed mutagenesis system (Promega). Oligonucleotides used in site-directed mutagenesis are listed in Table 1. Four tandem copies of transcription factor-binding sequences (Table 1) were synthesized and inserted into the sites upstream of the minimal simian virus 40 (SV40) promoter in a reporter plasmid, pGL3-promoter (Promega), which generated pGL3(AP-1)4, pGL3(AP-2)4, and pGL3(NF-κB)4. All plasmids constructed were verified by DNA sequencing.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Position | Sequence (5′ to 3′)a |
|---|---|---|
| Primers for site-directed mutagenesis | ||
| Egr-1m | −169 to −160 | TCCCCGCCCAGATATGGAGAAGAAAC |
| Etsm | −116 to −112 | CACTACCGCTCTTTCCAGATGAGCTC |
| AP-1/CREm | −106 to −99 | TTCCTCCAGATCTGTGCATGGGTTTC |
| NF-κBm | −97 to −88 | ATGAGCTCATGGATATCTCCACCAAGG |
| AP-1m | −65 to −59 | TTTCCGCTGGTGTAAGGATTCTTTCCC |
| SP-1m | −52 to −45 | TGATTCTTTCCGGACCCTCCTCTCGCC |
| AP-2m | −36 to −28 | CCTCCTCTCGCCTCATGGACATATAAG |
| Tandem copies of transcription factor-binding elements | ||
| (AP-1)4 | −65 to −59 | ctgaacgcgtTGAATGATGAATGATGAATGATGAATGActcgagagtc |
| (AP-2)4 | −36 to −28 | ctgaacgcgtCCCCAGGGACCCCAGGGACCCCAGGGACCCCAGGGActcgcgcgtc |
| (NF-κB)4 | −97 to −88 | ctgaacgcgtGGGTTTCTCCGGGTTTCTCCGGGTTTCTCCGGGTTTCTCCctcgcgcgtc |
| Probes for EMSA | ||
| AP-1 | −65 to −59 | TTTCCGCTGGTTGAATGATTCTTTCCC |
| AP-2 | −36 to −28 | CCTCCTCTCGCCCCAGGGACATATAAG |
| Egr-1 | −169 to −160 | TCCCCGCCCCCGCGATGGAGAAGAAAC |
| NF-κB | −97 to −88 | ATGAGCTCATGGGTTTCTCCACCAAGG |
| AP-1m | −65 to −59 | TTTCCGCTGGTGTAAGGATTCTTTCCC |
| AP-2m | −36 to −28 | CCTCCTCTCGCCTCATGGACATATAAG |
Binding sites are shown in bold type. The linker sequences at the ends of the oligomers are indicated by lowercase letters.
Stable transfection.
U937 cells at a concentration of 1.6 × 106 cells/0.2 ml were mixed with 5 μg of p3′SS encoding the LacI repressor gene (Promega) and pulsed twice for 5 ms at 150 V with an electroporator apparatus, Electro Square Porator T820 (BTX, San Diego, Calif.). Transfectants were selected in medium containing 300 μg of hygromycin B (Roche Diagnostics, Mannheim, Germany) per ml and examined for expression of Lac repressor by Western blotting assay with rabbit anti-LacI antiserum (Stratagene). LacI-positive clones were further transfected with pOPRSV1-N8NS or pOPRSV1CAT by the electroporation method, and transfectants were selected with 300 μg of Geneticin (Wako, Osaka, Japan) per ml. A number of Geneticin-resistant NS1 clones and chloramphenicol acetyltransferase (CAT) clones were isolated by a limiting dilution method. U937/NS17 and U937/NS13 were examined as representative clones of high and low NS1 producers, respectively. U937/CAT12 was used as a representative clone of control cell lines.
Mouse anti-NS1 antibody.
Plasmid pQE44NS/F, provided by A. Gigler and A. Von Poblozki (Universität Regensburg) (56), contains the B19 genome coding for the C-terminal region of NS1 protein (amino acids 493 to 671) and nucleotides coding for six histidine residues at the 5′ end of the NS1 gene. pQE44NS/F was expressed in Escherichia coli DH5α, and the bacterial lysate was passed through a column of Ni-nitrilotriacetic acid agarose resin (Qiagen, Tokyo, Japan) to purify the His-tagged truncated NS1, NS/F. BALB/c mice were immunized with NS/F, and their spleen cells were used to establish hybridomas. Hybridoma clones were isolated by a limiting dilution method, and the ability of clones to secrete monoclonal antibodies was evaluated by enzyme-linked immunosorbent assay (ELISA) using NS/F as a target antigen. Consequently, a single hybridoma clone, F440, was established. A BALB/c mouse sensitized with 2,6,10,14-tetramethylpentadecane (Wako) was peritoneally inoculated with F440 cells for 7 to 10 days, and the ascitic fluid was removed. Anti-NS1 antibody was purified by passing the ascitic fluid through a protein G affinity chromatography column (MabTrapG) (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). The subclass IgG2b antibody, named mNS440, detects NS/F on a membrane in Western blotting analysis and NS1 protein in B19-infected erythroblastoid KU812 cells in immunofluorescence analysis.
Real-time RT-PCR.
U937/NS17, U937/NS13, and U937/CAT12 cells at a concentration of 2 × 105 cells/ml were treated with isopropyl-β-d-thiogalactopyranoside (IPTG) (Wako) at various concentrations and incubated at 37°C for various periods (concentrations and incubation times are shown in the figures). Cells were incubated in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) at various concentrations for 24 h prior to IPTG treatment in some experiments. Total RNA was extracted from the cells twice with ISOGEN-LS (Nippon Gene, Toyama, Japan) to reduce traces of DNA. The RNA samples were subjected to quantitation of B19 NS1 mRNA, TNF-α mRNA, and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The quantity of each mRNA was determined by a method based on the TaqMan protocol (Applied Biosystems, Foster City, Calif.). This method allows for the precise quantitation of mRNA loads over 7 orders of magnitude (28). Reverse transcription and PCR were performed in a one-step reaction (RT-PCR). The reaction mixture contained 1 U of RiverTra Ace (Toyobo, Osaka, Japan) and 1.25 U of AmpliTaqGold polymerase (Applied Biosystems) in a 50-μl aliquot of a solution containing 5.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3) (at room temperature), 10 mM EDTA, 60 nM Passive Reference 1 (Applied Biosystems), 0.2 mM (each) deoxynucleoside triphosphate, 0.2 μM (each) primer, 0.1 μM 6-carboxyfluorescein (FAM)- and 6-carboxy-N,N,N′,N′-tetramethylrhodamine (TAMRA)-labeled probe, and each RNA sample. The primers and probes used were NS1 forward primer (5′-AAAGCCATTTTAGGCGGGC-3′), NS1 reverse primer (5′-TCCAGGCACAGCTACACTTCC-3′), NS1 probe (5′FAM-CCCACCAGGGTAGATCAAAAAATGCG-TAMRA-3′), TNF-α forward primer (5′-CTGCCCCAATCCCTTTATT-3′), TNF-α reverse primer (5′-CCCAATTCTCTTTTTTGAGCC-3′), TNF-α probe (5′FAM-CCTCCTTCAGACACCCTCAACCTCT-TAMRA-3′), GAPDH forward primer (5′-GAAGGTGAAGGTCGGAGTC-3′), GAPDH reverse primer (5′-GAAGATGGTGATGGGATTTC-3′), and GAPDH probe (5′FAM-CAAGCTTCCCGTTGTCAGCC-TAMRA3′). Thermal cycling parameters were as follows: (i) 30 min at 60°C; (ii) 10 min at 95°C; and (iii) 35 cycles, with 1 cycle consisting of 15 s at 95°C and 1 min at 60°C. Amplification and detection were performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The target quantity was determined from the standard curve and expressed relative to GAPDH as recommended by the manufacturer (Applied Biosystems user bulletin no. 2).
ELISA.
U937/NS17, U937/NS13, and U937/CAT12 cells at a concentration of 106 cells/ml were treated with IPTG at 37°C for various periods of time. The amount of human TNF-α in the culture supernatant was measured with an ultrasensitive ELISA kit (BioSource International, Inc., Camarillo, Calif.). The minimum detectable dose of TNF-α was <0.09 pg/ml, according to the manufacturer (as given in the instruction manual for the kit).
Luciferase assay.
U937/NS17, U937/NS13, and U937/CAT12 cells were transiently transfected with 3 μg of firefly luciferase reporter plasmids and 0.1 μg of a Renilla luciferase reporter plasmid by a DEAE-dextran method (50 μg of DEAE-dextran in 0.5 ml of phosphate-buffered saline). After 24 h of incubation in culture media, 3 × 105 to 4 × 105 cells/ml of cells were treated with 10 mM IPTG and harvested after various periods of incubation at 37°C. Luciferase activity was determined as light units (LU) using a Dual Luciferase Assay Kit (Promega) and a luminometer (model 1254; BioOrbit, Oslo, Norway). Firefly luciferase activity indicated the promoter activity tested, and Renilla luciferase activity showed transfection efficiency. Protein concentration was determined with a protein assay kit (Bio-Rad, Hercules, Calif.) and used to normalize the luciferase activity.
Nuclear extract preparation and electrophoretic mobility shift assay (EMSA).
After U937/NS17, U937/NS13, and U937/CAT12 cells (5 × 106 cells) were treated with 10 mM IPTG for 24 h, nuclear extracts were prepared by the method of Andrews and colleagues (3). Nuclear extract (5 to 10 μg) was incubated with 20 pmol of fluorescein (FITC)-labeled oligonucleotide probe containing the putative binding sequence of TNF-α promoter for AP-1, AP-2, NF-κB, or Egr-1 (Table 1) in a 10-μl binding mixture containing 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, 10 mM Tris-HCl (pH 7.5) (at room temperature), and 5 ng of poly(dI-dC) (Amersham Pharmacia Biotech); this incubation lasted 30 min and was done at room temperature. The probe bound by nuclear proteins was separated from free probe in a 4% polyacrylamide gel, and the fluorescence image was scanned by an FM-BIO II (Hitachi, Tokyo, Japan). For the competition assay, unlabeled oligonucleotide was added to the binding buffer prior to the addition of FITC-labeled probe. For the inhibition assay, mouse monoclonal antibody against human c-Jun, AP-2α, p65 subunit of NF-κB, or Egr-1 (Santa Cruz Biotechnology, Santa Cruz, Calif.) or mouse monoclonal anti-NS1 antibody (mNS440; described above) was incubated with nuclear extract overnight at 4°C, and then the probe was added.
RESULTS
Expression of NS1 gene in stably transfected U937 cells.
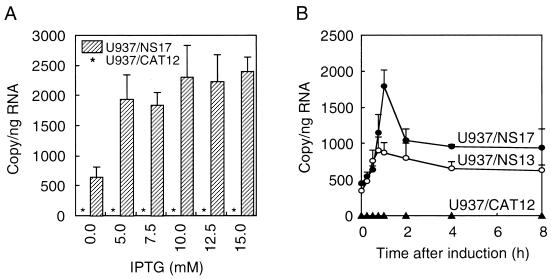
U937 cells were stably transfected by the NS1 gene that is regulated by the Lac repressor. The most effective concentration of IPTG for inactivating Lac repressor was examined by evaluating the synthesis of NS1 mRNA in the cells of an NS1 producer, U937/NS17, and a control clone, U937/CAT12. The RT-PCR method used in the present study detected 610 copies of NS1 mRNA per ng of total RNA from U937/NS17 cells even under noninducible conditions (Fig. 1A). It is notable that PCR with the RNA samples from U937/NS17 cells did not amplify any NS1 sequence. Repeated extraction by ISOGEN-LS seemed to completely remove contaminating DNA from the samples. Treatment of U937/NS17 cells for 1 h with 5 to 15 mM IPTG induced a four- to fivefold increase of NS1 mRNA, whereas U937/CAT12 cells did not express NS1 mRNA, irrespective of IPTG treatment. On the basis of these results, 10 mM IPTG was used to induce NS1 gene expression in experiments. In order to examine the association of NS1 and TNF-α expression, the kinetics of mRNA production in U937/NS17, U937/NS13, and U937/CAT12 cells were analyzed after IPTG induction. There were 498 copies of NS1 mRNA per 106 copies of GAPDH mRNA in U937/NS17 cells and 388 copies of NS1 mRNA in U937/NS13 cells before induction in a representative experiment (Fig. 1B). The quantity of NS1 mRNA in U937/NS17 cells increased by fourfold 1 h after induction and then dropped to half of the peak level after 2 h. U937/NS13 cells showed a 2.6-fold increase of NS1 mRNA. Other U937/NS clones gave results similar to those of U937/NS17 and U937/NS13 cells (data not shown). U937/CAT12 cells did not produce mRNA at any time before or after induction.
FIG. 1.
Expression of NS1 gene. (A) Induction by IPTG. Cells (2 × 105 cells/ml) were treated with the indicated concentrations of IPTG for 1 h. Total RNA from the cells was prepared, and the amount of NS1 mRNA was determined by real-time RT-PCR as described in Materials and Methods. The hatched bars indicate the copy numbers of NS1 mRNA in 1 ng of total RNA of U937/NS17 cells, and asterisks indicate the absence of NS1 mRNA in U937/CAT12 cells. (B) Kinetics of NS1 mRNA production. U937/NS17, U937/NS13, and U937/CAT12 cells were treated with 10 mM IPTG for the indicated periods of time. Total RNA from cells was prepared, and the amount of NS1 mRNA was determined as a copy number per nanogram of total RNA. The means (± standard deviations [error bars]) of three wells are shown.
Enhanced expression of TNF-α gene.
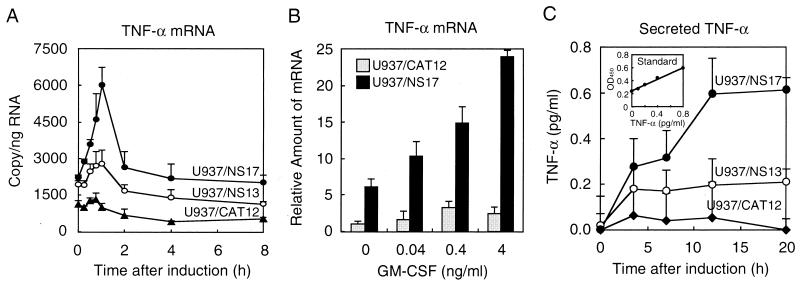
U937/CAT cells showed constitutive production of TNF-α mRNA under noninducible conditions (Fig. 2A). U937/NS13 and U937/NS17 cells produced 1.7- and 2.3-fold-more TNF-α mRNA, respectively, than U937/CAT cells did under noninducible conditions. The levels of TNF-α mRNA increased 2.2-fold in U937/NS17 cells, 1.4-fold in U937/NS13 cells, and 1.2-fold in U937/CAT cells 45 min to 1 h after induction. Maximum levels were 4.5-fold higher in U937/NS17 cells and 2.1-fold in U937/NS13 cells than in U937/CAT cells. TNF-α mRNA levels rapidly dropped to levels lower than those at 0 h. Treatment of U937/NS17 and U937/CAT cells with GM-CSF prior to IPTG induction stimulated cells to transcribe TNF-α mRNA in a concentration-dependent manner (Fig. 2B). However, the levels of stimulation were different in cell lines. GM-CSF at a concentration of 4 ng/ml resulted in almost 10-fold-more TNF-α mRNA in U937/NS cells than in U937/CAT cells. The kinetics of TNF-α protein production was examined by measuring secreted TNF-α in the culture medium of cells (Fig. 2C). Representative data showed an increase of TNF-α secretion from U937/NS17 cells, which reached a plateau of 0.6 pg/ml at 12 h. U937/NS13 cells also showed an increase in secretion at 0.2 pg/ml. U937/CAT12 cells secreted very low levels of TNF-α (below 0.05 pg/ml).
FIG. 2.
Activation of TNF-α gene expression. (A) Induction of TNF-α mRNA. Cells were treated with 10 mM IPTG for the indicated periods. Total RNA was prepared, and the amount of TNF-α mRNA was determined by real-time RT-PCR. The means (± standard deviations [error bars]) of three wells are shown. (B) TNF-α mRNA induction after treatment with GM-CSF. Cells were treated with various concentrations of recombinant human GM-CSF (PeproTech EC Ltd., London, England) for 24 h and then induced with 10 mM IPTG for 1 h. Cellular RNA was processed as described above for panel A. (C) Secretion of TNF-α. Cells (106 cells/ml) were treated with 10 mM IPTG for the indicated periods. Supernatants from wells were collected at the times indicated. TNF-α levels in supernatants were measured by ELISA. The insert shows a standard curve (OD450, optical density at 450 nm).
NS1-responsive elements in the TNF-α promoter.
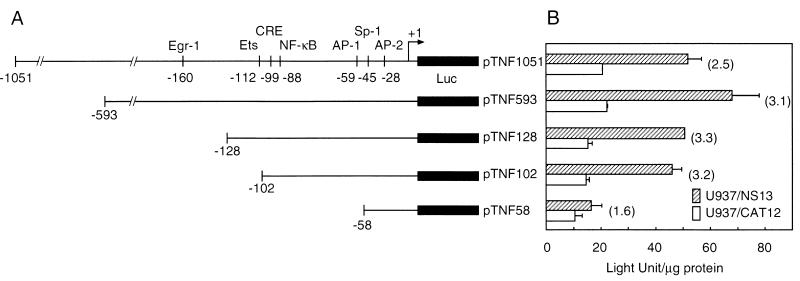
The activation of TNF-α production at mRNA and protein levels was associated with NS1 gene expression. We confirmed these findings by reporter assays with a luciferase reporter plasmid containing the full-length TNF-α promoter region (pTNF1051). Since the basal levels of NS1 expression are undesirable for evaluating the role of NS1, we compared IPTG-treated U937/NS13 cells to that of similarly treated U937/CAT cells. U937/NS13 cells showed reproducible increases in luciferase activity (2.5- to 2.8-fold) compared to that of U937/CAT cells (Fig. 3). The results indicate that the activation of TNF-α production occurs at the transcriptional level. To define the 5′ boundary of NS1-responsive elements in the TNF-α promoter region, U937/NS13 and U937/CAT cells were transiently transfected with a series of plasmids containing progressive truncations of the sequence between positions −1052 and −58 relative to the transcription initiation site (Fig. 3). Deletion of the sequence upstream of position −594, which removes two NF-κB sites at positions −867 and −622, slightly increased luciferase activity in the NS1-expressing cells. Removal of a region between positions −593 and −129, which contains two NF-κB sites at −585 and −204 and the Egr-1 site at −160, reduced both basal and NS1-induced activities, indicating no significant effect on NS1 inducibility. Deletion to position −103, which removed the Ets site at −112 and disturbed the AP-1/CRE site between −99 and −106, had no significant effect on basal or NS1-induced luciferase activities. Finally, deletion to position −59, which removed the NF-κB site at −88, the cEBP/β site at −74, and the AP-1 site at −59, dramatically reduced NS1-induced promoter activity, suggesting that transcription elements activated by NS1 expression were located in this region.
FIG. 3.
Effects of deletion mutations on the transcription activity of the TNF-α promoter in response to NS1 expression. (A) Schematic presentation of the TNF-α promoter region and its 5′ truncated forms. Putative cis-acting regulatory motifs in the reporter constructs are denoted, and the numbers identify the position relative to the transcription initiation site. Luc, luciferase. (B) Luciferase activities in cells transfected with the truncated reporter constructs and pRL-TK. The transfected cells were cultured for 24 h in the presence of 10 mM IPTG, and then cell lysates were assayed for firefly and Renilla luciferase activities. Protein concentrations of cell lysates were determined by the Bradford method using bovine serum albumin as the standard protein, and firefly luciferase activity was normalized to light units (LU) per microgram of protein. Hatched bars indicate the promoter activity in U937/NS13 cells, and white bars indicate that in U937/CAT12 cells. Firefly luciferase activity in U937/NS13 cells transfected with the positive-control construct pGL3-control was 902.2 ± 105.0 LU/μg of protein, and that in U937/CAT12 cells was 850.2 ± 185.9 LU/μg of protein, whereas the activity from the negative-control construct pGL-Basic in U937/NS13 cells was 0.5 ± 1.3 LU/μg of protein and that in U937/CAT12 cells was 0.4 ± 1.3 LU/μg of protein. Numbers in parentheses besides the pairs of bars indicate the increased induction (fold). Transfection efficiency, assessed by Renilla luciferase activity from pRL-TK, was similar in each assay. The means (± standard deviations [error bars]) of three separate experiments are shown.
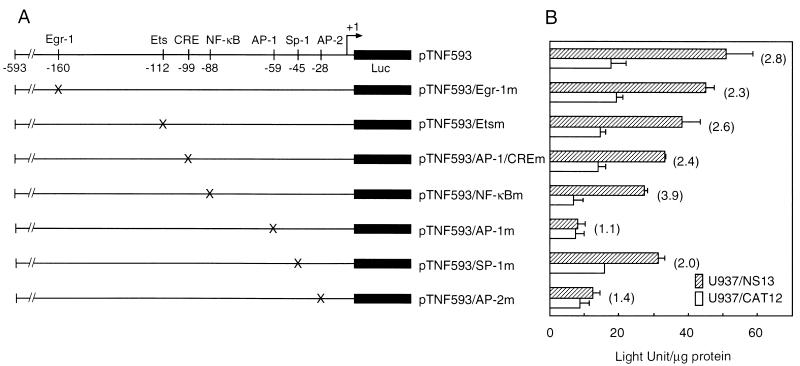
To determine the NS1-responsive motifs in the TNF-α promoter region, reporter plasmids with specific site-directed mutations were examined. Substitution mutations in the AP-1 (−59) and AP-2 (−28) sites markedly reduced NS1 activation of the TNF-α promoter (Fig. 4). In contrast, mutations in the Egr-1 (−160) site had no effect on basal or NS1-induced levels of TNF-α promoter activity. Mutations in the Ets (−112), AP-1/CRE (−99), and NF-κB (−88) sites reduced both basal and NS1-induced promoter activities. These results are consistent with the results of deletion constructs. U937/NS17 cells provided similar results (data not shown).
FIG. 4.
Effects of site-directed mutations. (A) Schematic presentation of reporter constructs with site-directed mutations in regulatory motifs. The positions of mutations (X) are marked. Luc, luciferase. (B) Luciferase activity in cells transfected with the site-directed reporter constructs and pRL-TK. Firefly luciferase activity in U937/NS13 cells transfected with pGL3-control was 872.3 ± 117.1 LU/μg of protein and that in U937/CAT12 was 963.1 ± 140.4 LU/μg of protein, whereas the activity of pGL3-basic in U937/NS13 was 0.8 ± 2.0 LU/μg of protein and that in U937/CAT12 was 0.9 ± 1.2 LU/μg of protein. Renilla luciferase activity was similar in each assay. The means (± standard deviations [error bars]) of three separate experiments are shown.
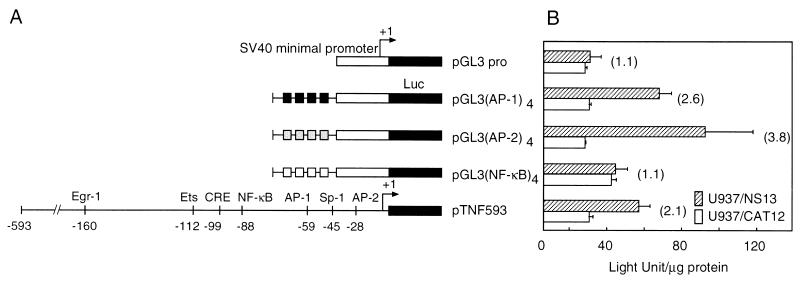
The contribution of the transcription factor-binding motif to NS1 induction of TNF-α expression was assessed by examining the activity of the minimal SV40 promoter with four copies of the motif (Fig. 5). Higher luciferase activity was obtained in U937/NS13 cells than in U937/CAT12 cells after transfection of reporter plasmid carrying the AP-1 or AP-2 motif, but not the NF-κB motif, suggesting that AP-1 and AP-2 sites play a major role in the induction of TNF-α expression by NS1.
FIG. 5.
Effect of four tandem copies of a regulatory motif. (A) Schematic representation of reporter constructs with four tandem copies of a regulatory motif. The repeated sequence of the AP-1, AP-2, or NF-κB binding motif was inserted upstream of the minimal SV40 promoter of a reporter vector pGL3 promoter. Luc, luciferase. (B) Luciferase activity in cells transfected with the constructs of repeated motifs and pRL-TK. Firefly luciferase activity in U937/NS13 cells transfected with pGL3-control was 952.6 ± 155.2 LU/μg of protein, and that in U937/CAT12 was 1,053.9 ± 135.8 LU/μg of protein, whereas the activity pGL-basic in U937/NS13 was 0.6 ± 1.0 LU/μg of protein and that in U937/CAT12 was 0.6 ± 1.7 LU/μg of protein. Renilla luciferase activity was similar in each assay. The means (± standard deviations [error bars]) of three separate experiments are shown.
Nuclear factors that bind AP-1 and AP-2 sites in the TNF-α promoter.
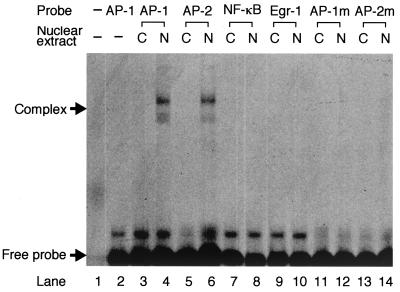
The experiments described above suggest that the AP-1 and AP-2 sites could be responsive to NS1 in the regulation of the TNF-α promoter. EMSA was performed to determine whether the AP-1 and AP-2 motifs were bound by nuclear factors in an NS1-dependent manner (Fig. 6). FITC-labeled oligonucleotide probe containing AP-1 binding sequence efficiently bound nuclear proteins extracted from U937/NS13 cells, but not those from U937/CAT12 cells. Similar results were obtained with AP-2 probe but not with Egr-1 probe, NF-κB probe, mutated AP-1 (AP-1m) probe, or mutated AP-2 (AP-2m) probe.
FIG. 6.
EMSA with various probes. U937/NS13 and U937/CAT12 cells were treated for 24 h with 10 mM IPTG. Nuclear extracts were prepared and incubated with FITC-labeled oligonucleotides (probes) containing the putative transcription factor-binding sequences. The formation of complexes between nuclear proteins and a probe was analyzed by running them through a polyacrylamide gel. A similar EMSA was performed using probes with mutations in the AP-1 (AP-1m) and AP-2 (AP-2m) motifs. Nuclear extract from U937/CAT12 cells (C) and U937/NS13 cells (N) was used, as indicated above the gel. Lanes 1 and 2 are controls and contain no (−) probe and/or no (−) nuclear extract.
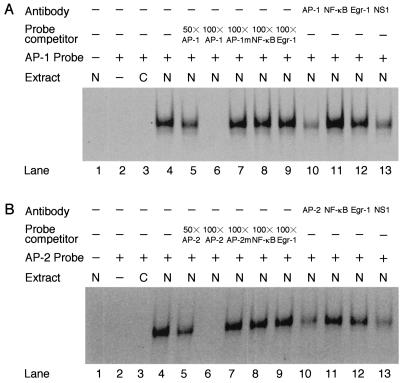
Binding specificity was shown by incubating nuclear extract with an excess amount of unlabeled AP-1 competitor fragment prior to binding with the labeled AP-1 probe (Fig. 7A). Dose-dependent inhibition occurred with 50-fold and 100-fold excess amounts of competitors. Such inhibition was not observed by competitors of Egr-1, NF-κB, and AP-1m. Similar results were obtained with the AP-2 probe (Fig. 7B). These results suggest that NS1-mediated binding is highly specific for AP-1 and AP-2 motifs on the TNF-α promoter. Anti-c-Jun antibody inhibited specific binding of AP-1, whereas anti-p65 (NF-κB) and anti-Egr-1 antibodies did not inhibit binding. Anti-AP-2α antibody had a similar effect on the AP-2 reaction. Thus, AP-1 and AP-2 were clearly identified as nuclear factors that bind to the TNF-α promoter in an NS1-mediated manner.
FIG. 7.
Inhibition of EMSA by competitors and antibodies. EMSA with the AP-1 (A) and AP-2 (B) probes. The nuclear extract from IPTG-treated U937/NS13 cells was incubated with 50- (50×) to 100-fold (100×) excess amounts of unlabeled AP-1 or AP-2 oligonucleotide or with 100-fold excess amounts of AP-1m or AP-2m, NF-κB, or Egr-1 oligonucleotide prior to binding with the FITC-labeled AP-1 or AP-2 probe. The nuclear extract was also incubated with antibodies against AP-1 or AP-2, NF-κB, Egr-1, or NS1 prior to the binding reaction. EMSA was performed as described in the legend to Fig. 6. The presence (+) or absence (−) of antibody, probe competitor, probe, and nuclear extract from U937/CAT12 cells (C) and U937/NS13 cells (N) is indicated above the gel. In both panels, lane 2 is a control and does not contain nuclear extract.
Role of NS1 in activating transcription factors to bind the corresponding elements on the TNF-α promoter.
To examine the role of NS1 protein in the binding reaction, nuclear extract of U937/NS13 cells was treated with anti-NS1 antibody mNS440 prior to the assays. Complexes between AP-1 and AP-1 probe or AP-2 and AP-2 probe were effectively inhibited by the anti-NS1 antibody (Fig. 7A, lane 13, and B, lane 13), demonstrating the participation of NS1 protein in the activation of AP-1 and AP-2.
DISCUSSION
B19 NS1 has the ability to activate its own promoter (p6), as well as other genes of infected cells (15, 18, 31, 46). In the present study, we have demonstrated that NS1 induced up-regulation of TNF-α promoter in promonocytic U937 cells, which occurred through activation of AP-1 and AP-2.
NS1 is cytotoxic to infected cells (32, 36, 47, 48). The cytotoxic effect of NS1 has hampered studies on mechanisms associated with NS1 expression and activation of TNF-α and other cytokine genes. This prompted us to employ an inducible expression system for establishing permanent monocytic U937 cell lines with the NS1 gene. NS1 gene-transduced cell clones were positive for low-level expression of NS1 mRNA in the uninduced state by a sensitive quantitative RT-PCR. This was not unexpected, since the vector plasmids used in this study were known to express low levels of transduced gene even in the repressed state (Stratagene catalog no. 217450).
Inactivation of the repressor by treating the cells with IPTG induced a rapid increase in NS1 mRNA synthesis. The increase in NS1 mRNA was consistently followed by the synthesis of TNF-α mRNA. Enhancement of reporter activity by using the full-length TNF-α promoter also suggests a role for NS1 in the activation of TNF-α transcription. This is supported by our data from preliminary studies indicating that a ribozyme with the ability to cleave NS1 mRNA specifically inhibits up-regulation of TNF-α expression in an NS1 gene-transduced U937 system (data not shown). These findings indicate that NS1 participates in the regulation of TNF-α transcription in promonocytic cell line U937. TNF-α was secreted into the culture medium with kinetics similar to those of U937 and THP-1 cells stimulated with bacterial components (1, 42). The level of TNF-α secreted by U937/NS17 cells was ∼0.6 pg/ml. The low level of secretion may be partly due to low-level expression of NS1 in these cells. Namely, NS1 synthesis was incompletely repressed in NS1 gene-transduced U937 clones in an uninduced state. The leakage of cytotoxic NS1 might affect the maintenance of permanent cell lines, resulting in the selection of low-level producers of NS1 in the cloning procedure. Another factor may be that U937 is an immature monocyte and does not secrete TNF-α, even after treatment with lipopolysaccharide. GM-CSF is frequently used to induce differentiation of peripheral monocytes and U937 and is endowed with the ability to produce TNF-α mRNA in response to lipopolysaccharide (10). In our system, addition of GM-CSF before IPTG induction markedly enhanced the expression of TNF-α mRNA. These results suggest that the level of TNF-α secreted from B19-infected macrophages might be much higher than those from NS1 gene-transduced U937 cells.
Here we have shown that two cis-acting regulatory sequences, the AP-1 motif at position −59 and the AP-2 motif at position −28, were required for the activation of TNF-α promoter in NS1 gene-transduced U937 cells. These sites are also essential for the expression of TNF-α in phorbol myristate acetate- or GM-CSF-stimulated U937 cells (39-41). Previous studies, however, did not address whether trans-acting AP-1 and AP-2 elements actually bind to the corresponding motifs. Here we have shown in an EMSA that AP-1 and AP-2 do form complexes with AP-1 and AP-2 probes, respectively. This was confirmed by inhibition of the formation of complexes using anti-AP-1 and anti-AP-2 antibodies. Inhibition by anti-NS1 antibody implies that NS1 induces expression of TNF-α through activation of AP-1 and AP-2. We have also demonstrated that either the AP-1 or AP-2 site at the 5′ end of the minimal promoter was adequate for the NS1-mediated promoter activation, whereas the AP-1/Cre site at position −99 (26) was not. This study and others showed that the NF-κB site at −88 (52, 61) has a role in the basal-level transcription of TNF-α. The TNF-α promoter may be regulated by a multicomponent complex of regulatory elements that includes AP-1, AP-2, AP1/CRE, and NF-κB sites, as predicted by a model of B19 p6 promoter bridged by the NS1 protein (18).
AP-1 proteins are dimers of proteins from the Jun, Fos, and ATF families (5). Nonstimulated cells express low levels of these proteins, but they are not phosphorylated (4, 14). After exposure to various stimuli, the component proteins are rapidly phosphorylated by mitogen-activated protein kinases and acquire high transcriptional activity (21, 30). Thus, the rapid increase of TNF-α transcription after the induction of NS1 expression may be partly due to the phosphorylation of AP-1 components by the action of NS1 protein or NS1-activated mitogen-activated protein kinases. Few studies have investigated the viral factors that induce regulation of the TNF-α promoter. The human immunodeficiency virus Tat protein stimulates TNF-α expression in monocytes and macrophages through a mechanism dependent on NF-κB (11). The adenoviral protein E1A 13S trans activates the TNF-α promoter in U937 cells through the AP-1, AP-2, and CRE sites (41), where E1A may activate viral and cellular transcription through modification of cellular factors by phosphorylation or by direct interaction with factors that do bind to promoter consensus sequences (13, 27). Comparison of the protein structure and cellular factors that interact with NS1 and E1A may provide clues to understanding the mechanism of AP-1 and AP-2 activation by viral factors.
It is worth discussing the proposed mechanism in relation to B19 infection in vivo. B19 DNA has been detected in circulating mononuclear cells and macrophages after B19 infection (25, 38). B19 DNA was also detected in the synovium of patients with rheumatoid arthritis as well as healthy subjects (22, 43, 45). Further, both VP antigen and B19 RNA were positive in macrophages, follicular dendritic cells, or T and B cells in tonsils (33) and in rheumatoid synovium (50), where TNF-α plays a central role in causing the lesion (6, 8, 20). Recent studies showed elevated levels of TNF-α in serum in subjects with acute and chronic B19 infections (23, 34, 57). In this paper, we have presented evidence demonstrating that NS1 protein activates the TNF-α promoter, providing an explanation for the mechanism of TNF-α production in vivo. Further analysis using the model presented, NS1 gene-transduced U937 cells, will provide more information to understand the mechanisms associated with the induction of TNF-α synthesis following B19 infection of monocytes/macrophages in the inflammatory process in vivo.
Acknowledgments
Y. Fu and K. K. Ishii contributed equally to this work.
We thank K. Umene (Kyushu University), T. Nunoue (Nakamura Gakuen University), A. Gigler and A. Von Poblotzki (Universität Regensburg) for generously providing the plasmids used and E. Morita and K. Sugamura (Tohoku University School of Medicine) for helpful suggestions.
This work was supported in part by a Grant-in-Aid for Exploratory Research from the Ministry of Education, Science, Sports and Culture of Japan and by The Hiromi Medical Trust Fund (K.K.I.).
REFERENCES
- 1.Abramson, T., H. Kedem, and D. A. Relman. 2001. Proinflammatory and proapoptotic activities associated with Bordetella pertussis filamentous hemagglutinin. Infect. Immun. 69:2650-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. J., P. G. Higgins, L. R. Davis, J. S. Willman, S. E. Jones, I. M. Kidd, J. R. Pattison, and D. A. Tyrrell. 1985. Experimental parvovirus infection in humans. J. Infect. Dis. 152:257-265. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins for limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angel, P., K. Hattori, T. Smeal, and M. Karin. 1988. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 55:875-885. [DOI] [PubMed] [Google Scholar]
- 5.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 6.Arend, W. P., and J.-M. Dayer. 1990. Cytokines and cytokine inhibitors or antagonists in rheumatoid arthritis. Arthritis Rheum. 33:305-315. [DOI] [PubMed] [Google Scholar]
- 7.Brass, C., L. M. Elliott, and D. A. Stevens. 1982. Academy rash, a probable epidemic of erythema infectiosum (fifth disease). JAMA 248:568-572. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, F. M., D. Chantry, A. Jackson, R. N. Maini, and M. Feldmann. 1989. Inhibitory effect of TNF-α antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet 340:244-247. [DOI] [PubMed] [Google Scholar]
- 9.Brown, T., A. Anand, L. D. Ritchie, J. P. Clewley, and T. M. Reid. 1984. Intrauterine parvovirus infection associated with hydrops fetalis. Lancet 2:1033-1034. [DOI] [PubMed] [Google Scholar]
- 10.Cannistra, S. A., A. Rambaldi, D. R. Spriggs, F. Herrmann, D. Kufe, and J. D. Griffin. 1987. Human granulocyte-macrophage colony-stimulating factor induces expression of the tumor necrosis factor gene by the U937 cell line and by normal human monocytes. J. Clin. Investig. 79:1720-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, P., M. Mayne, C. Power, and A. Nath. 1997. The Tat protein of HIV-1 induces tumor necrosis factor-α production. Implications for HIV-1-associated neurological disease. J. Biol. Chem. 272:22385-22388. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, B. J., M. M. Buckley, J. P. Clewley, V. E. Jones, A. H. Puttick, and R. K. Jacoby. 1986. Human parvovirus infection in an early rheumatoid and inflammatory arthritis. Ann. Rheum. Dis. 45:832-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta, S., C.-J. Soong, D. M. Wang, and M. L. Harter. 1991. A purified adenovirus 289-amino-acid E1A protein activates RNA polymerase III transcription in vitro and alters transcription factor TFIIIC. J. Virol. 65:5297-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devary, Y., R. A. Gottlieb, L. F. Lau, and M. Karin. 1991. Rapid and preferential activation of the c-jun gene during the mammalian UV response. Mol. Cell. Biol. 11:2804-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doerig, C., B. Hirt, J.-P. Antonietti, and P. Beard. 1990. Nonstructural protein of parvovirus B19 and minute virus of mice controls transcription. J. Virol. 64:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman, M., P. Taylor, E. Paleolog, F. M. Brennan, and R. N. Maini. 1998. Anti-TNF alpha therapy is useful in rheumatoid arthritis and Crohn's disease: analysis of the mechanism of action predicts utility in other diseases. Transplant. Proc. 30:4126-4127. [DOI] [PubMed] [Google Scholar]
- 17.Frickhofen, N., J. L. Abkowitz, M. Safford, J. M. Berry, J. Antunez-de-Mayolo, A. Astrow, R. Cohen, I. Halperin, L. King, D. Mintzer, et al. 1990. Persistent B19 parvovirus infection in patients infected with human immunodeficiency virus type 1 (HIV-1): a treatable cause of anemia in AIDS. Ann. Intern. Med. 113:926-933. [DOI] [PubMed] [Google Scholar]
- 18.Gareus, R., A. Gigler, A. Hemauer, M. Leruez-Ville, F. Morinet, H. Wolf, and S. Modrow. 1998. Characterization of cis-acting and NS1 protein-responsive elements in the p6 promoter of parvovirus B19. J. Virol. 72:609-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gran, J. T., V. Johnsen, G. Myklebust, and S. A. Nordbo. 1995. The variable clinical picture of arthritis induced by human parvovirus B19. Scand. J. Rheumatol. 24:174-179. [DOI] [PubMed] [Google Scholar]
- 20.Haworth, C., F. M. Brennan, D. Chantry, M. Turner, R. N. Maini, and M. Feldman. 1991. Expression of granulocyte-macrophage colony-stimulating factor in rheumatoid arthritis: regulation by tumor necrosis factor-α. Eur. J. Immunol. 21:2575-2579. [DOI] [PubMed] [Google Scholar]
- 21.Karin, M. 1995. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270:16483-16486. [DOI] [PubMed] [Google Scholar]
- 22.Kerr, J. R., J. P. Carton, M. D. Curran, J. E. Moore, J. R. M. Elliott, and R. A. B. Mollan. 1995. A study of the role of parvovirus B19 in rheumatoid arthritis. Br. J. Rheumatol. 34:809-813. [DOI] [PubMed] [Google Scholar]
- 23.Kerr, J. R., F. Barah, D. L. Mattey, I. Laing, S. J. Hopkins, I. V. Hutchinson, and D. A. J. Tyrrell. 2001. Circulating tumor necrosis factor-α and interferon-γ are detectable during acute and convalescent parvovirus B19 infection and are associated with prolonged and chronic fatigue. J. Gen. Virol. 82:3011-3019. [DOI] [PubMed] [Google Scholar]
- 24.Kurtzman, G. J., K. Ozawa, B. Cohen, G. Hanson, R. Oseas, and N. S. Young. 1987. Chronic bone marrow failure due to persistent parvovirus infection. N. Engl. J. Med. 317:287-294. [DOI] [PubMed] [Google Scholar]
- 25.Kurtzman, G. J., P. Gascon, M. Caras, B. Cohen, and N. S. Young. 1988. B19 parvovirus replicates in circulating cells of acutely infected patients. Blood 71:1448-1454. [PubMed] [Google Scholar]
- 26.Leitman, D. C., R. C. Ribeiro, E. R. Mackow, J. D. Baxter, and B. L. West. 1991. Identification of a tumor necrosis factor-responsive element in the tumor necrosis factor alpha gene. J. Biol. Chem. 266:9343-9346. [PubMed] [Google Scholar]
- 27.Liu, F., and M. R. Green. 1990. A specific member of ATF transcription factor family can mediate transcription by the adenovirus E1a protein. Cell 61:1217-1224. [DOI] [PubMed] [Google Scholar]
- 28.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at the opposite ends provide a quenched prove system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 29.MacNaul, K. L., N. I. Hutchinson, J. N. Persons, E. K. Bayne, and M. J. Tocci. 1990. Analysis of IL-1 and TNF-α gene expression in human rheumatoid synoviocytes and normal monocytes by in situ hybridization. J. Immunol. 145:4154-4166. [PubMed] [Google Scholar]
- 30.Minden, A., A. Lin, T. Smeal, B. Dérijard, M. Cobb, R. Davis, and M. Karin. 1994. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol. Cell. Biol. 14:6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffatt, S., N. Tanaka, K. Tada, M. Nose, M. Nakamura, O. Muraoka, T. Hirano, and K. Sugamura. 1996. A cytotoxic nonstructural protein NS1 of human parvovirus B19 induces activation of interleukin-6 gene expression. J. Virol. 70:8485-8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffatt, S., N. Yaegashi, K. Tada, N. Tanaka, and K. Sugamura. 1998. Human parvovirus B19 nonstructural protein NS1 induces apoptosis in erythroid lineage cells. J. Virol. 72:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murai, C., Y. Munakata, Y. Takahashi, T. Ishii, S. Shibata, T. Muryoi, T. Funato, M. Nakamura, K. Sugamura, and T. Sasaki. 1999. Rheumatoid arthritis after human parvovirus B19 infection. Ann. Rheum. Dis. 58:130-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nigro, G., V. Bastianon, V. Colloridi, F. Ventriglia, P. Gallo, G. D'Amati, W. C. Koch, and S. P. Adler. 2000. Human parvovirus B19 infection in infancy associated with acute and chronic lymphocytic myocarditis and high cytokine levels: report of 3 cases and review. Clin. Infect. Dis. 31:65-69. [DOI] [PubMed] [Google Scholar]
- 35.Nikkari, S., A. Roivainen, P. Hannonen, T. Möttönen, R. Luukkainen, T. Yli-Jama, and P. Toivanen. 1995. Persistence of parvovirus B19 in synovial fluid and bone marrow. Ann. Rheum. Dis. 54:597-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozawa, K., J. Ayub, S. Kajigaya, T. Shimada, and N. Young. 1988. The gene encoding the nonstructural protein of B19 (human) parvovirus may be lethal in transfected cells. J. Virol. 62:2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pattison, J. R., S. E. Jones, J. Hodgson, L. R. Davis, J. M. White, C. E. Stroud, and L. Murtaza. 1981. Parvovirus infections and hypoplastic crisis in sickle-cell anaemia. Lancet 1:664-665. [DOI] [PubMed] [Google Scholar]
- 38.Porter, H. J., A. M. Quantrill, and K. A. Fleming. 1988. B19 parvovirus infection of myocardial cells. Lancet i:535-536. [DOI] [PubMed]
- 39.Rhoades, K. L., S. H. Golub, and J. S. Economou. 1992. The regulation of the human tumor necrosis factor α promoter region in macrophage, T cell, and B cell lines. J. Biol. Chem. 267:22102-22107. [PubMed] [Google Scholar]
- 40.Rhoades, K. L., S. Cai, and J. S. Economou. 1995. Granulocyte-macrophage colony-stimulating factor and interleukin-4 differentially regulate the human tumor necrosis factor-α promoter region. Cell. Immunol. 161:125-131. [DOI] [PubMed] [Google Scholar]
- 41.Rhoades, K. L., S. H. Golub, and J. S. Economou. 1996. The adenoviral transcription factor, E1A 13S, trans-activates the human tumor necrosis factor-α promoter. Virus Res. 40:65-74. [DOI] [PubMed] [Google Scholar]
- 42.Rutault, K., A. C. A. Hazzalin, and L. C. Mahadevan. 2001. Combination of ERK and p38 MARK inhibitors ablate tumor necrosis factor-α (TNF-α) mRNA induction. J. Biol. Chem. 276:6666-6674. [DOI] [PubMed] [Google Scholar]
- 43.Saal, J. G., M. Stendle, H. Einsele, C. A. Muller, P. Fritz, and J. Zacher. 1992. Persistence of B19 parvovirus in synovial membranes of patients with rheumatoid arthritis. Rheumatology 12:147-151. [DOI] [PubMed] [Google Scholar]
- 44.Shade, R. O., M. C. Blundell, S. F. Cotmore, P. Tattersall, and C. R. Astell. 1986. Nucleotide sequence and genome organization of human parvovirus B19 isolated from the serum of a child during aplastic crisis. J. Virol. 58:921-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soderlund, M., R. von Essen, J. Haapasaari, U. Kilstala, O. Kiviluoto, and K. Hedman. 1997. Persistence of parvovirus B19 DNA in synovial membranes of young patients with and without chronic arthropathy. Lancet 349:1063-1065. [DOI] [PubMed] [Google Scholar]
- 46.Sol, N., F. Morinet, M. Alizon, and U. Hazan. 1993. Trans-activation of the long terminal repeat of human immunodeficiency virus type 1 by the parvovirus B19 NS1 gene product. J. Gen. Virol. 74:2011-2014. [DOI] [PubMed] [Google Scholar]
- 47.Sol, N., L. Le Junter, I. Vassias, J. M. Freyssinier, A. Thomas, A. F. Frigent, B. B. Rudkin, S. Fichelson, and F. Morinet. 1999. Possible interactions between the NS-1 protein and tumor necrosis factor alpha pathways in erythroid cell apoptosis induced by human parvovirus B19. J. Virol. 73:8762-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivastava, A., E. Bruno, R. Briddell, R. Cooper, C. Srivastava, K. Van Besien, and R. Hoffman. 1990. Parvovirus B19-induced perturbation of human megakaryocytopoiesis in vitro. Blood 76:1997-2004. [PubMed] [Google Scholar]
- 49.Sundstrom, C., and K. Nilsson. 1976. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 15:565-577. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi, Y., C. Murai, S. Shibata, Y. Munakata, T. Ishii, K. Ishii, T. Saitoh, T. Sawai, K. Sugamura, and T. Sasaki. 1998. Human parvovirus B19 as a causative agent for rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 95:8227-8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takashiba, S., L. Shapira, S. Amar, and T. E. Van Dyke. 1993. Cloning and characterization of human TNF-α promoter region. Gene 131:307-308. [DOI] [PubMed] [Google Scholar]
- 52.Trede, N. S., A. V. Tsytsykova, T. Chatila, A. E. Goldfeld, and R. S. Geha. 1995. Transcriptional activation of the human TNF-α promoter by superantigen in human monocytic cells: role of NF-κB. J. Immunol. 155:902-908. [PubMed] [Google Scholar]
- 53.Tyndall, A., W. Jelk, and H. H. Hirsch. 1994. Parvovirus B19 and erosive polyarthritis. Lancet 343:480-481. [DOI] [PubMed] [Google Scholar]
- 54.Umene, K., and T. Nunoue. 1990. The genome type of human parvovirus B19 strains isolated in Japan during 1981 differs from types detected in 1986 to 1987: a correlation between genome type and prevalence. J. Gen. Virol. 71:983-986. [DOI] [PubMed] [Google Scholar]
- 55.Vassalli, P. 1992. The pathophysiology of tumor necrosis factors. Annu. Rev. Immunol. 10:411-452. [DOI] [PubMed] [Google Scholar]
- 56.Von Poblotzki, A., A. Gigler, B. Lang, H. Wolf, and S. Modrow. 1995. Antibodies to parvovirus B19 NS-1 protein in infected individuals. J. Gen. Virol. 76:519-527. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe, M., Y. Shimamoto, M. Yamaguchi, S. Inada, S. Miyazaki, and H. Sato. 1994. Viral-associated haemophagocytosis and elevated serum TNF-α with parvovirus-related pancytopenia in patients with hereditary spherocytosis. Clin. Lab. Haematol. 16:179-182. [DOI] [PubMed] [Google Scholar]
- 58.Webb, D. S., Y. Shimizu, G. A. Van Seventer, S. Shaw, and T. L. Gerrard. 1990. LFA-3, CD44, and CD45: physiologic triggers of human monocyte TNF and IL-1 release. Science 249:1295-1297. [DOI] [PubMed] [Google Scholar]
- 59.White, D. G., A. D. Woolf, P. P. Mortimer, B. J. Cohen, D. R. Blake, and P. A. Bacon. 1985. Human parvovirus arthropathy. Lancet i:419-421. [DOI] [PubMed] [Google Scholar]
- 60.Woolf, A. D., G. V. Campion, A. Chishick, S. Wise, B. J. Cohen, P. T. Klouda, O. Caul, and P. A. Dieppe. 1989. Clinical manifestations of human parvovirus B19 in adults. Arch. Intern. Med. 149:1153-1156. [PubMed] [Google Scholar]
- 61.Yao, J., N. Mackman, T. S. Edgington, and S. T. Fan. 1997. Lipopolysaccharide induction of the tumor necrosis factor-α promoter in human monocytic cells. Regulation by Egr-1, c-Jun, and NF-κB transcription factors. J. Biol. Chem. 272:17795-17801. [DOI] [PubMed] [Google Scholar]
- 62.Yurochko, A. D., D. Y. Liu, D. Eierman, and S. Haskill. 1992. Integrins as a primary signal transduction molecule regulating monocyte immediate-early gene induction. Proc. Natl. Acad. Sci. USA 89:9034-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]